Highlights
► First systems biology review on Trichoderma reesei. ► First ‘omics comparison of Trichoderma reesei and other Trichoderma spp. ► The ‘omics results offer novel understanding of biomass hydrolyzing enzymes. ► Routes for further research are illustrated.
Keywords: Trichoderma, Genomics, Transcriptomics, Proteomics, Metabolomics, Cellulase, Hemicellulase
Abstract
Recent progress and improvement in “-omics” technologies has made it possible to study the physiology of organisms by integrated and genome-wide approaches. This bears the advantage that the global response, rather than isolated pathways and circuits within an organism, can be investigated (“systems biology”). The sequencing of the genome of Trichoderma reesei (teleomorph Hypocrea jecorina), a fungus that serves as a major producer of biomass-degrading enzymes for the use of renewable lignocellulosic material towards production of biofuels and biorefineries, has offered the possibility to study this organism and its enzyme production on a genome wide scale. In this review, I will highlight the use of genomics, transcriptomics, proteomics and metabolomics towards an improved and novel understanding of the biochemical processes that involve in the massive overproduction of secreted proteins.
1. Introduction
The plant cell wall consists of the β-(1,4)-linked glucose polymer cellulose, hemicellulose polysaccharides of varying composition, and lignin. The former two are formed by plants at an annual production rate of about 7.2 and 6 × 1010 tons, respectively (Gilbert, 2010). They thus represent the largest reservoir of renewable carbon sources on earth which could be used for the production of biofuels and other biorefinery products, thereby replacing products derived from fossile carbon components (Kubicek, 2012). While chemical and enzymatic processes for the hydrolysis of these polymers are known, enzymatic hydrolysis is preferred because it produces no inhibitory by-products and is thus environmentally compatible.
The Sordariomycete Trichoderma reesei (teleomorph Hypocrea jecorina) is most widely used for the industrial production of cellulolytic and hemicellulolytic enzymes and has become a paradigm for research on these enzymes (Xu et al., 2009). Consequently, many details about the structure and function of these cellulases and hemicellulases have been elucidated, and several aspects of the regulation of their expression and secretion into the medium have also been described (Kubicek, 2012).
The sequencing of the T. reesei genome (Martinez et al., 2008) and the advances in the various -omics technologies has recently established the fundament for a systems biological (i.e. genome wide and holistic) approach towards an understanding of cellulase and hemicellulase production. Such knowledge could then provide new strategies for strain improvement. While only a few of these studies have already been published (or will soon be), they have already considerable aided to our understanding of this process and led to the identification of genes critical to it.
In this paper I will therefore the main findings obtained by these approaches and show how they have already been contributed to strain improvement.
2. The genomic inventory of cellulases and hemicellulases in T. reesei
The T. reesei genome (34.1 Mbp) comprises 9143 genes (Martinez et al., 2008), which is a relatively small number compared to most other ascomycetes. This number is even smaller than that of other Trichoderma spp. such as Trichoderma virens (12518 genes) and Trichoderma atroviride (11865 genes; Kubicek et al., 2011, 2012), and likely due to the operation of repeat-induced point mutation (RIP), a gene silencing mechanism that occurs in sexually reproducing mycelia. While genes essential for the operation of RIP have indeed been found in all three Trichoderma spp., the stronger operation in T. reesei can be explained by the fact that – in contrast to T. virens and T. atroviride – T. reesei is almost exclusively found only as the teleomorph in nature (Druzhinina et al., 2010).
Yet, what was most puzzling to the scientific community, was the comparatively small number of cellulase and hemicellulase genes present in the T. reesei genome. With a total of 200 glycosyl hydrolase (GH) encoding genes, it actually ranks at the bottom of Pezizomycota GHs. However, T. virens and T. atroviride, who evolved earlier than T. reesei (Kubicek et al., 2011, 2012), rank among the top (260 and 257, respectively). Since the innate nature of Trichoderma is that of a mycoparasite, whereas T. reesei has specialized towards saprophytism on predecayed wood (Druzhinina et al., 2011), it is likely that the ancestors of T. reesei also possessed these genes but T. reesei lost them during speciation. This in turn implies that these additional genes are not needed for efficient breakdown of predigested lignocelluloses. Banerjee et al. (2010) showed that the specific activity of an optimized mixture of eleven purified T. reesei cellulases and cellulase-stimulating proteins equaled that of contemporary commercial enzymes (Accellerase 1000 or Spezyme CP) in in vitro hydrolysis experiments. The enzymes that are lacking in T. reesei may therefore be beneficial to the success of plant pathogens and mycoparasites in their natural habitat, but – if the core set of enzymes is expressed in the appropriate proportions – not be further stimulating cellulose and hemicelluloses hydrolysis.
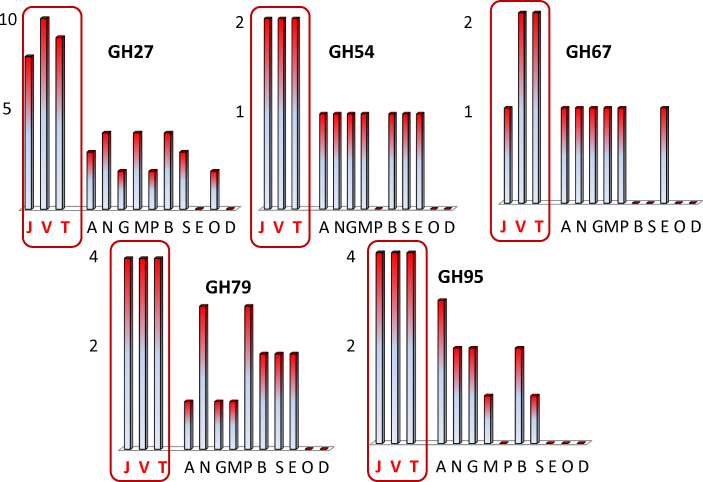
Despite of the small number of cellulases in T. reesei, all three Trichoderma spp. are enriched in some hemicellulolytic components, such as GH27 α-galactosidases, GH43 α-arabinofuranosidases/β-xylosidases, GH67 and GH79 α-methyl-glucuronidases and α-fucosidases (Fig. 1). Also, a comparison of the cellulase and xylanase inventory reveals that the Trichoderma spp. rank the highest in GH30 glucuronyl-xylanases (Table 1). These data therefore indicate that Trichoderma has apparently specialized towards efficient hydrolysis of some hemicelluloses side chains. Several of these enzymes are encoded by genes that share no homologues in other fungi but in bacteria, and may have been obtained by horizontal gene transfer (although this has not yet been rigorously tested) which would further stress that they are important in the natural habitat of Trichoderma.
Fig. 1.
CAZys, indicated by GH-number, that are amplified in Trichoderma. Numbers on the vertical axes of the plots indicate number of gene paralogues. Abbreviations: J, T. reesei; V, T. virens; T, T. atroviride; A, Aspergillus nidulans; N, Aspergillus niger; G, Gibberella zeae; M, Magnaporthe grisea; P, Podospora anserina; B, Botryodiplodia fuckeliana; S, Sclerotium sclerotiorum; E, Neurospora crassa; O, Rhizopus nigricans; D, Batrachochytrium dendrobatidis. Data were compiled from Martinez et al. (2008), Eastwood et al. (2011), Goodwin et al. (2011), and Kubicek et al. (2011, 2012).
Table 1.
