Abstract
We used a two-chamber system to study transcytosis of Enterococcus faecalis across monolayers of human colon carcinoma-derived T84 cells, which show structural resemblance to the native intestine. Among 16 E. faecalis isolates from different sources, the well-characterized strain OG1RF and 8 other isolates (2 endocarditis isolates, 1 urine isolate, and all 5 fecal isolates) showed translocation in this assay, while 6 clinical isolates (3 endocarditis and 3 urine isolates), the recipient strain JH2-2, and the control, Escherichia coli DH5α, had no detectable translocation. Of two OG1RF mutants involving the previously studied epa (enterococcal polysaccharide antigen) gene cluster, known to be needed for virulence and resistance to killing by polymorphonuclear leukocytes, one epa mutant (TX5179) was unable to translocate, while TX5180, with an epa disruption farther downstream, showed a moderate decrease in translocation relative to that of the wild-type strain OG1RF (P < 0.01), indicating that the epa gene cluster is important for translocation across a T84 monolayer. This observation was confirmed by complementation of the epa mutant (TX5179) with epa genes and restoration of its translocation ability. In conclusion, we have demonstrated translocation of at least some strains of E. faecalis across T84 monolayers, although strains differ considerably in this ability, and we have demonstrated that epa mutations can cause marked changes in successful translocation. These results suggest that this model may be a useful in vitro system for studying the process of translocation from the intestinal tract.
Enterococci are part of the normal flora in human intestines and are also a leading cause of nosocomial infections (5, 12). These organisms seem to be able to migrate from the gastrointestinal tract into the bloodstream and cause systemic infections such as bacteremia and even endocarditis. The passage of bacteria from the gastrointestinal tract to extraintestinal sites is called translocation (1, 22). Wells and colleagues have presented evidence that Enterococcus faecalis can translocate across the mouse intestinal tract (23). In their study, following E. faecalis overgrowth (elicited by metronidazole and streptomycin treatment after oral inoculation of E. faecalis resistant to these agents), coccal bacteria appeared in the intestinal lumen and were observed within vacuoles in the cytoplasm of epithelial cells (23). By immunofluorescent microscopy, E. faecalis cells were localized within columnar epithelial cells, lamina propria, submucosa, and muscularis externa. Thus, E. faecalis cells appear to be able to translocate across an intact intestinal tract, although the enterococcal traits involved in the process were not determined. Runkel et al. also reported that morphine, a known inhibitor of myoelectric activity in the intestines of rats that prolongs gut transit time, increases intestinal microflora levels and promotes bacterial translocation from the intestinal tract to extraintestinal sites (16).
An in vitro transcytosis assay has been established to study the interaction of Salmonella spp. and other gram-negative bacteria with intestinal cells (2, 3, 6, 8, 10, 13). The transcytosis assay involves a two-chamber system for growing a monolayer of tissue culture cells on the top chamber side of a filter, followed by addition of bacteria to the top chamber and, at the end of the experiment, counting of the bacteria in the lower chamber. A similar assay has also been used by others to study gram-positive bacteria; for example, pneumococci were shown to translocate through the endothelial cell line EC 219, which is a rat brain microvessel-derived cell line, and group B streptococci were found to penetrate through a chorion cell line without disruption of intercellular junctions (15, 24).
A human colon carcinoma cell line, T84, has been widely used for in vitro models of transport studies (3, 4, 8, 9). These cells have the ability to form a tall columnar epithelial monolayer and to develop tight or occluding intercellular junctions when grown on permeable supports, with the basolateral surface attached to the support and the apical surface exposed to the outside (4). Transepithelial electrical resistance through the polarized monolayer is frequently used as an index of tight junction permeability and monolayer integrity (4, 9).
