Abstract
The N and W-Beijing families of Mycobacterium tuberculosis are phylogenetically closely related. The ability of the W-Beijing family to rapidly cause widespread disease is well described; however, few outbreaks involving the N family have been reported outside the New York City, N.Y., area. During 2002 to 2003, Seattle, Wash., experienced a rapidly expanding tuberculosis outbreak involving 38 persons in a 23-month period. The outbreak strain, SBRI9, exhibited the genotypic properties of the N family. Its IS6110 restriction fragment length polymorphism pattern was identical or nearly identical to those of two N family strains that were responsible for clusters of tuberculosis cases, including a large nosocomial outbreak, in New York City and New Jersey from 1989 to 1990. It was also identical to strains involved in late 1990s tuberculosis cases in Michigan, Maryland, and Arkansas. Further monitoring of the N family may show that it shares with the W-Beijing family the propensity to spread rapidly, suggesting that this characteristic evolved prior to the divergence of the two genetic lineages.
Genotypic analysis of Mycobacterium tuberculosis isolates has been used to confirm tuberculosis (TB) outbreaks (17), to investigate TB transmission in communities (3, 24), to identify cases of exogenous reinfection (23), to confirm cross-contamination in the laboratory (10), and to study clonal expansion of strains (14). Clonal expansion has been studied most extensively in the W-Beijing family of M. tuberculosis strains (25). A review of the epidemiology and phylogenetic reconstruction of the W-Beijing family was published recently (2).
The widespread geographic distribution and frequent TB outbreaks associated with the W-Beijing family suggest that these strains are successful human pathogens that spread more aggressively than other strains (2, 16, 20, 25). The factors responsible for this success are not yet known. Studies have documented clonal expansion of multidrug-resistant (MDR) as well as pan-susceptible W family strains across the United States (4, 14). Characterization of the development of MDR during dissemination of one clone, strain W14 (14), improved our understanding of how MDR M. tuberculosis develops and how it can be controlled. By understanding how the W-Beijing family evolved, we may better understand the specific characteristics that make some strains of M. tuberculosis spread more rapidly than others.
W and Beijing strains share certain genotypic characteristics, and they are viewed as members of the same phylogenetic lineage. These characteristics are (i) an IS6110 restriction fragment length polymorphism (RFLP) pattern similar to that of strain W, with 15 to 26 IS6110 copies; (ii) a spoligotype pattern, S00034, characterized by the absence of spacers 1 to 34 and positive hybridization to spacers 35 to 43 in the direct repeat region; (iii) the occurrence of two mutations, one in katG codon 463 (Leu) and one in gyrA codon 95 (Thr), that define principal genetic group 1; (iv) an IS6110 insertion in the origin of replication between the dnaA and dnaN genes; and (v) one or two IS6110 insertions in the NTF chromosomal region (Beijing strains have one insertion, whereas W MDR strains from New York City, N.Y., have two insertions in a head-to-tail orientation) (3, 4, 14, 18, 21).
In contrast to the case for the W-Beijing family, the public health importance of closely related groups has not been extensively documented. Doing so would help us to understand the characteristics that make the strains of the W-Beijing family spread so aggressively and how they evolved. One ancestral branch related to the W-Beijing family, the N family, was described by Kurepina et al. (14). N family strains lack IS6110 insertions in the NTF chromosomal region and have IS6110-based RFLP patterns distinct from those of W-Beijing strains, they but share other genotypic characteristics with W-Beijing strains.
In 2002, Public Health Seattle and King County, Wash., reported a 100% increase in TB among its homeless population, suggesting that an outbreak was occurring. Strain typing of M. tuberculosis isolates from homeless TB patients identified an outbreak cluster, which was determined to be caused by an N family strain that is closely related or identical to strains responsible for TB cases in New York City, New Jersey, and Michigan. This is the first report of N family clusters outside the New York City area.
MATERIALS AND METHODS
Strain typing of isolates from the Seattle outbreak.