Inventory of genes encoding cellulolytic and xylanolytic in Trichoderma spp. and other fungi.a
| Cellulases |
β-Glucosidases |
PMOb | Xylanases |
β-Xylosidase | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GH6 | GH7 | GH12 | GH45 | GH1 | GH3 | GH61 | GH10 | GH11 | GH30 | GH54 | ||
| Stagonospora nodorum | 4 | 5 | 4 | 3 | NI | 16 | 30 | 7 | 7 | NI | 1 | 77 |
| Nectria haematococca | 1 | 3 | 6 | 1 | 5 | 38 | 12 | 3 | 3 | 0 | 1 | 73 |
| Podospora anserina | 4 | 6 | 2 | 2 | 1 | 11 | 33 | 8 | 6 | 0 | 0 | 73 |
| Magnaporthe grisea | 3 | 6 | 3 | 1 | 2 | 19 | 17 | 5 | 5 | 1 | 1 | 63 |
| Fusarium graminearum | 1 | 2 | 4 | 1 | 3 | 22 | 15 | 5 | 3 | 0 | 1 | 57 |
| Aspergillus nidulans | 2 | 3 | 1 | 1 | 3 | 20 | 9 | 3 | 2 | 0 | 1 | 45 |
| Aspergillus niger | 2 | 2 | 4 | 0 | 3 | 17 | 7 | 1 | 4 | 1 | 1 | 42 |
| Neurospora crassa | 3 | 5 | 1 | 1 | 1 | 9 | 14 | 4 | 2 | 1 | 1 | 42 |
| Botrytis cinerea | 1 | 3 | 1 | 2 | 5 | 14 | 10 | 2 | 2 | NI | 1 | 41 |
| Trichoderma virens | 1 | 2 | 4 | 2 | 2 | 17 | 3 | 1 | 4 | 2 | 2 | 40 |
| Trichoderma atroviride | 1 | 2 | 3 | 1 | 4 | 14 | 3 | 1 | 4 | 2 | 2 | 37 |
| Penicillium chrysogenum | 1 | 2 | 3 | 0 | 3 | 17 | 4 | 3 | 1 | 1 | 1 | 36 |
| Sclerotinia sclerotiorum | 1 | 3 | 2 | 2 | 1 | 12 | 9 | 2 | 3 | NI | 1 | 36 |
| Trichoderma reesei | 1 | 2 | 2 | 1 | 2 | 13 | 3 | 2 | 4 | 2 | 2 | 34 |
| Mycospherella graminearum | 0 | 1 | 1 | 1 | NI | NI | 2 | 2 | 1 | NI | 1 | 9 |
| Blumeria graminis var. hordei | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | NI | 0 | 2 |
Data taken from Martinez et al. (2008), Kubicek et al. (2011, 2012), Eastwood et al. (2011) and Goodwin et al. (2011); GH numbers specify the CAZy glycosyl hydrolase classification. Note that GH5 is not listed, because it includes – in addition to endo-β-1,4-glucanases – also many other β-glycanases. GH1 and GH3 are given, though, although it must be born in mind that not all of them are indeed β-glucosidases. NI, no information available.
PMO, polysaccharide monooxygenases; former termed GH61 “endo-β-1,4-glucanases”.
One must also note that gene prediction from genome sequence data is notoriously difficult for fungal species. Arvas et al. (2010) therefore used sparse arrays and identified further 23 genes in T. reesei that were not detected by Martinez et al. (2008). All of them were found to be expressed at high levels, and represented orphan, i.e. lineage-specific genes that are only found in T. reesei or its close relatives. Interestingly, many of them are located next to regulatory genes such as IRE1 (involved in the unfolding protein response; Valkonen et al., 2004) or the carbon catabolite repressor CRE1 (Strauss et al., 1995; Ilmen et al., 1996).
3. Biased distribution of CAZyme genes in the T. reesei genome
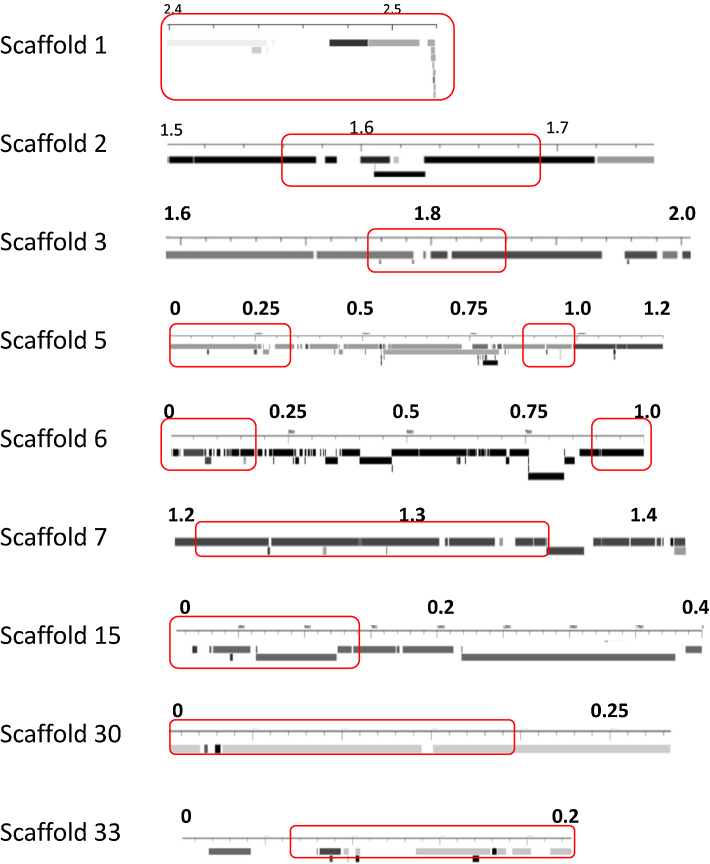
Another intriguing feature that distinguishes the genomes of T. reesei (and also T. atroviride and T. virens; unpublished data) from that of other Pezizomycota is the occurrence of the cellulase and hemicellulase genes in clusters (Martinez et al., 2008). In T. reesei, 130 of the 316 (41%) of the GH and other carbohydrate active genes (such as glycosyl transferases, carbohydrate esterases, and carbohydrate-binding proteins; further termed “CAZyme” genes) were reported to occur in 25 discrete regions ranging from 14 kb to 275 kb in length at an average density of fivefold greater than the expected density for randomly distributed genes. Since these data were only manually obtained, we reassessed this with REEF (“Regionally Enriched Features”), a software that identifies genomic regions enriched in specific features using a statistic test based on the hypergeometric distribution by using a sliding window approach and adopting the false discovery rate for controlling multiplicity (Coppe et al., 2006). Thereby, we only tested CAZy genes that are upregulated during growth on the cellulase inducing vcarbon sources lactose or cellulose. In this way, we confirmed that indeed 49 genes (25%) of the CAZyme encoding genes that are expressed on lactose are in fact clustered in the T. reesei genome (Table 2). Further, most of these clusters occurred near the end of the respective T. reesei scaffolds. Since the ends of the scaffolds may either be ends of chromosomes (a chromosome map for T. reesei is not yet available), or regions consisting of long repeat sequences arising from chromosome rearrangements, this suggests that these CAZymes are either located near the chromosomal ends or in dynamic regions of the genome. It is thereby intriguing that the CAZyme genes are located within or near nonsyntenic genomic areas (Fig. 2). Also Martinez et al. (2008) reported that 95 (73%) clustered CAZyme genes are located in non-syntenic areas of the genome. Interestingly, only few CAZyme paralogues are co-located in the clusters that lie within the non-syntenic areas illustrating that gene relocation after duplication is responsible for the formation of the CAZyme clusters. Also of interest is the finding that genes encoding cellulose-binding proteins are exclusively found in the syntenic gaps (Martinez et al., 2008).
Table 2.