In the present study, the T84 cell line was used in a two-chamber transcytosis system to mimic the translocation of enterococci across intestinal epithelial cells. We chose T84 cells in part because of their phenotypic similarity to colonic crypt cells and in part because our preliminary studies showed successful translocation of T84 monolayers by E. faecalis in vitro. Fourteen E. faecalis human isolates, two well-characterized strains (OG1RF and JH2-2), and mutants with disruptions in the epa gene cluster, previously shown to be involved in the synthesis of a major antigenic polysaccharide of E. faecalis, were studied in this model. The results demonstrated considerable differences in the abilities of clinical isolates and mutants to translocate across the T84 monolayers.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
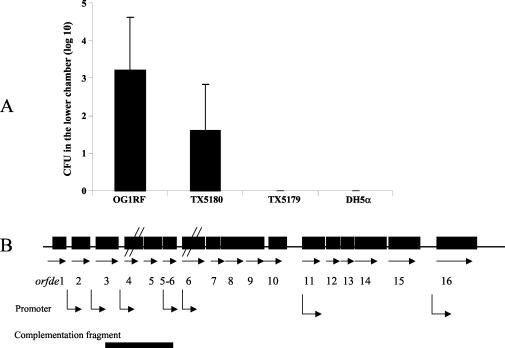
Bacterial isolates and plasmids used in this study are listed in Table 1. They included nine clinical isolates (five endocarditis and four urine isolates), five fecal isolates, OG1RF, and JH2-2. E. faecalis isolates were grown at 37°C in brain heart infusion (BHI) broth or agar (Difco, Sparks, Md.). The epa mutants, TX5179 and TX5180 (locations of mutations are shown in Fig. 1B), have previously been shown to be deficient in a major polysaccharide component (20, 25) and were grown in BHI broth or agar supplemented with kanamycin at 2,000 μg/ml. Escherichia coli DH5α, a strain previously shown to be unable to translocate Caco-2 monolayers (2), was grown in Luria-Bertani (Difco) broth or agar.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Host strain | Stratagene, La Jolla, Calif. |
| E. faecalis | ||
| OG1RF (TX4002) | Well characterized strain; Rifr FAr Gel+ | 19 |
| TX5179 | OG1RF with orfde4 disruption; Kanr | 27 |
| TX5180 | OG1RF with orfde6 disruption; Kanr | 27 |
| TX5179.1 | TX5179 with pTEX5175.1; Kanr Eryr | This study |
| Plasmids | ||
| pAT18 | Shuttle vector derived from pUC18; Eryr | 21 |
| pTEX5175 | 3.2-kb fragment which includes part of orfde3, intact orfde4 and orfde5, and half of orfde5-6 in pBluescript SK (−); Ampr | 26 |
| pTEX5175.1 | 3.2-kb insert of pTEX5175 recloned into pAT18; Eryr | This study |
Amp, ampicillin; Ery, erythromycin; FA, fusidic acid; Gel, gelatinase; Kan, kanamycin; Rif, rifampin.
FIG. 1.
(A) Translocation of bacteria across a T84 monolayer. Results of four combined experiments are shown. Six Transwells were used for each strain in each experiment. Bacterial cell counts (CFU) in the lower chamber at 6 h are shown. No TX5179 or DH5α bacteria were recovered in the lower chamber. The Mann-Whitney U test was used for statistical comparison. A P value of <0.01 was obtained for TX5180 versus OG1RF (combined results). (B) Diagram of the epa gene cluster. Horizontal arrows, open reading frames; bent arrows, predicted and determined promoters. It has been shown that orfde4 and orfde6 are in different transcripts, orfde6 to orfde10 are in one transcript, and orfde11 and other downstream genes are in different transcripts from orfde6 to orfde10 (27). orfde4 is disrupted in TX5179, and orfde6 is disrupted in TX5180.
Growth and maintenance of T84 cells.
T84 cells seeded into 75-cm2 tissue culture flasks were grown in Dulbecco's modified Eagle medium-F-12 medium (1:1 mix; Cellgro, Herndon, Va.) supplemented with 10% fetal calf serum (FCS) (Invitrogen, Grand Island, N.Y.) and 1% antibiotic-antimycotic (Invitrogen) and were incubated at 37°C under 5% CO2. When the monolayer reached confluence or near-confluence, the cells were detached from the bottom of the flask by using trypsin-EDTA (Invitrogen) and were split 1:4 into new flasks.
Translocation experiment.
Translocation experiments were performed by methods described previously, with slight modifications (8, 11, 15). Briefly, 105 T84 cells from passages 65 to 75 were seeded into a 12-well Transwell system with 3.0-μm-pore-size polycarbonate membranes (Corning Costar Corp., Cambridge, Mass.). This pore size allows bacteria, but not T84 cells, to penetrate the membrane. One and two milliliters of cell growth medium were added to the upper and lower Transwell chambers, respectively, and the medium was changed every 3 days. The developing progress of T84 tight junctions was monitored by Millicell-ERS measurement (Millipore, Billerica, Mass.). The translocation experiments were performed when the electrical resistance of the T84 monolayer reached about 1,850 Ω/cm2 or higher, which usually occurred in 10 to 12 days.