M. tuberculosis isolates from patients suspected to be involved in the Seattle, Wash., outbreak were sent to the Seattle Biomedical Research Institute for genotyping from 1 August 2002 through 1 January 2003. Standard spoligotyping methods (12) were used to rapidly assess whether isolates were likely related to the outbreak strain. Subsequently, standard IS6110-based RFLP methods (22) were performed on all isolates, with priority given to isolates with matching spoligotype patterns. Further genetic analysis was performed on 16 isolates with identical spoligotype and RFLP patterns. Matching spoligotypes were assigned designations according to standard nomenclature (9). RFLP fingerprint images were captured by using an Expression 1680 instrument (Epson) and analyzed by using BioNumerics software (BioSystematica) on a Dell workstation.
Genotypic analysis of the Seattle outbreak strain.
Strains were analyzed by PCR for the presence of IS6110 in the dnaA-dnaN intergenic region of the origin of replication. Primers were originally designed for use as a PCR-generated probe for region A1 in dnaA-dnaN (region Af [5′-CGCATCCGTCAGCGCTCCAA] and region Ar [5′-GCCAACTCTTGTCGTAGCCGC]) (14). Solutions consisting of 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs) (200 μM each), 1 μM primer (each), 50 ng of DNA template, and 1 U of Taq polymerase (Sigma Chemical Co., St. Louis, Mo.) were prepared for a final reaction volume of 50 μl. PCR cycling times were (i) 4 min at 94°C, (ii) 30 cycles of 30 s at 94°C, 30 s at 60°C, and 2 min at 72°C], and (iii) 7 min at 72°C. Amplification products were analyzed on a 1% agarose gel. A 537-bp product was expected in samples that did not have an IS6110 insertion in the dnaA-dnaN region, compared to a ∼2,000-bp product expected in those that carried the insertion. Amplification products were confirmed by automated sequencing. Products were purified with the QIAquick Spin PCR purification kit (Qiagen, Valencia, Calif.) and sequenced with an ABI PRISM 377 DNA sequencer (Applied Biosystems, Foster City, Calif.). Sequences were analyzed by BLAST comparison to the National Center for Biotechnology Information nonredundant sequence database (www3.ncbi.nlm.nih.gov/Blast/).
The principal genetic group of the outbreak strains was determined by PCR amplification and DNA sequencing of target-specific regions within the genes katG and gyrA. Primers katGf (5′-TCGACTTGCCGCCCTGACG)-katGr (5′-ACAGCCACCGAGCACGAC) and gyrAf (5′-CACCTACATCGACTATGCGA)-gyrA (5′-GGGCTTCGGTGTACCTCAT) were used as previously described (13). The PCR mixture contained 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs (200 μM each), 500 nM primer (each), 50 ng of DNA template, and 1 U of Taq polymerase (Sigma), for a final volume of 50 μl. PCR cycling times for katG were as follows: (i) 4 min at 94°C; (ii) 35 cycles of 1 min at 94°C, 1 min at 60°C, and 2 min at 72°C; and (iii) 15 min at 72°C. Those for gyrA were as follows: (i) 4 min at 94°C; (ii) 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C; and (iii) 15 min at 72°C. Products were analyzed on a 1% agarose gel, and the appropriate product sizes were detected (katG, 1,429 bp; gyrA, 320 bp). The products were then purified and sequenced as described above.
A multiplex PCR approach was used to determine whether there was an IS6110 in the NTF-1 region of the outbreak strains of M. tuberculosis. PCR primers and parameters were previously described by Plikaytis et al. (18). Briefly, a 50-μl reaction mixture containing 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs (200 μM each), 1.0 μM primer (each), 50 ng of DNA template, and 1 U of Taq polymerase was amplified with the following parameters: (i) 4 min at 94°C; (ii) 30 cycles of 1.5 min at 94°C, 1.75 min at 60°C, and 2.5 min at 72°C; and (iii) 7 min at 72°C. Products were analyzed on a 1.5% agarose gel. The expected band sizes were 523 and 223 bp for Beijing family strains and 523 bp for the positive control (H37Rv).
Seattle outbreak investigation.