Analysis of potential clustering of CAZyme encoding genes in the T. reesei genome.a
| Scaffold | Total scaffold length (Mbp) | Position of cluster (Mbp) | Distanceb | Clustered genes | p-value | Cellulose | p-Value | Lactose | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.75 | 2.3–2.47 | 0.45 | 4 | 3.30E−03 | 4 | <0.05 | 3 | <0.05 |
| 2.4–2.5 | 0.25 | 3 | 3.10E−03 | 3 | <0.05 | ||||
| 2 | 2 | 1.56–1.69 | 0.44 | 3 | 1.90E−03 | ||||
| 3 | 1.91 | 1.74–1.86 | 0.17 | 3 | 2.50E−03 | ||||
| 5 | 1.72 | 0–0.27 | 0.27 | 7 | 1.20E−03 | ||||
| 0.95–1.08 | 0.55 | 3 | 3.30E−03 | ||||||
| 6 | 1.45 | 0.01–0.12 | 0.14 | 3 | 1.30E−03 | ||||
| 0.04–0.21 | 0.12 | 4 | 2.30E−03 | 3 | <0.05 | ||||
| 0.89–1.06 | 0.56 | 3 | 1.50E−03 | ||||||
| 7 | 1.42 | 1.21–1.35 | 0.21 | 3 | 3.00E−03 | ||||
| 15 | 0.84 | 0–0.14 | 0.14 | 3 | 2.30E−03 | ||||
| 30 | 0.24 | 0–0.21 | 0.03 | 7 | 2.50E−03 | 3 | <0.05 | 3 | <0.05 |
| 33 | 0.2 | 0.06–0.2 | 0.06 | 3 | 1.20E−03 | ||||
| Total gene number | 49 | 13 | 6 | ||||||
Clustering of genes was tested by REEF, a software that applies a statistic test based on the hypergeometric distribution by using a sliding window approach and adopting the false discovery rate for controlling multiplicity (http://telethon.bio.unipd.it/bioinfo/reef/). To use it for T. reesei, the scaffolds were treated as chromosomes. A window width of 100 kb, a shift of 10 kb and an FDR-corrected p-value of <0.05 were used. A minimum number of 3 clustered genes was used as a threshold for the analysis. CAZyme genes expressed during cultivation on cellulose or lactose were taken from Tisch et al. (2012) and Ivanova et al. (2012).
Distance to either end of the scaffold.
Fig. 2.
Genomic synteny of areas containing cellulase clusters, as determined by REEF. Synteny data were retrieved from the synteny browser at http://genome.jgi-psf.org/Trire2/Trire2.home.html, using the genome of T. atroviride as a comparison. Interrupted bars indicate lack of synteny. The different degrees of fillings of the bars (from black to grey) indicate occurrence of the respective areas on different scaffolds in T. atroviride. Numbers indicate the respective position on the scaffold in Mbp's. Areas containing cellulase genes are indicated by red boxes.
Several of the regions of high CAZyme gene density also contain genes encoding synthases of non-ribosomal peptides (NRPS) and polyketides (PKS), which supports the hypothesis that the CAZymes have a function in the ability of Trichoderma to compete with other organisms in its natural environment, and that this function is supported by the formation of antagonistic metabolites. Since the genes responsible for secondary metabolite synthesis in other fungi (particularly Aspergillus spp.) have been demonstrated to be regulated by the putative protein methyltransferase LaeA (Keller and Hohn, 1997), which is believed to reverse the repression of gene expression at the level of heterochromatin structure (for review see Strauss and Reyes-Dominguez, 2011), we tested whether cellulase expression may be regulated by a T. reesei LaeA orthologue (LAE1). In fact, we could show that in the lae1 knock-out strains the production of cellulases and other CAZymes was reduced almost to zero, whereas it is dramatically increased in strains overexpressing lae1. Thus, LAE1 is essential for cellulase and hemicellulase formation in T. reesei (Kubicek et al., 2012; Seiboth et al., 2012). However, examination of the histone methylation pattern around the cellulase genes showed that these genes are accessible for regulatory proteins, independently of LAE1 function (Seiboth et al., 2012). The mechanism responsible for this effect is more complex than would be anticipated from the data obtained with Aspergillus spp. Arvas et al. (2011) recently proposed that GCN5-acetyltransferases may also be involved in the coordinated expression of clustered cellulase genes in T. reesei.
4. Sequencing of T. reesei mutants genomes identifies genes of potential impact on cellulase production
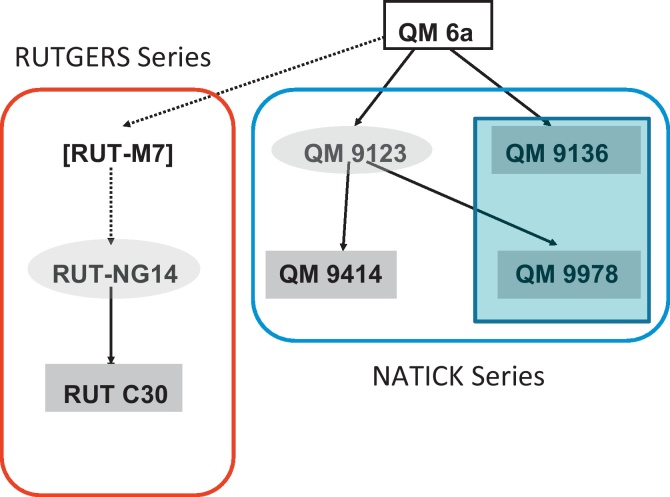
The recent progress in high-throughput massive parallel sequencing technologies made it possible to sequence the genomes of several mutant strains that are either improved or defective in cellulase production (Fig. 3). Le Crom et al. (2009) compared the genome sequence of T. reesei QM 6a (the wild-type isolate sequenced by Martinez et al., 2008) with two isolates of the Rutgers lineage, NG14 and RUT C30 (Eveleigh and Montenecourt, 1979). NG14 was derived from QM6a in two steps: the first involved UV mutagenesis (leading to strain M7 which is no more available) and the second N-nitrosoguanidine mutagenesis. Strain RUT C30 was produced from NG14 by UV mutagenesis and screening for increased cellulose hydrolysis with concomitant resistance to 2-deoxyglucose to eliminate catabolite repression. T. reesei RUT C30 has become a reference strain among T. reesei high cellulase producers, and been used in numerous studies, particularly by researchers from VTT (cf. Arvas et al., 2006, 2011; Rautio et al., 2006). Three mutations (a truncation of the cre1 gene encoding the carbon catabolite repressor CRE1, Ilmen et al., 1996; a frameshift mutation in the glucosidase II alpha subunit gene gls2 involved in protein glycosylation which increases protein secretion, Geysens et al., 2005; and an 85-kb deletion that eliminated 29 genes, including transporters, transcription factors, and primary metabolic enzymes; Seidl et al., 2008) had previously been described to occur in strain RUT C30. Comparison of NG14 and RUT C30 with QM6a identified further 18 and additional 25, respectively, non-synonymous mutations in them (Table 3). Nine of the 18 mutated genes in T reesei NG14 are involved either in RNA metabolism (3 genes), protein secretion/vacuolar targeting (3 genes), or transcriptional regulation (3 genes). Genes affected in RUT C30 partially fall into the same categories, but also comprise genes involved in sugar transport and general metabolism (8 genes), probably due to the selection for growth on glycerol in the presence of 2-deoxyglucose (Eveleigh and Montenecourt, 1979).
Fig. 3.
Pedigree of H. jecorina mutant strains whose genome has been sequenced. The parent strain, whose genome sequence is already available is boxed; the cellulase negative mutants are located within the blue box.
Table 3.