To prepare bacteria for translocation, overnight bacterial cultures (with appropriate antibiotics) were inoculated 1:100 into fresh BHI or Luria-Bertani medium (without antibiotics) and incubated at 37°C for about 2.5 h (optical density at 600 nm, ≈0.2). The bacterial cells were collected, washed twice with HBSS (Hanks balanced salt solution without Ca2+ and Mg2+) (Fisher, Herndon, Va.), and adjusted to 2 × 108 CFU/ml in the tissue culture medium described above (without antibiotic-antimycotic).
On the day of the experiment, the filters were washed twice with Dulbecco's modified Eagle medium-F-12 medium supplemented with 10% FCS without antibiotics. One milliliter of fresh medium (with 10% FCS and no antibiotics) was added to the lower chamber, and about 8 × 107 bacteria in 400 μl of tissue culture medium were added to the upper chamber; this inoculum is consistent with that used by others (2, 3, 11) and with the density of intestinal enterococci in some settings. Transepithelial electrical resistance was monitored at the beginning and end of the experiment. Viable bacteria in lower chambers were counted at 0, 4, and 6 h by removing 100-μl aliquots, serially diluting, and plating on BHI agar plates; viable bacteria in upper chambers were similarly counted at 6 h. For each strain, four to six Transwells were used and the experiments were repeated at least three times. The Mann-Whitney U test was used for statistical comparison of OG1RF derivatives to concurrent results with OG1RF. For clinical isolates, because of some fluctuations in translocation seen with different passages of cells used over the span of the experiments, results are presented as percentages relative to concurrently determined results for OG1RF.
Complementation of TX5179.
DNA manipulations, transformation, and PCR were performed by methods described previously (17). Previously, in our laboratory, a 3.2-kb epa fragment containing intact orfde4 and orfde5 genes was cloned into pBluescript SK(−) (26). In the present study, the 3.2-kb fragment was released from pBluescript SK(−) by EcoRI/HindIII digestion and cloned into the pAT18 shuttle vector to produce pTEX5175.1. pTEX5175.1 was then electroporated into TX5179, and 10 μg of erythromycin/ml was used to select for colonies with the plasmid. Following overnight growth at 37°C, erythromycin-resistant colonies were restreaked onto Todd-Hewitt agar plates with 2,000 μg of kanamycin/ml and 10 μg of erythromycin/ml, and complementation was confirmed by PCR.
RESULTS
Translocation of E. faecalis across T84 monolayers.
When E. faecalis OG1RF and E. coli DH5α were used in the translocation model (four individual experiments; six wells per experiment), DH5α was not detected in the lower chamber of any of the 24 wells at 6 h, while 102 to 104 OG1RF cells were detected in the lower chambers of 22 out of 24 wells (Fig. 1A). Comparable counts for the two strains were obtained in the upper chambers at 6 h (about 109 at the end of the experiments). Without the T84 monolayer, both species were able to cross the 3.0-μm-pore-size polycarbonate membrane (data not shown). Transepithelial electrical resistance remained high at 6 h and also at 8 h, indicating that the T84 monolayers were intact throughout the experiments (data not shown).
Translocation of other E. faecalis isolates across T84 monolayers.
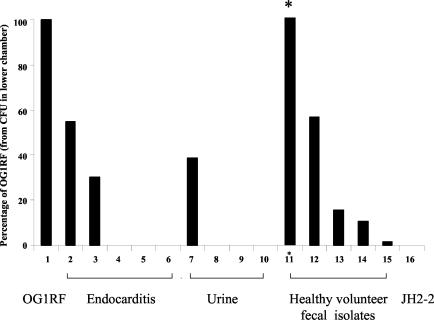
Fifteen additional E. faecalis isolates were then tested in the T84 monolayer model (Fig. 2). All of these grew well in T84 medium (to about 109 CFU in the upper chamber at 6 h). Two endocarditis isolates showed translocation across the T84 monolayers, similar to that seen with OG1RF (102 to 104 CFU in the lower chamber), and three endocarditis strains did not show translocation. Among urine strains, only one of four showed detectable translocation, while all five fecal strains showed translocation, ranging from 2 to 267% that seen with OG1RF. The widely used recipient strain JH2-2 showed no detectable translocation across the T84 monolayers.
FIG. 2.
Translocation of E. faecalis strains across a T84 monolayer, expressed as a percentage of concurrently determined results for OG1RF. Results shown are for three combined experiments. Four Transwells were used for each strain in each experiment. For six human isolates and JH2-2, no bacteria were recovered in the lower chamber. Levels of recovery of the other eight isolates ranged from 2 to 267%. Asterisk indicates 267%.