The medical charts of all homeless persons diagnosed with TB in 2002 were reviewed to determine each patient's site of disease and results of the sputum examination and human immunodeficiency virus (HIV) test. Contact investigations were reviewed to determine epidemiological associations among patients and results of tuberculin skin tests in contacts. Drug susceptibility testing was performed on all M. tuberculosis isolates.
Identification of related strains involved in TB clusters in other states.
Spoligotyping and RFLP images were independently compared to those in the Centers for Disease Control and Prevention (CDC) 1996 to 2000 National TB Genotyping and Surveillance Network (NTBGSN) database (7) and the Public Health Research Institute (PHRI) TB Center database in Newark, N.J.
RESULTS
Genotypic characteristics of the Seattle outbreak strain.
Spoligotyping and RFLP analysis were performed on 123 isolates from King County. Spoligotype analysis of 16 isolates from epidemiologically linked homeless persons (K. H. Lofy, M. McConnell, S. Goldberg, N. Mills, T. Kuss, G. Cangelosi, J. Milan, L. Diem, J. Pang, K. Hawthorne, and P. McElroy, presented at the International Meeting of the American Thoracic Society, Seattle, Wash., 2003) revealed positive hybridization to spacers 35 to 43 of the direct repeat locus (spoligopattern S0034). IS6110 RFLP analysis of these 16 isolates showed a matching 15-band pattern. The number of bands is at the low end of range of the IS6110 copy numbers expected for W-Beijing family isolates (Table 1). The Seattle outbreak strain RFLP pattern was designated SBRI9.
TABLE 1.
Genotypic characteristics of M. tuberculosis strains from the W-Beijing family and the N family and of Seattle outbreak strain SBRI9
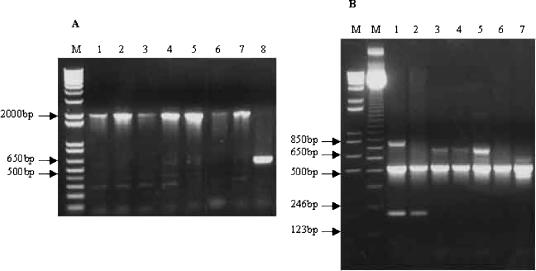
As shown by PCR analysis, strain SBRI9 has an IS6110 insertion in the origin of replication, between the dnaA and dnaN genes (Fig. 1A). Sequence analysis showed that it has the mutations in the katG and gyrA genes (Kat463Leu and Gyr95Thr, respectively) that categorize it as a member of principal genetic group 1. These characteristics are consistent with the W-Beijing family (2). However, strain SBRI9 does not have an IS6110 insertion in the NTF region (Fig. 1B). Taken together, these characteristics are consistent with the N family (Table 1).
FIG. 1.
Genotypic analysis by PCR. (A) A1 IS6110 insertion in the dnaA-dnaN intergenic region (14). Lanes 1 to 7, Seattle outbreak isolates. Lane 8, control M. tuberculosis strain H37Ra. A ∼640-bp product is seen in the H37Ra control, and a ∼2,000-bp product is seen in the Seattle outbreak isolates, indicating an IS6110 insertion in the origin of replication. (B) Multiplex PCR for IS6110 insertions in the NTF region (18). Lanes 1 and 2, Beijing family control strains. Lanes 3 to 5, Seattle outbreak isolates. Lanes 6 and 7, non-W-Beijing control strains KY and H37Ra, respectively. Beijing strains are expected to have one IS6110 insertion in the NTF region, resulting in a 223-bp PCR product. W strains, which were not included in this experiment, have a second insertion resulting in a 175-bp product. The 500-bp product is an internal PCR control, and larger bands are nonspecific products, which were seen previously in some strains analyzed under these conditions (18).
Characteristics of the Seattle outbreak.