Proteins, whose genes show a non-silent mutation in T. reesei RUT C30, NG14, QM 9414 and QM 9123.a
| Protein ID | Annotation |
|---|---|
| NG14 | |
| 3027 | Pso2 (Snm1) protein family involved in DNA interstrand crosslink repair |
| 67658 | A/G-specific adenine DNA glycosylase |
| 60243 | AMP1, medium subunit of the adaptor protein complex AP-1 of clathrin-coated vesicles |
| 69437 | B-type cyclin |
| 4921 | C2H2 transcriptional regulator |
| 64866 | DSBA-like thioredoxin domain-containing protein |
| 105874 | FAD-dependent (isoamyl?) oxidase |
| 55358 | Flavin-containing monooxygenase, putative |
| 79405 | HET-containing protein, unknown |
| 40758 | Homocysteine methyltransferase |
| 104161 | Importin beta-5 subunit, putative |
| 104599 | Mandelate racemase/muconate lactonizing enzyme |
| 28409 | MFS permease |
| 58561 | MFS permease |
| 64882 | MFS permease |
| 109320 | Orphan protein |
| 109432 | Orphan protein |
| 45456 | Protein kinase |
| 124172 | Protein kinase NPKA [E. nidulans], probably involved in DNA damage signal transduction |
| 75072 | Snf7 family protein, possibly involved in vesicular trafficking |
| 78320 | Sulfatase |
| 3501 | Unknown protein |
| 6014 | Unknown protein |
| 22841 | Unknown protein |
| 53492 | Unknown protein |
| 67030 | Unknown protein |
| 69181 | Unknown protein |
| 74570 | Unknown protein |
| 109285 | Unknown protein |
| 74765 | Unknown protein with C-terminal bromodomain |
| 65104 | Vacuolar protein sorting-associated protein Vps13 |
| 70071 | Zn2Cys6 transcriptional regulator |
| 77513 | Zn2Cys6 transcriptional regulator |
| 107601 | Zn-finger protein, Tim8, probable subunit of mitochondrial import inner membrane translocase |
| RUT C30 | |
| 1751 | FAD monooxygenase |
| 2583 | Imidazoleglycerol-phosphate synthase subunit H |
| 5403 | Enoyl-CoA hydratase/isomerase |
| 22294 | Nuclear transport factor 2 |
| 22994 | AAA-ATPase Cdc48, ER-resident protein degradation |
| 26255 | Zn2Cys6 transcriptional regulator |
| 54157 | Unknown protein |
| 54511 | Unknown protein, contains BTB/POZ domain (=protein binding) |
| 55887 | Unknown protein, secreted |
| 56077 | Zn2Cys6 transcriptional regulator |
| 56726 | Branched chain alpha-keto acid dehydrogenase complex, alpha subunit |
| 58073 | Uroporphyrinogen decarboxylase |
| 58790 | Glycerol-3-phosphate phosphatase, putative |
| 59146 | Polyketide synthase |
| 59388 | MSF permease |
| 59801 | Unknown protein |
| 60458 | NRPS |
| 63702 | Unknown protein |
| 63935 | Isoleucyl-tRNA synthetase, class Ia. |
| 65106 | CN_hydrolase |
| 65773 | Unknown protein |
| 73912 | Unknown protein |
| 75105 | Unknown protein |
| 76515 | Vacuolar protein sorting-associated protein Vps33 |
| 78158 | Importin subunit beta-3 |
| 78268 | Unknown protein |
| 79014 | Vacuolar ATP synthase 16 kDa proteolipid subunit |
| 81149 | Aquaglyceroporin |
| 82499 | Integral membrane protein, putative |
| 102776 | Unknown protein |
| 103061 | Unknown protein |
| 105391 | Unknown protein |
| 106009 | Zn2Cys6 transcriptional regulator |
| 107078 | Unknown protein |
| 108133 | Unknown protein |
| 110882 | Unknown protein |
| 111109 | Unique, no reference in JGI |
| 119768 | Myb transcriptional regulator |
| 120117 | C2H2 transcriptional regulator |
| 122689 | Unknown protein |
| 123786 | NRPS |
| QM 9123 | |
| 2439 | Arc15 (N. crassa) ortholog |
| 5645 | G_glu_transpeptGamma-glutamyltranspeptidase |
| 5645 | G_glu_transpeptGamma-glutamyltranspeptidase |
| 35386 | Actin-interacting protein AIP3 |
| 43191 | Unknown protein |
| 54781 | Peptidyl-tRNA-hydrolase |
| 57940 | Alternative oxidase aox1 |
| 58910 | Unknown protein |
| 68956 | d-Aspartate oxidase |
| 68956 | d-Aspartate oxidase |
| 70546 | Poly(A) polymerase, RNA-binding region |
| 76018 | CysK Cysteine synthase |
| 81136 | Serine/threonine phosphatase 2C, PTC2 |
| 104335 | Unknown protein |
| 107460 | Unknown protein |
| 108645 | Unknown protein |
| 108962 | Unknown protein |
| 111849 | GH30 |
| 120127 | Transcriptional regulator GATA-type zinc finger protein ASD-4 (ascospore development protein 4) |
| 124022 | Zn2Cys6 transcriptional regulator |
| QM 9414 | |
| 23028 | Ca2+ permeable channel, related to N. crassa NCU02762.1 |
| 33895 | Peptidylprolyl isomerase, FKBP-type |
| 52499 | C2H2 transcriptional regulator |
| 60508 | Unknown protein |
| 62633 | Unknown protein |
| 65021 | Oxidoreductase, short-chain dehydrogenase/reductase family |
| 65141 | Cytochrome P450 monooxygenase |
| 65232 | Short chain oxidoreductase/dehydrogenase, putative |
| 77661 | Oxysterol binding protein-like protein |
| 78306 | Rhodanese-like domain-containing protein |
| 79813 | Flavoprotein monooxygenases |
| 102776 | Unknown protein |
| 103044 | Unique, no reference in JGI |
| 104175 | Unique protein |
| 104333 | Unknown protein |
| 108540 | Unknown protein |
| 109278 | GH24 |
| 109313 | NmrA-like family protein |
| 111645 | Unique protein |
| 122036 | Unknown protein |
| 124295 | Cell wall protein with a CFEM domain. |
Only mutations that arose specifically in the strain listed, are given; i.e. mutations already present in NG14 are not again listed in RUT C30, and the same accounts for QM 9123 and QM 9414.
Vitikainen et al. (2010), using tiling arrays and comparative genomic hybridization, identified additional 12 and 4 mutations in NG14 and RUT C30, respectively, which besides unknown genes comprised a transcription factor and a clathrin complex subunit in the former, and another transcription factor and a Gβ-WD40 protein in the latter (Table 3). While it is unclear at the moment whether any of these mutations in RUT C30 is causally related to the increased cellulase forming ability (or whether its higher production can be solely explained by the cre1 and gls2 mutation only), it is clear that one of the mutations found in NG14 must have caused a doubling of cellulase production. The 85-kb gap in RUT C30 has been shown to be not related to cellulase formation (Vitikainen et al., 2010).
Vitikainen et al. (2010) also compared the genomes of the two early strains of the Natick mutagenesis program (Reese, 1976) to their parent QM 6a. They detected 20 mutations in QM 9123, of which 16 were preserved in QM 9414, and additional 7 in the latter (Table 3). Interestingly, three genes mutated in QM 9414 (81136, 124295 and 108540) encode putative cell wall or plasma membrane proteins. It is possible that an altered cell wall/membrane structure may have been beneficial for spore survival and germination during mutagenesis (radiation), but it cannot be ruled out that these genes are also relevant for improved cellulase production. Such an assumption would be supported by two other mutated genes involved in cytoskeleton function (2439 encoding a ARP2/3 complex protein; and 35386 encodes actin interacting protein 3), and lend to hypothesize that the improvement of cellulase production in QM 9414 is due to mutations in cellulase trafficking and secretion.
The genomes of three cellulase-negative T. reesei mutants (QM 9136, QM 9978, QM 9979; cf. Fig. 3) have recently also been sequenced and their evaluation is currently in progress (S.E. Baker and S. Le Crom, personal information). One of them bears a frameshift mutation in the gene encoding the transcriptional activator of cellulase and hemicellulase gene expression, xyr1, and reintroduction of the native gene restores its ability to produce cellulases (S.E. Baker, B. Seiboth and colleagues, unpublished data). However, xyr1 is intact in the other two mutants and the identification of the gene(s) responsible for the cellulase-negative phenotype will likely identify yet unknown regulators of cellulase production.
5. The T. reesei transcriptome
Formation of cellulases and hemicellulases by T. reesei is dependent on induction by cellulose, lactose or sophorose, and they are not formed constitutively. This is managed by the interplay of positive (XYR1, ACE2) and negative (CRE1 and ACE1) transcription factors (for review see Kubicek et al., 2009). Consequently, comparison of the transcriptional pattern between induced and non-induced T. reesei cultures can be used to identify genes whose expression correlates with cellulase and hemicellulase gene expression. In a pioneering study, Foreman et al. (2003) partially sequenced over 5100 random T. reesei cDNA clones from cultures growing on lactose. Among the sequences whose predicted gene products had significant similarity to known proteins, 12 were identified that encode previously unknown enzymes that likely function in biomass degradation, thereby significantly enhancing the number of cellulase and hemicellulase genes known until then. They then constructed microarrays from these genes to investigate the expression levels of each of the sequences under different conditions known to induce cellulolytic enzyme synthesis (growth on lactose, replacement on sophorose) in the wild-type QM6a and a hyperproducer, RL-P37. Most of the genes encoding known and putative biomass-degrading enzymes were found to be transcriptionally co-regulated. Moreover, despite the fact that several of these enzymes are not thought to degrade cellulase directly, they were coordinately overexpressed in a cellulase overproducing strain. They also found that the expression of genes involved in protein processing and secretion was not significantly different under cellulase producing conditions.