Effect of epa genes on translocation of E. faecalis across T84 monolayers.
Two epa mutants of OG1RF, TX5179 and TX5180, were compared with wild-type OG1RF in the translocation assay, and consistent results were obtained from the four individual experiments performed. OG1RF was not detected in the lower chamber at 0 h; at 4 h, OG1RF cells were detected in the lower chambers of 3 of 24 wells versus 0 of 24 wells for TX5179 and TX5180. At 6 h, TX5179 was not recovered from the lower chamber of any of the 24 wells, while translocation of TX5180 across the T84 monolayer (10 to 102 CFU of TX5180 in 15 of 22 wells) was lower than that of wild-type OG1RF (102 to 104 CFU of OG1RF in 20 of 23 wells) (P = 0.004, 0.22, 0.23, and 0.01 for four individual experiments; P < 0.01 for combined results) (Fig. 1A). In each experiment, about 50 translocated colonies of TX5180 were streaked onto BHI agar with 2,000 μg of kanamycin/ml, and all grew well, indicating that the epa mutant TX5180 was stable during the experiment. All strains tested reached similar cell densities in the top chamber (ca. 2 × 109 to 3 × 109 CFU) at 6 h.
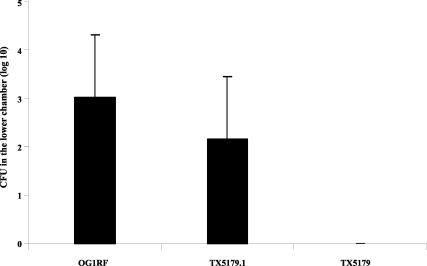
To further confirm the importance of epa genes for translocation, the epa mutant with a disruption in orfde4, TX5179, was complemented with extrachromosomal orfde4 and its downstream gene orfde5 (both genes are likely to be in the same operon [27]). The polysaccharide content of the complemented strain was compared with that of the mutant (TX5179) and wild-type OG1RF, and the polysaccharide smear missing in the mutant TX5179 appeared in the complemented strain, which showed a polysaccharide pattern similar to that of the wild-type strain OG1RF; the pAT18 shuttle vector did not affect the polysaccharide component (data not shown). As in the experiments described above, TX5179 could not be detected in any of the lower chambers of 12 wells at 6 h, while complemented cells (TX5179.1) were detected in the lower chambers of 10 of 12 wells versus 11 of 12 wells for wild-type OG1RF, indicating restoration of translocation capability by complementation (Fig. 3). The number of CFU recovered is shown in Fig. 3; the slight difference between OG1RF and TX5179.1 was not statistically significant. About 50 translocated colonies of TX5179.1 were streaked onto BHI agar with 2,000 μg of kanamycin/ml and 10 μg of erythromycin/ml, and all grew well, indicating that the insert and complementing plasmid were stably maintained during the experiment.
FIG. 3.
Translocation of OG1RF, TX5179, and TX5179.1 (complemented strain) across a T84 monolayer. Three combined experiments were performed, and four Transwells were used for each strain in each experiment. Bacterial cell counts (CFU) in the lower chamber at 6 h are shown. No TX5179 bacteria were recovered in the lower chamber.
DISCUSSION
It has been shown previously in animal models in which the intestinal epithelium was not damaged that some indigenous bacteria are able to translocate across the intestine intracellularly (through the epithelial cells) rather than extracellularly (through tight junctions in between epithelial cells), as bacteria were seen inside the epithelial cells during the translocation process (1). Although E. faecalis is included among the organisms that have shown translocation in animals, and such translocation is often assumed to occur in humans also, this phenomenon has not, to our knowledge, been demonstrated in an in vitro transcytosis system; indeed, the single study we could find on this subject reported failure of strain OG1X to show translocation across epithelial cell monolayers, and the authors concluded that the intact epithelial cell layer serves as a barrier for enterococci in vitro (18). That paper and others have, however, shown internalization of some E. faecalis organisms by T84 and other intestinal cell lines (14, 18). In the present study, we first demonstrated that the E. faecalis strain OG1RF, unlike E. coli DH5α, was able to translocate across a T84 monolayer, a model for intestinal epithelium transcytosis. Since tight junctions were intact in the monolayer throughout the experiment, as indicated by the maintenance of high electrical resistance, we suspect that the bacteria crossed through the T84 monolayer, presumably by entry into the T84 cells following initial attachment; survived within these cells; and subsequently escaped from the T84 cells on the opposite side.