Seventeen (57%) of 30 TB cases among homeless people diagnosed in 2002 were linked by molecular strain typing to strain SBRI9. As of 14 November 2003, 21 (78%) of 27 TB cases among homeless people diagnosed in 2003 matched the outbreak strain, bringing the total number of cases involving this strain to 38 in a 23-month period. Further molecular and epidemiological analysis is in progress. Isolates from all outbreak cases have been susceptible to isoniazid, rifampin, ethambutol, pyrazinamide, and streptomycin. Seven (41%) of the patients diagnosed in 2002 were HIV infected.
Of 242 high-risk outbreak contacts who completed tuberculin skin testing and were entered in the database as of 15 April 2003, 95 (39%) had positive skin tests with ≥5 mm of induration (Lofy et al., presented at the International Meeting of the American Thoracic Society). In contrast, during the first 10 months of 2002, before program resources were diverted from routine screening to intensive screening activities, routine screening resulted in only 52 positive results (7%) (≥10 mm) among 798 homeless persons in Seattle completing tuberculin testing. One outbreak case (HIV negative) had previously been treated successfully for active disease with a different M. tuberculosis strain that exhibited a 3-band IS6110 RFLP, indicating that this patient was exogenously reinfected with the 15-band SBRI9 strain (data not shown).
Related strains involved in TB clusters in other states.
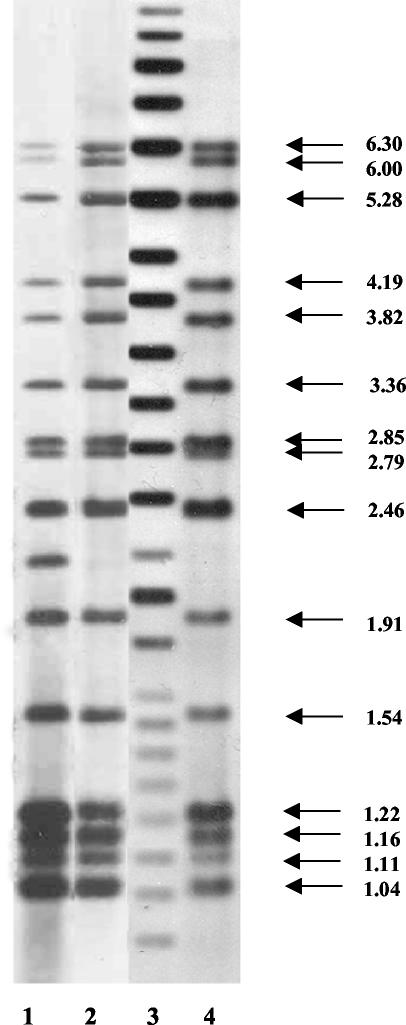
Comparison of the 15-band SBRI9 RFLP pattern to those in the PHRI TB Center database found strain SBRI9 to be identical to a pan-susceptible N family strain, N4, that has been isolated from 24 patients in New York City (n = 22) and New Jersey (n = 2) between 1992 and 2002 (Fig. 2). Strain SBRI9 was almost identical by IS6110 RFLP pattern to another N family isolate, an MDR strain designated N2 (14). This N2 strain caused a significant MDR TB outbreak at St. Luke's Roosevelt Hospital in New York City in 1989 to 1990 (8). The PHRI TB Center database contains 77 N2 strains isolated from TB patients between 1992 and 1998 (n = 75) and in 2001 (n = 2). The SBRI9 and N2 RFLP patterns were identical, with the exception of a 2.25-kb fragment harboring IS6110 in strain N2 that is absent in strain SBRI9 (Fig. 2).
FIG. 2.
IS6110-based RFLP genotyping patterns of N family strains isolated in New York City and Seattle. Lanes 1 and 2, New York City strains N2 and N4. Lane 4, Seattle outbreak strain SBR19. Lane 3, molecular size standards (in kilobases).
Comparison of the SBRI9 RFLP pattern to those in the NTBGSN database found 10 RFLP pattern matches with isolates obtained from TB patients diagnosed during 1996 to 2000 in Michigan (n = 7), Arkansas (n = 1), Maryland (n = 1), and New Jersey (n = 1); the national fingerprint number assigned by the NTBGSN to this fingerprint pattern is 00787. All 10 cases included pulmonary disease. Six (diagnosed in 1996 to 1997) of the seven Michigan cases, including two in infants, were in a single zip code. Of the seven cases, it was apparent that in two cases adults transmitted the disease to one or two of their younger relatives, accounting for five of the seven cases. The additional two cases did not have enough epidemiological information to link them to any of the other cases.