Whole-genome microarray analyses of the T. reesei transcriptome formed during growth on cellulose or lactose have now been performed in several laboratories (Portnoy et al., 2012; Häkkinen et al., 2012; Ivanova et al., 2012), but not yet been published as full papers. In my group, the transcriptome of T. reesei cultures grown on lactose, glucose and glycerol was compared. This showed that the lactose grown cultures not only displayed enhanced transcription of all cellulase genes, but also of most hemicellulase genes (Ivanova et al., 2012). Also the transcriptome upregulated on lactose comprised a high number of genes encoding putative transporters of 12-helix major solute facilitator type that may be responsible for the uptake of the cellulose and hemicellulose degradation products. A comparison with the T. reesei transcriptome on cellulose (Tisch et al., 2012; C.P. Kubicek, unpublished data) revealed that the transcriptome on cellulose is richer in the diversity of xylanases, polygalacturonases and chitinases, but otherwise qualitatively similar to that on lactose.
Arvas et al. (2011) studied the effect of the growth rate of T. reesei on genome-wide events related for cellulase formation and protein production in general, using chemostat cultures. They found that the expression of many genes of secreted proteins and secondary metabolism, as well as various lineage specific, mostly unknown genes, were all positively correlated with the specific extracellular protein production rate, which in turn was highest at the lowest growth rate (0.03 h−1). The major biosynthetic activities, in contrast, negatively correlated with the extracellular protein production. Transcriptomic and proteomic analysis suggested that the growth rate and the cell density strongly influence the flux through upper glycolysis or the TCA cycle. At low specific growth rates, this results in a low flux to biomass, and this could be the fundamental determining factor of protein production, by induction of lineage specific genes and regulatory factors for it.
They also noted a significant nonrandom distribution of significantly expressed genes in the genome, which agrees with the above discussed clustering of CAZyme expression (vide supra). Based on the presence of GCN5-related acetyltransferase genes in the upregulated transcriptome, they discussed that a general opening of lineage specific chromosomal regions or more specifically promoters of individual lineage specific genes through histone acetylation could be a major inducing factor in the transcription of Zn2Cys6 transcription factors (which were also abundant in the transcriptome), and these could then induce further actual lineage specific, often neighboring, enzyme genes.
A similar approach has been used by Portnoy et al. (2011), who studied the role of the specific growth rate on carbon catabolite repression in T. reesei. Using chemostat cultures on glucose as a carbon source and a recombinant strain defective in the function of the CRE1 carbon catabolite repressor (delta-cre1), they noted a complex interplay between carbon catabolite repression and the growth rate. They also provided evidence for a CRE1-independent repression of gene expression by high specific growth rates, which most strongly affected genes involved in C1 and carbohydrate metabolism. In this regards it is important to stress that the results of Arvas et al. (2011) were obtained with the T. reesei mutant RUT C30 which has a nonfunctional cre1 gene, and the effects of the specific growth rates on the transcriptome must therefore be viewed with some caution. Interestingly, none of the cellulase genes was significantly (>log 1.5-fold) expressed on glucose in the delta-cre1 strain and at low growth rates, indicating that induction is essentially for their expression (Portnoy et al., 2012).
Protein production at high levels, or production of heterologous proteins, in T. reesei and other fungi has been shown to stimulate the unfolded protein response (UPR), a mechanism that enables the cell to rapidly remove misfolded proteins (Valkonen et al., 2004). Arvas et al. (2006) used an EST-based approach to identify the genes that accompany this induction of UPR. They obtained two new insights: one was that secretion stress induces the histone genes H2A and H4 in an UPR-like manner. The second was that CPC1, the master regulator of the general amino acid control (=the yeast GCN4 orthologue; Hinnebusch, 2005), is upregulated during UPR. While similar findings had already been published for S. cerevisiae Gcn4p and the mouse homologue ATF4 (Harding et al., 2003), it was of interest to learn that in T. reesei – like in mouse but unlike in yeast – CPC1 seems to induce genes involved in glutathione biosynthesis (including the T. reesei homologue of human microsomal glutathione S-transferase 3, MGST3) to alleviate the lack of reducing power. The possibility that induction of cpc1 during UPR could simply be due to amino acid starvation due to massive protein overproduction was ruled out by demonstrating sufficiently high concentrations of 15 amino acids inside of the T. reesei cells.
The soon publication of full details on the transcriptome of T. reesei wild type strains and mutants will show how these findings are reflected in enzyme producing fermentations on cellulose and lactose, and likely lead to the identification of new genes whose modulation impacts cellulase and hemicellulase formation by T. reesei. In fact, researchers at VTT have recently patented 11 and 4 genes whose overexpression or deletion, respectively, increased cellulase and hemicellulase production in T. reesei QM6a (Table 4; Pakula et al., 2011a,b,c). According to the protocols supplied, these genes were selected from those whose transcriptomic expression pattern paralleled that of cellulase genes in RUT C30 during growth on Avicel cellulose. Most of them comprise Zn2Cys6 transcription factors, but two others GCN5-related acetyltransferases, thus supporting the histone acetylation hypothesis proposed by Arvas et al. (2011).
Table 4.
Novel genes affecting cellulase and xylanase production.a
| Cellulase | Xylanase | Notes | ||
|---|---|---|---|---|
| 77513 | Zn2Cys6 transcriptional regulator | Up | Up | Lower biomass formation |
| 80291 | Zn2Cys6 transcriptional regulator | Up | Up | |
| 41573 | bZIP-transcriptional regulator | Up | (Up) | |
| 74765 | Unknown bromodomain-containing protein | (Up) | (Up) | |
| 64608 | Unknown protein with WD 40 domains | Up | (Up) | |
| 123668 | GCN5-related N-acetyltransferase | Up | (Up) | |
| 66966 | Unknown protein with WD 40 repeats | Up | (Up) | |
| 122523 | Zn2Cys6 transcriptional regulator | Up | No effect | |
| 120120 | GCN5-related N-acetyltransferase | Up | (Up) | |
| 112524 | Zn2Cys6 transcriptional regulator | No effect | Up | |
| 108381 | Zn2Cys6 transcriptional regulator | Down | Down | |
| 81972 | Cellular retinaldehyde-binding/triple function | Down | Down | |
| 62244 | Zn2Cys6 transcriptional regulator | Down | Down | |
| 76677 | Zn2Cys6 transcriptional regulator | Down | Down | |
| 47317 | Zn2Cys6 transcriptional regulator | Down | No effect | |
Extracted from Pakula et al. (2011a,b,c). “up” means upregulation in a strain overexpressing the gene, “down” means downregulation in the overexpressing strain. Up in brackets indicates only a weak effect (<30%).
6. The T. reesei secretome
Proteomic approaches related to cellulase and hemicellulase formation in T. reesei mostly concentrated on the analysis of the extracellular proteins secreted during growth on cellulose or lactose, i.e. the secretome. Chundawat et al. (2011) explored the protein composition of several commercial cellulase and xylanase preparations from T. reesei using a proteomics approach with high-throughput quantification using liquid chromatography–tandem mass spectrometry (LC–MS/MS). As expected, Cel7A (former CBH1) was the predominant cellulase in all major commercial enzymes, followed by Cel6A (former CBH 2).The endoglucanases and hemicellulases made up for less then 10% of total secreted protein. Also the accessory proteins swollenin and CIP1 were present in high amounts (up to 4% and 3%, respectively). Also the xyloglucanase GH 74 was present at high levels, which is consistent with transcriptomic data (Ivanova et al., 2012; Tisch et al., 2012). Interestingly, the Cel7A/Cel6A ratio varied considerably (between 1.8 and 3.3), suggesting that protein accumulation of these two cellulases in the medium does not correlate with the well established coregulation seen for these two enzymes at the mRNA-level.