Having established that E. faecalis can show translocation in this model, we then investigated whether this model could show differences between OG1RF and derivatives; for this purpose, we chose to evaluate the effect of mutations in a gene cluster (epa) that our group has previously demonstrated is involved in polysaccharide biosynthesis in E. faecalis (Fig. 1B) and that is important for virulence in mice and for resistance to phagocytosis and/or killing by polymorphonuclear leukocytes (7, 20, 25-27). The epa gene cluster has been shown to be widespread among E. faecalis strains (20), and Epa is thought to be a species-specific polysaccharide (7). While we have not characterized Epa biochemically, Hancock and Gilmore were able to detect three different cell wall carbohydrates in E. faecalis: a type-specific capsular polysaccharide, a rhamnopolysaccharide (possibly Epa), and a polymer likely representing an integral cell wall teichoic acid (7). Their model, in agreement with our earlier observations (27), suggests that the Epa polysaccharide is not on the cell surface, at least in vitro; however, Epa may be important for the overall integrity of membrane and/or cell wall structures, or conceivably, it could be exposed on the surface in vivo. Our present results showed that there was no detectable translocation of the epa mutant TX5179 across the T84 monolayers and that the translocation efficiency of the epa mutant TX5180 was lower (about 10-fold) than that of the wild-type strain OG1RF, indicating that some epa genes, especially orfde4, are important for successful translocation. This conclusion was further confirmed by complementation of TX5179 with intact orfde4 and orfde5 genes (likely in the same operon).
In the epa gene cluster, orfde6 (encoding a putative glucose-1-phosphate thymidyltransferase) is located downstream of orfde4 (encoding a putative glycosyltransferase), and each gene appears to have an individual promoter for expression and to be on a separate transcript, as indicated by the complementation in E. coli of the transposon insertion in orfde4 by pTEX5175, which does not have an intact copy of orfde6 (Fig. 1B) (26, 27). Since both mutants lack the Epa polysaccharide (20), the different phenotypes suggest that other, as yet undefined differences exist between these mutants. For example, the gene product of orfd4 might be used in other pathways that are important for translocation, and/or some function of orfde6 may be partially complemented in E. faecalis by another gene(s). Further characterization of these mutants and the epa gene cluster will be helpful for understanding these differences.
We next evaluated the abilities of 14 human isolates to translocate across T84 monolayers and found considerable differences among strains (Fig. 2), with 8 strains showing translocation and 6 strains failing to show detectable translocation. The reason for the difference in strains is not known. The previous study that failed to show translocation by E. faecalis in a similar in vitro system used OG1X, a nitrosoguanidine-generated derivative of OG1 (which is also the parent of OG1RF [19]) which has an unknown number of uncharacterized mutations (18); we also found that OG1X did not show translocation in our assay (unpublished data). In any case, our results with OG1RF and other isolates indicate that at least some E. faecalis isolates, including isolates of different geographic and clinical origins (endocarditis, urine, and community-derived feces), can translocate across T84 monolayers and that the ability to translocate varies among these strains. We also note that all five fecal isolates showed translocation as opposed to three of nine clinical isolates, suggesting the possibility that translocation ability may be a common feature of organisms recently derived from feces; however, the numbers are too small to allow any conclusion.
In summary, an in vitro model with T84 cells was established to study enterococcal translocation across intestine-derived cells. In this work, we showed that OG1RF and 8 of 15 other E. faecalis isolates, but not E. coli DH5α, were able to translocate and that the epa polysaccharide biosynthesis gene cluster, especially orfde4, is important for the successful translocation of E. faecalis. This study suggests that polarized T84 cell models of translocation may be suitable for studying the migration of E. faecalis from the intestinal tract, a process that may lead to transient bacteremia and subsequent endocarditis.
Acknowledgments
We thank Karen D. Jacques-Palaz for outstanding technical assistance.
This work was supported by grant NIH R37 AI47923 from the Division of Microbiology and Infectious Diseases, NIAID, to B.E.M.