DISCUSSION
This is the first report of a large TB outbreak due to the M. tuberculosis N family outside the New York City area. Initial characterization of the N family showed that it is very closely related to the W-Beijing strain family (Table 1). Kurepina et al. (14) originally identified 10 N family strains, 3 of which, strains N, N2, and N3, were recovered from a total of 197 patients as of 1998. Strains N and N3 were both pan-susceptible; however, strain N2 was an MDR strain responsible for a nosocomial outbreak in 1989 to 1990 (8). Since 1992, 77 N2 strains have been entered into the PHRI TB Center database. Strain N4, which shares an identical IS6110 RFLP pattern with strain SBRI9 and is pan-susceptible like the Seattle strain, has been isolated from two or three cases per year in New York City and New Jersey from 1992 to 2002 (n = 24). Our findings show that this strain family has spread to other states as well (Table 2).
TABLE 2.
TB case clusters associated with the N family
| Strain | Location | No. of cases confirmed by strain genotype | Time period |
|---|---|---|---|
| N4 | New York City and New Jersey | 24 | 1992-2002 |
| N2 | New York City | 77 | 1992-2001 |
| Abcedef | Michigan | 7 | 1996-2000 |
| SBRI9 | Seattle | 38 | 2002-2003 |
Due to the significant public health threat posed by the MDR W strain identified in New York City in the early 1990s (4), the clonal expansion and molecular nature of this group have been extensively characterized (3, 14, 18). The global distribution of the Beijing family has also been extensively reported (2). However, most reports identified Beijing family strains based on subsets of the characteristics in Table 1, along with other characteristics relevant to their studies. For example, the W-Beijing genotype is commonly defined on the basis of its spoligotype pattern, the high-copy-number IS6110 pattern, and in some cases the presence of the IS6110 insertion(s) in the dnaA-dnaN region (5, 16, 24, 25). These genetic markers are also common to members of the N family. It is the presence or absence of the IS6110 insertions(s) in the NTF chromosomal region, combined with distinct IS6110 RFLP patterns, that distinguishes the W-Beijing and N family strain lineages. Given that most studies do not distinguish this branch from the W-Beijing strains, it is possible that members of the N family have caused case clusters that were attributed to the W-Beijing family. In an attempt to refine the molecular epidemiology of W-Beijing strains and related lineages, a DNA “polyprobe” which can identify and distinguish members of the W-Beijing family and related strain families, including the N family, has been developed (N. Kurepina, E. Shashkina, B. Mathema, E. Likhoshvay, J. Bifani, K. Kremer, D. van Soolingen, and B. Kreiswirth, Abstr. 3rd Meet. Concerted Action Project, Prague, Czech Republic, p. 40, 2003).
Prior to the present report, it was not known whether the N family was involved in TB cases outside the metropolitan New York City area. It now appears that this family has been responsible for TB cases in at least six states (New York, New Jersey, Michigan, Maryland, Arkansas, and Washington). It is not known whether there are epidemiological links between N family cases in other states and the Seattle outbreak. None of the Michigan patients with isolates matching the Seattle outbreak strain were homeless.