Similar finding were also reported by Herpoël-Gimbert et al. (2009), who compared the secretome of T. reesei RUT C30 and the hyperproducer CL847 (Durand et al., 1988) on lactose by 2-dimensional electrophoresis (2DE). They found that the 2DE profiles of CL847 and Rut-C30 grown on lactose were very different both in terms of spot numbers and protein composition: CL847 2DE reveals many more protein spots that RUT-C30, especially in minor spots corresponding to less than 0.5% of total spots volume, and presence of several Cel7A isoforms for CL847. Also, the relative amount of β-glucosidase BGLI, of endo-β-1,4-glucanase CEL12A and several xylanases is much higher in CL847. A drawback of the study by Herpoël-Gimbert et al. (2009), however, is that they detected only a small number of proteins whose genes are clearly expressed on lactose. Jun et al. (2011) studied the composition of the secretome upon cultivation of T. reesei RUT C30 on two different cellulase inducing media (spent hydrolysate model medium; and a lactose-based standard medium), using 2DE followed by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) and electrospray ionization liquid chromatography tandem mass spectrometry (ESI-LC MS/MS). Fifty two proteins were detected, most of them related to the degradation of lignocellulosic biomass. The enzyme production profiles on the two media were found to be similar, with the exception of β-glucosidase and β-galactosidase which were only produced on the lactose containing medium.
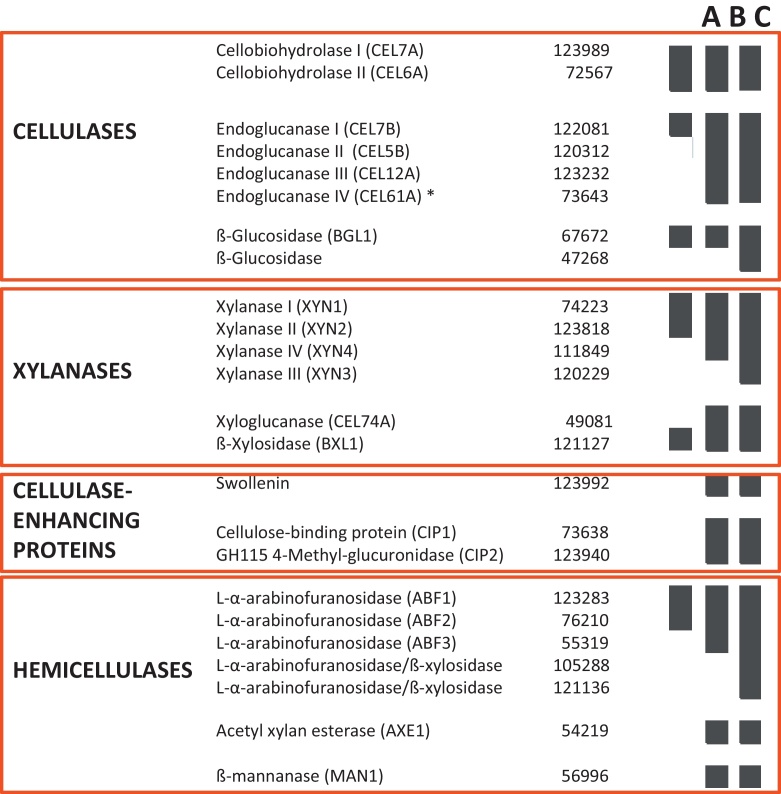
The most comprehensive analysis to date has been performed by Adav et al. (2011), who analyzed the profile of extracellular enzymes in T. reesei QM6a, QM 9414, RUT C30 and a QM 9414-MG5 during growth on cellulose at different pH by liquid chromatography–tandem mass spectrometry (LC–MS/MS). They identified a total of 50 cellulases, hemicellulases and cellulase-accessory proteins, respectively. A comparison of the proteins detected in the studies by Herpoël-Gimbert et al. (2009), Jun et al. (2011) and Adav et al. (2011) is presented in Fig. 4.
Fig. 4.
Extracellular CAZymes identified by proteomic approaches by Jun et al., 2011 (A), Herpoël-Gimbert et al., 2009 (B) and Adav et al., 2011 (C). Black boxes indicate that the protein was detected, numbers are the protein IDs in the T. reesei genome database. Note that the endoglucanase marked with an * is actually a cellulose monooxygenase.
Adav et al. (2011) also detected the presence of several proteins putatively representing laccases, glyoxal oxidase, peroxin 11C, peroxidase/catalase, bifunctional catalase-peroxidase, glutathione transferase, cytochrome oxidase, and cytochrome c peroxidase. Glyoxal oxidase, a H2O2 generating, copper metalloenzyme plays an important role in lignin degradation catalyzed by lignin- and manganese peroxidases in white rot fungi (Hammel and Cullen, 2008). The fact that all these enzymes are formed during growth on cellulose raises the speculation that the breakdown of the predigested cellulose by T. reesei may be aided by oxidative enzymes. It is thus challenging to test their effect on in vitro cellulose hydrolysis by T. reesei cellulases. Also several peptidases and amidases were found.
Another interesting detail of the proteomic studies by Jun et al. (2011) and Adav et al. (2011) is that they detected several intracellular enzymes (such as dehydrogenases) in the culture filtrate. This indicates that enzyme secretion by T. reesei is accompanied by considerable autolysis or mycelia fragmentation, whose role for high enzyme production has not yet been investigated.
7. The T. reesei metabolome
Metabolomic approaches have so far not been strongly introduced to study cellulase or hemicellulase formation by T. reesei. We recently studied the intracellular accumulation of galactose-containing oligosaccharides (“galactoglycome”) during growth of T. reesei on lactose. The rationale for this was the finding that cellulase production on lactose needs both an enzymatically active version of the galactokinase encoding gene gal1 (Hartl et al., 2007), as well as the aldose reductase XYL1 that can convert galactose to galactitol and which is essential for a second pathway of galactose catabolism that is particularly active during growth on lactose (Seiboth et al., 2007). We therefore wondered whether metabolites from both pathways would form oligosaccharides that could act as inducers of cellulase formation (P.J. Punt, C.P. Kubicek and colleagues, in preparation). The data indeed revealed that the intracellular concentration of some oligogalactosides statistically correlates with cellulase gene expression and these could thus be inducers for lactose-promoted cellulase formation. Yet a knock-out of several putative β-galactosyltransferases of T. reesei did not result in any effect on cellulase formation on lactose (M. Mikus, B. Seiboth and C.P. Kubicek, unpublished data), and this hypothesis has thus first to be verified by detection of the enzyme responsible for the formation of this oligogalactoside.
Towards a quantitative description of metabolic fluxes during cellulase formation, Rautio et al. (2006) used a metabolomic C13 partitioning analysis for quantifying fluxes through the carbon catabolic and anabolic pathways. They found that the induction of cellulase gene expression with sophorose did not result in significant changes in the ratio of anabolic and oxidative activities of the TCA cycle. Also, the relative requirements of proteinogenic amino acids were not altered, which they interpreted by the fact that the amino acid composition of the T. reesei cellulases and hemicellulases do not significantly differ from that of the total intracellular protein. However, the requirement for increased biosynthetic amino acid supply may become critical under production of high amounts of cellulases like with industrial strains.
Rautio et al. (2006) also found that the respiratory pathway in T. reesei is not regulated by the carbon catabolite repressor CRE1, and only minor differences were observed in the ratio of the relative anaplerotic flux to the respirative pathway flux of the wild type and the Δcre1 T. reesei strains. This indicates that CRE1 does not mediate carbon source dependent metabolic state alterations in the central carbon metabolism of T. reesei, what is consistent with the transcriptomic data by Portnoy et al. (2011; vide supra).
8. Concluding remarks
As I have attempted to highlight above, the application of several “-omics” to T. reesei and its formation of biomass degrading enzymes have led to a number of new and exciting insights into this process and its regulation, and many more findings will be published in the next future. In addition, tools for high throughput molecular genetics work with T. reesei have been considerably expanded over the last years, and recipient strains with almost 100% integration into the homologous locus, a high throughput system for gene disruption, methods for marker removement, several dominant selection markers, and mating type partners for sexual crossing are now available (for review see Seidl and Seiboth, 2010; Schuster et al., 2012; Seiboth et al., 2011). It is therefore possible to tackle all important questions related to cellulase expression and secretion directly in this organism, and consequently there is no need to perform these investigations in other “model fungi”. The only disadvantage, which remains and which probably will not be rapidly overcome, is the comparatively small community of researchers working on the molecular biology of T. reesei which has so far prevented ambitious projects like the establishment of a genome-wide collection of knock-out strains, which would further speed up our understanding of this important biotechnological area and likely broaden the research groups involved in it.