REFERENCES
- 1.Berg, R. D. 1995. Bacterial translocation from the gastrointestinal tract. Trends Microbiol. 3:149-154. [DOI] [PubMed] [Google Scholar]
- 2.Bras, A. M., and J. M. Ketley. 1999. Transcellular translocation of Campylobacter jejuni across human polarised epithelial monolayers. FEMS Microbiol. Lett. 179:209-215. [DOI] [PubMed] [Google Scholar]
- 3.Burns, J. L., A. Griffith, J. J. Barry, M. Jonas, and E. Y. Chi. 2001. Transcytosis of gastrointestinal epithelial cells by Escherichia coli K1. Pediatr. Res. 49:30-37. [DOI] [PubMed] [Google Scholar]
- 4.Dharmsathaphorn, K., and J. L. Madara. 1990. Established intestinal cell lines as model systems for electrolyte transport studies. Methods Enzymol. 192:354-389. [DOI] [PubMed] [Google Scholar]
- 5.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 6.Finlay, B. B., and S. Falkow. 1990. Salmonella interactions with polarized human intestinal Caco-2 epithelial cells. J. Infect. Dis. 162:1096-1106. [DOI] [PubMed] [Google Scholar]
- 7.Hancock, L. E., and M. S. Gilmore. 2002. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. USA 99:1574-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopper, S., J. S. Wilbur, B. L. Vasquez, J. Larson, S. Clary, I. J. Mehr, H. S. Seifert, and M. So. 2000. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect. Immun. 68:896-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madara, J. L., S. P. Colgan, A. Nusrat, C. Delp, and C. A. Parkos. 1992. A simple approach to measurement of electrical parameters of cultured epithelial monolayer: use in assessing neutrophil epithelial interaction. J. Tissue Culture Methods 14:209-216. [Google Scholar]
- 10.McCormick, B. A., P. M. Hofman, J. Kim, D. K. Carnes, S. I. Miller, and J. L. Madara. 1995. Surface attachment of Salmonella typhimurium to intestinal epithelia imprints the subepithelial matrix with gradients chemotactic for neutrophils. J. Cell Biol. 131:1599-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monteville, M. R., and M. E. Konkel. 2002. Fibronectin-facilitated invasion of T84 eukaryotic cells by Campylobacter jejuni occurs preferentially at the basolateral cell surface. Infect. Immun. 70:6665-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray, B. E. 1998. Enterococci, p. 1723-1730. In S. L. Gorbach, J. G. Bartlett, and N. R. Blacklow (ed.), Infectious diseases, 2nd ed. W. B. Saunders Company, Philadelphia, Pa.
- 13.Nassif, X., and M. So. 1995. Interaction of pathogenic neisseriae with nonphagocytic cells. Clin. Microbiol. Rev. 8:376-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olmsted, S. B., G. M. Dunny, S. L. Erlandsen, and C. L. Wells. 1994. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J. Infect. Dis. 170:1549-1556. [DOI] [PubMed] [Google Scholar]
- 15.Ring, A., J. N. Weiser, and E. I. Tuomanen. 1998. Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J. Clin. Investig. 102:347-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runkel, N. S., F. G. Moody, G. S. Smith, L. F. Rodriguez, Y. Chen, M. T. Larocco, and T. A. Miller. 1993. Alterations in rat intestinal transit by morphine promote bacterial translocation. Dig. Dis. Sci. 38:1530-1536. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Sartingen, S., E. Rozdzinski, A. Muscholl-Silberhorn, and R. Marre. 2000. Aggregation substance increases adherence and internalization, but not translocation, of Enterococcus faecalis through different intestinal epithelial cells in vitro. Infect. Immun. 68:6044-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 20.Teng, F., K. D. Jacques-Palaz, G. M. Weinstock, and B. E. Murray. 2002. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect. Immun. 70:2010-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 22.Wells, C. L. 1996. Colonization and translocation of intestinal bacterial flora. Transplant. Proc. 28:2653-2656. [PubMed] [Google Scholar]
- 23.Wells, C. L., R. P. Jechorek, and S. L. Erlandsen. 1990. Evidence for the translocation of Enterococcus faecalis across the mouse intestinal tract. J. Infect. Dis. 162:82-90. [DOI] [PubMed] [Google Scholar]
- 24.Winram, S. B., M. Jonas, E. Chi, and C. E. Rubens. 1998. Characterization of group B streptococcal invasion of human chorion and amnion epithelial cells in vitro. Infect. Immun. 66:4932-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu, Y., L. Jiang, B. E. Murray, and G. M. Weinstock. 1997. Enterococcus faecalis antigens in human infections. Infect. Immun. 65:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu, Y., B. E. Murray, and G. M. Weinstock. 1998. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect. Immun. 66:4313-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu, Y., K. V. Singh, X. Qin, B. E. Murray, and G. M. Weinstock. 2000. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect. Immun. 68:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]