A region in the United States is defined by the CDC as having a low incidence of TB if it has ≤3.5 cases per 100,000 (11). The Seattle metropolitan area is a moderate-incidence region, with case rates of 6 per 100,000 throughout the 1990s (6). One case associated with the Seattle 2002 outbreak was an exogenous reinfection of an HIV-negative individual, something that is rarely reported for moderate-incidence settings (19, 23). Bandera et al. (1) reported exogenous reinfection in a region of Italy described as low incidence. However, that region had a case rate of 17.5 per 100,000, five times higher than the CDC definition of low incidence in the United States. Sixteen percent of case patients in that study with recurrent TB (n = 5 of 32) had exogenous reinfection, and 60% of the reinfection strains (n = 3 of 5) were also responsible for case clusters within the community. It is difficult to discern the extent to which host factors, rather than pathogen characteristics, contribute to exogenous reinfection and aggressive expansion of TB outbreaks. However, these observations are consistent with highly virulent strains of M. tuberculosis. Recent data have shown that a W-Beijing strain, HN878, was more virulent in mice and rabbits than strains CDC1551 and H37Rv (15; G. Kaplan, presented at the Keystone Symposium Tuberculosis: Integrating Host and Pathogen Biology, Taos, N.Mex., 26 January 2003). Analysis of the N family may show that it shares similar characteristics, which would indicate that these characteristics developed prior to the divergence of the N and W-Beijing family lineages. Such observations, combined with comparative genomic approaches, may help to identify the genetic basis for the rapid geographic spread of certain M. tuberculosis strains.
Acknowledgments
This work was supported by the Seattle Biomedical Research Institute, the Seattle-King County Department of Public Health, the Washington State Department of Health, the Centers for Disease Control and Prevention, and grant AI25767 from the National Institutes of Health (to G.A.C.).
BioNumerics software was a kind gift of the Firland Foundation. We thank Kaeryn Lewis, Linda Haba, Jeanette Frazier, Carolyn Wallis, Christy Huynh, and the Washington State Department of Health, especially Trang Kuss, Kim Field, and Joseph Aharchi.
REFERENCES
- 1.Bandera, A., A. Gori, L. Catozzi, A. Degli Esposti, G. Marchetti, C. Molteni, G. Ferrario, L. Codecasa, V. Penati, A. Matteelli, and F. Franzetti. 2001. Molecular epidemiology study of exogenous reinfection in an area with a low incidence of tuberculosis. J. Clin. Microbiol. 39:2213-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. Mathema, Z. Liu, S. L. Moghazeh, B. Shopsin, B. Tempalski, J. Driscol, R. Frothingham, J. M. Musser, P. Alcabes, and B. N. Kreiswirth. 1999. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA 282:2321-2327. [DOI] [PubMed] [Google Scholar]
- 4.Bifani, P. J., B. B. Plikaytis, V. Kapur, K. Stockbauer, X. Pan, M. L. Lutfey, S. L. Moghazeh, W. Eisner, T. M. Daniel, M. H. Kaplan, J. T. Crawford, J. M. Musser, and B. N. Kreiswirth. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452-457. [PubMed] [Google Scholar]
- 5.Caminero, J. A., M. J. Pena, M. I. Campos-Herrero, J. C. Rodriguez, I. Garcia, P. Cabrera, C. Lafoz, S. Samper, H. Takiff, O. Afonso, J. M. Pavon, M. J. Torres, D. van Soolingen, D. A. Enarson, and C. Martin. 2001. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am. J. Respir. Crit. Care Med. 164:1165-1170. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. August 2000, posting date. Reported tuberculosis in the United States, 1999. [Online.] http://www.cdc.gov/nchstp/tb/surv/surv99/surv99.htm.