Acknowledgements
The author acknowledges support by the Austrian Science Foundation (projects P19690, P21266 and P23202), and by several International Companies. Work within ACIB is supported by the Federal Ministry of Economy, Family and Youth (BMWFJ), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol and ZIT – Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG.
References
- Adav S.S., Ravindran A., Chao L.T., Tan L., Singh S., Sze S.K. Proteomic analysis of pH and strains dependent protein secretion of Trichoderma reesei. Journal of Proteome Research. 2011;10:4579–4596. doi: 10.1021/pr200416t. [DOI] [PubMed] [Google Scholar]
- Arvas M., Haiminen N., Smit B., Rautio J., Vitikainen M., Wiebe M., Martinez D., Chee C., Kunkel J., Sanchez C., Nelson M.A., Pakula T., Saloheimo M., Penttilä M., Kivioja T. Detecting novel genes with sparse arrays. Gene. 2010;467:41–51. doi: 10.1016/j.gene.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvas M., Pakula T., Lanthaler K., Saloheimo M., Valkonen M., Suortti T., Robson G., Penttilä M. Common features and interesting differences in transcriptional responses to secretion stress in the fungi Trichoderma reesei and Saccharomyces cerevisiae. BMC Genomics. 2006;7:32. doi: 10.1186/1471-2164-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvas M., Pakula T., Smit B., Rautio J., Koivistoinen H., Jouhten P., Lindfors E., Wiebe M., Penttilä M., Saloheimo M. Correlation of gene expression and protein production rate – a system wide study. BMC Genomics. 2011;12:616. doi: 10.1186/1471-2164-12-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee G., Car S., Scott-Craig J.S., Borrusch M.S., Bongers M., Walton J.D. Synthetic multi-component enzyme mixtures for deconstruction of lignocellulosic biomass. Bioresource Technology. 2010;101:9097–9105. doi: 10.1016/j.biortech.2010.07.028. [DOI] [PubMed] [Google Scholar]
- Chundawat S.P., Lipton M.S., Purvine S.O., Uppugundla N., Gao D., Balan V., Dale B.E. Proteomics-based compositional analysis of complex cellulase-hemicellulase mixture. Journal of Proteome Research. 2011;10:4365–4372. doi: 10.1021/pr101234z. [DOI] [PubMed] [Google Scholar]
- Coppe A., Danieli G.A., Bortoluzzi S. REEF: searching REgionally Enriched Features in genomes. BMC Bioinformatics. 2006;7:453. doi: 10.1186/1471-2105-7-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhinina I.S., Komoń-Zelazowska M., Atanasova L., Seidl V., Kubicek C.P. Evolution and ecophysiology of the industrial producer Hypocrea jecorina (Anamorph Trichoderma reesei) and a new sympatric agamospecies related to it. PLoS One. 2010;5:e9191. doi: 10.1371/journal.pone.0009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhinina I.S., Seidl-Seiboth V., Herrera-Estrella A., Horwitz B.A., Kenerley C.M., Monte E., Mukherjee P.K., Zeilinger S., Grigoriev I.V., Kubicek C.P. Trichoderma: the genomics of opportunistic success. Nature Reviews Microbiology. 2011;16:749–759. doi: 10.1038/nrmicro2637. [DOI] [PubMed] [Google Scholar]
- Durand H., Clanet M., Tiraby G. Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzyme and Microbial Technology. 1988;10:341–346. [Google Scholar]
- Eastwood D.C., Floudas D., Binder M., Majcherczyk A., Schneider P., Aerts A., Asiegbu F.O., Baker S.E., Barry K., Bendiksby M., Blumentritt M., Coutinho P.M., Cullen D., de Vries R.P., Gathman A., Goodell B., Henrissat B., Ihrmark K., Kauserud H., Kohler A., LaButti K., Lapidus A., Lavin J.L., Lee Y.H., Lindquist E., Lilly W., Lucas S., Morin E., Murat C., Oguiza J.A., Park J., Pisabarro A.G., Riley R., Rosling A., Salamov A., Schmidt O., Schmutz J., Skrede I., Stenlid J., Wiebenga A., Xie X., Kües U., Hibbett D.S., Hoffmeister D., Högberg N., Martin F., Grigoriev I.V., Watkinson S.C. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- Eveleigh D.E., Montenecourt B.S. Increasing yields of extracellular enzymes. Advances in Applied Microbiology. 1979;25:57–74. doi: 10.1016/s0065-2164(08)70146-1. [DOI] [PubMed] [Google Scholar]
- Foreman P.K., Brown D., Dankmeyer L., Dean R., Diener S., Dunn-Coleman N.S., Goedegebuur F., Houfek T.D., England G.J., Kelley A.S., Meerman H.J., Mitchell T., Mitchinson C., Olivares H.A., Teunissen P.J., Yao J., Ward M. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. Journal of Biological Chemistry. 2003;278:31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- Geysens S., Pakula T., Uusitalo J., Dewerte I., Penttilä M., Contreras R. Cloning and characterization of the glucosidase II alpha subunit gene of Trichoderma reesei: a frameshift mutation results in the aberrant glycosylation profile of the hypercellulolytic strain Rut-C30. Applied and Environment Microbiology. 2005;71:2910–2924. doi: 10.1128/AEM.71.6.2910-2924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert H.J. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiology. 2010;153:444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S.B., M’barek S.B., Dhillon B., Wittenberg A.H., Crane C.F., Hane J.K., Foster A.J., Van der Lee T.A., Grimwood J., Aerts A., Antoniw J., Bailey A., Bluhm B., Bowler J., Bristow J., van der Burgt A., Canto-Canché B., Churchill A.C., Conde-Ferràez L., Cools H.J., Coutinho P.M., Csukai M., Dehal P., De Wit P., Donzelli B., van de Geest H.C., van Ham R.C., Hammond-Kosack K.E., Henrissat B., Kilian A., Kobayashi A.K., Koopmann E., Kourmpetis Y., Kuzniar A., Lindquist E., Lombard V., Maliepaard C., Martins N., Mehrabi R., Nap J.P., Ponomarenko A., Rudd J.J., Salamov A., Schmutz J., Schouten H.J., Shapiro H., Stergiopoulos I., Torriani S.F., Tu H., de Vries R.P., Waalwijk C., Ware S.B., Wiebenga A., Zwiers L.H., Oliver R.P., Grigoriev I.V., Kema G.H. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genetics. 2011;7:e1002070. doi: 10.1371/journal.pgen.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häkkinen M., Arvas M., Oja O., Aro N., Penttilä M.E., Saloheimo M., Pakula T. Transcriptional analysis of Trichoderma reesei cultivated in the presence of different lignocellulose substrates. ECFG 11, book of abstracts; Marburg, FRG, Poster 766; 2012. [Google Scholar]
- Hammel K.E., Cullen D. Role of fungal peroxidases in biological ligninolysis. Current Opinion in Plant Biology. 2008;11:349–355. doi: 10.1016/j.pbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Molecular Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hartl L., Kubicek C.P., Seiboth B. Induction of the gal pathway and cellulase genes involves no transcriptional inducer function of the galactokinase in Hypocrea jecorina. Journal of Biological Chemistry. 2007;282:18654–18659. doi: 10.1074/jbc.M700955200. [DOI] [PubMed] [Google Scholar]
- Herpoël-Gimbert I., Margeot A., Dolla A., Jan G., Mollé D., Lignon S., Mathis H., Sigoillot J.C., Monot F., Asther M. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnology for Biofuels. 2009;1:18. doi: 10.1186/1754-6834-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annual Review of Microbiology. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Ilmen M., Thrane C., Penttilä M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Molecular and General Genetics. 1996;251:451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- Ivanova C., Bååth J.A., Seiboth B., Kubicek C.P. Lactose induces all genes related to plant biomass hydrolysis and corresponding mono- and oligosaccharide transporters in Trichoderma reesei. ECFG 11, book of abstracts; Marburg, FRG, abstract 605; 2012. [Google Scholar]
- Jun H., Kieselbach T., Jönsson L.J. Enzyme production by filamentous fungi: analysis of the secretome of Trichoderma reesei grown on unconventional carbon source. Microbial Cell Factories. 2011;10:68. doi: 10.1186/1475-2859-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N.P., Hohn T.M. Metabolic pathway gene clusters in filamentous fungi. Fungal Genetics and Biology. 1997;21:17–29. [PubMed] [Google Scholar]
- Kubicek C.P., Mikus M., Schuster A., Schmoll M., Seiboth B. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnology for Biofuels. 2009;2:19. doi: 10.1186/1754-6834-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek C.P. Wiley Blackwell; 2012. Fungi and Lignocellulosic Biomass. 304 pp. ISBN: 978-0-470-96009-7. [Google Scholar]
- Kubicek C.P., Herrera-Estrella A., Seidl-Seiboth V., Martinez D.A., Druzhinina I.S., Thon M., Zeilinger S., Casas-Flores S., Horwitz B.A., Mukherjee P.K., Mukherjee M., Kredics L., Alcaraz L.D., Aerts A., Antal Z., Atanasova L., Cervantes-Badillo M.G., Challacombe J., Chertkov O., McCluskey K., Coulpier F., Deshpande N., von Döhren H., Ebbole D.J., Esquivel-Naranjo E.U., Fekete E., Flipphi M., Glaser F., Gómez-Rodríguez E.Y., Gruber S., Han C., Henrissat B., Hermosa R., Hernández-Oñate M., Karaffa L., Kosti I., Le Crom S., Lindquist E., Lucas S., Lübeck M., Lübeck P.S., Margeot A., Metz B., Misra M., Nevalainen H., Omann M., Packer N., Perrone G., Uresti-Rivera E.E., Salamov A., Schmoll M., Seiboth B., Shapiro H., Sukno S., Tamayo-Ramos J.A., Tisch D., Wiest A., Wilkinson H.H., Zhang M., Coutinho P.M., Kenerley C.M., Monte E., Baker S.E., Grigoriev I.V. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biology. 2011;12:R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek, C.P., Seiboth, B., Linke, R., Karimi, R.A., 2012. LaeA from Trichderma reesei. International Patent Application WO2011/161063A1.