- 7.Crawford, J. T., C. R. Braden, B. A. Schable, and I. M. Onorato. 2002. National Tuberculosis Genotyping and Surveillance Network: design and methods. Emerg. Infect. Dis. 8:1192-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edlin, B. R., J. I. Tokars, M. H. Grieco, J. T. Crawford, J. Williams, E. M. Sordillo, K. R. Ong, J. O. Kilburn, S. W. Dooley, K. G. Castro, et al. 1992. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N. Engl. J. Med. 326:1514-1521. [DOI] [PubMed] [Google Scholar]
- 9.Filliol, I., J. R. Driscoll, D. Van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, G. Kallenius, E. Kassa-Kelembho, T. Koivula, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. De Waard, C. Sola, and N. Rastogi. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jasmer, R. M., M. Roemer, J. Hamilton, J. Bunter, C. R. Braden, T. M. Shinnick, and E. P. Desmond. 2002. A prospective, multicenter study of laboratory cross-contamination of Mycobacterium tuberculosis cultures. Emerg. Infect. Dis. 8:1260-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jereb, J. 2002. Progressing toward tuberculosis elimination in low-incidence areas of the United States. Morb. Mortal. Wkly. Rep. 51:1-16. [PubMed] [Google Scholar]
- 12.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapur, V., L. L. Li, M. R. Hamrick, B. B. Plikaytis, T. M. Shinnick, A. Telenti, W. R. Jacobs, Jr., A. Banerjee, S. Cole, K. Y. Yuen, et al. 1995. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch. Pathol. Lab. Med. 119:131-138. [PubMed] [Google Scholar]
- 14.Kurepina, N. E., S. Sreevatsan, B. B. Plikaytis, P. J. Bifani, N. D. Connell, R. J. Donnelly, D. van Sooligen, J. M. Musser, and B. N. Kreiswirth. 1998. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber. Lung Dis. 79:31-42. [DOI] [PubMed] [Google Scholar]
- 15.Manca, C., L. Tsenova, A. Bergtold, S. Freeman, M. Tovey, J. M. Musser, C. E. Barry III, V. H. Freedman, and G. Kaplan. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha /beta. Proc. Natl. Acad. Sci. USA 98:5752-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mokrousov, I., O. Narvskaya, T. Otten, A. Vyazovaya, E. Limeschenko, L. Steklova, and B. Vyshnevskyi. 2002. Phylogenetic reconstruction within Mycobacterium tuberculosis Beijing genotype in northwestern Russia. Res. Microbiol. 153:629-637. [DOI] [PubMed] [Google Scholar]
- 17.Narvskaya, O., T. Otten, E. Limeschenko, N. Sapozhnikova, O. Graschenkova, L. Steklova, A. Nikonova, M. L. Filipenko, I. Mokrousov, and B. Vyshnevskiy. 2002. Nosocomial outbreak of multidrug-resistant tuberculosis caused by a strain of Mycobacterium tuberculosis W-Beijing family in St. Petersburg, Russia. Eur. J. Clin. Microbiol. Infect. Dis. 21:596-602. [DOI] [PubMed] [Google Scholar]
- 18.Plikaytis, B. B., J. L. Marden, J. T. Crawford, C. L. Woodley, W. R. Butler, and T. M. Shinnick. 1994. Multiplex PCR assay specific for the multidrug-resistant strain W of Mycobacterium tuberculosis. J. Clin. Microbiol. 32:1542-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson, M., N. M. Carroll, E. Engelke, G. D. Van Der Spuy, F. Salker, Z. Munch, R. P. Gie, R. M. Warren, N. Beyers, and P. D. Van Helden. 2002. Multiple Mycobacterium tuberculosis strains in early cultures from patients in a high-incidence community setting. J. Clin. Microbiol. 40:2750-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson, M., S. W. van Lill, G. D. van der Spuy, Z. Munch, C. N. Booysen, N. Beyers, P. D. van Helden, and R. M. Warren. 2002. Historic and recent events contribute to the disease dynamics of Beijing-like Mycobacterium tuberculosis isolates in a high incidence region. Int. J. Tuberc. Lung Dis. 6:1001-1011. [PubMed] [Google Scholar]
- 21.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Rie, A., R. Warren, M. Richardson, T. C. Victor, R. P. Gie, D. A. Enarson, N. Beyers, and P. D. van Helden. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 341:1174-1179. [DOI] [PubMed] [Google Scholar]
- 24.van Rie, A., R. M. Warren, N. Beyers, R. P. Gie, C. N. Classen, M. Richardson, S. L. Sampson, T. C. Victor, and P. D. van Helden. 1999. Transmission of a multidrug-resistant Mycobacterium tuberculosis strain resembling “strain W” among noninstitutionalized, human immunodeficiency virus-seronegative patients. J Infect. Dis. 180:1608-1615. [DOI] [PubMed] [Google Scholar]
- 25.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]