- Le Crom S., Schackwitz W., Pennacchio L., Magnuson J.K., Culley D.E., Collett J.R., Martin J., Druzhinina I.S., Mathis H., Monot F., Seiboth B., Cherry B., Rey M., Berka R., Kubicek C.P., Baker S.E., Margeot A. Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16151–16156. doi: 10.1073/pnas.0905848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D., Berka R.M., Henrissat B., Saloheimo M., Arvas M., Baker S.E., Chapman J., Kubicek C.P., Han C.S., Ho I., Larrondo L.F., de Leon A.L., Magnuson J.K., Merino S., Misra M., Nelson B., Putnam N., Robbertse B., Salamov A.A., Schmoll M., Terry A., Thayer N., Westerholm-Parvinen A., Schoch C.L., Yao J., Barabote R., Nelson M.A., Detter C., Bruce D., Kuske C.R., Xie G., Richardson P., Rokhsar D.S., Lucas S.M., Rubin E.M., Dunn-Coleman N., Ward M., Brettin T.S. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn, Hypocrea jecorina) Nature Biotechnology. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- Pakula, T., Saloheimo, M., Häkkinen, M., Westernholm-Parvinen, A., Penttilä, M.E., Vitikainen, M., 2011. Method for improved protein production in filamentous fungi. International Patent WO2011/151513 A1.
- Pakula, T., Saloheimo, M., Häkkinen, M., Westernholm-Parvinen, A., Penttilä, M.E., Vitikainen, M., 2011. Method for protein production in filamentous fungi. International Patent WO2011/151512 A1.
- Pakula, T., Saloheimo, M., Häkkinen, M., Westernholm-Parvinen, A., Penttilä, M.E., Vitikainen, M., 2011. Improved production of proteins in filamentous fungi. International Patent WO2011/151515 A1.
- Portnoy T., Margeot A., Le Crom S., Linke R., Atanasova L., Fekete E., Sándor E., Karaffa L., Druzhinina I.S., Seiboth B., Kubicek C.P. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genomics. 2011;12:269. doi: 10.1186/1471-2164-12-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy T., Bidard-Michelot F., Seiboth B., Monot F., Baker S.E., Kubicek C.P., Le Crom S., Margeot A. Transcriptomic and genomic approaches to understand cellulase hyper-production in the filamentous fungus Trichoderma reesei. ECFG 11, book of abstracts; Marburg, FRG, Poster 793; 2012. [Google Scholar]
- Rautio J.J., Smit B.A., Wiebe M., Penttilä M., Saloheimo M. Transcriptional monitoring of steady state and effects of anaerobic phases in chemostat cultures of the filamentous fungus Trichoderma reesei. BMC Genomics. 2006;7:247. doi: 10.1186/1471-2164-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese E.T. History of the cellulase program at the U.S. army Natick Development Center. Biotechnology and Bioengineering Symposium. 1976;6:9–20. [PubMed] [Google Scholar]
- Schuster A., Bruno K.S., Collett J.R., Baker S.E., Seiboth B., Kubicek C.P., Schmoll M. A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnology for Biofuels. 2012;5:1. doi: 10.1186/1754-6834-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiboth B., Aghcheh R.K., Phatale P.A., Linke R., Hartl L., Sauer D.G., Smith K.M., Baker S.E., Freitag M., Kubicek C.P. The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Molecular Microbiology. 2012;84:1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiboth B., Gamauf C., Pail M., Hartl L., Kubicek C.P. The d-xylose reductase of Hypocrea jecorina is the major aldose reductase in pentose and d-galactose catabolism and necessary for beta-galactosidase and cellulase induction by lactose. Molecular Microbiology. 2007;66:890–900. doi: 10.1111/j.1365-2958.2007.05953.x. [DOI] [PubMed] [Google Scholar]
- Seiboth B., Ivanova C., Seidl-Seiboth V. Trichoderma reesei: a fungal enzyme producer for cellulosic biofuels. In: dos Santos Bernardes M.A., editor. Biofuel Production Recent Developments and Prospects. InTech; Rijeka: 2011. pp. 309–340. [Google Scholar]
- Seidl V., Gamauf C., Druzhinina I.S., Seiboth B., Hartl L., Kubicek C.P. The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics. 2008;9:327. doi: 10.1186/1471-2164-9-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl V., Seiboth B. Trichoderma reesei: genetic approaches to improving strain efficiency. Biofuels. 2010;1:343–354. [Google Scholar]
- Strauss J., Mach R.L., Zeilinger S., Hartler G., Stöffler G., Wolschek M., Kubicek C.P. Cre1, the carbon catabolite repressor protein from Trichoderma reesei. FEBS Letters. 1995;376:103–107. doi: 10.1016/0014-5793(95)01255-5. [DOI] [PubMed] [Google Scholar]
- Strauss J., Reyes-Dominguez D. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genetics and Biology. 2011;48:62–69. doi: 10.1016/j.fgb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisch D., Kubicek C.P., Schmoll M. The phosducin-like protein PhLP1 impacts regulation of glycoside hydrolases and light response in Trichoderma reesei. BMC Genomics. 2012;12:613. doi: 10.1186/1471-2164-12-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkonen M., Penttilä M., Saloheimo M. The ire1 and ptc2 genes involved in the unfolded protein response pathway in the filamentous fungus Trichoderma reesei. Molecular Genetics and Genomics. 2004;272:443–451. doi: 10.1007/s00438-004-1070-0. [DOI] [PubMed] [Google Scholar]
- Vitikainen M., Arvas M., Pakula T., Oja M., Penttilä M., Saloheimo M. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genomics. 2010;11:441. doi: 10.1186/1471-2164-11-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Singh A., Himmel M.E. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Current Opinion in Biotechnology. 2009;20:364–371. doi: 10.1016/j.copbio.2009.05.006. [DOI] [PubMed] [Google Scholar]