Abstract
We verified the analytical performance characteristics of a previously described real-time reverse transcription-PCR (RT-PCR) assay targeting the open reading frame (ORF) 1b region of the severe acute respiratory syndrome coronavirus (SARS-CoV) with RNA transcripts. We then compared it to a novel nucleocapsid gene real-time RT-PCR assay with genomic RNA. The assays differed only in the primer and probe sequences and final concentrations. A commercially available armored RNA (Ambion, Austin, Tex.) was evaluated as positive control for the ORF 1b assay. The analytical sensitivity, reproducibility, amplification efficiency, and dynamic range of the assays were similar. Both were specific for SARS-CoV as determined by testing against human CoV 229E and OC43, specimens from patients without SARS, and by BLAST searches of GenBank for primer and probe sequence homology. The armored RNA was found to be a suitable positive control for the ORF 1b assay that could be reliably recovered and amplified from a variety of clinical specimens.
In November 2002, an outbreak of atypical pneumonia characterized by fever, respiratory compromise, and a high fatality rate emerged in Guangdong province, China. It subsequently was termed severe acute respiratory syndrome (SARS) and rapidly spread across five continents. A novel coronavirus (CoV) was quickly identified as the etiologic agent based on electron microscopy, virus isolation, serology, and reverse transcription-PCR (RT-PCR) amplification (7). Several publications followed, all confirming the presence of this CoV in specimens from SARS patients (2, 3, 9).
Multiple independent and collaborative efforts led to the sequencing of the entire SARS-associated CoV (SARS-CoV) genome (4, 10). The genome shares characteristics with other members of the Coronaviridae family, but phylogenetic analysis showed that it is distinct from all previously recognized species and probably represents an early branch from group 2 CoVs (10). The World Health Organization (WHO) estimated that there were 8,439 cases of SARS, with 812 deaths at the end of the transmission cycle in 2003.
Should SARS return, a premium would be placed on early diagnosis because of the nonspecific clinical presentation, high mortality, and potential for epidemic spread. Currently, there is no effective treatment for the disease. Therefore, prompt and accurate diagnosis and institution of infection control measures remain the best hope for controlling future epidemics.
Several diagnostic modalities including virus culture, serologic testing, and nucleic acid amplification techniques are useful for SARS. However, culture requires special expertise and facilities, since the SARS-CoV is a biosafety level 3 pathogen. Serology has proven to be sensitive, but it may require up to 20 days for serologic conversion (6). Furthermore, if SARS reemerges and becomes endemic, a single serologic value may be difficult to interpret. Several reports have shown that conventional and real-time RT-PCR assays are very specific for SARS-CoV, but they may lack sensitivity depending on the assay, specimen, and time course of disease (6-8). Multiple primer and probe sets and assay formats have been described, but there is little data comparing the performance characteristics of the different assays. However, two first-generation WHO RT-PCR protocols were recently evaluated and found to have similar analytical and clinical performance characteristics (12). The development of SARS diagnostics has been hampered by a lack of appropriate positive-control material and clinical specimens.
Most of the published RT-PCR assays target the open reading frame (ORF) 1b of the RNA polymerase gene. Coronaviridae family members employ a characteristic replication strategy whereby the polymerase gene region is directly translated. The remainder of the genome is transcribed into subgenomic mRNAs in a nested fashion (11). The nucleocapsid, or N gene, is included in all of these transcripts and is consequently found at a higher intracellular concentration than ORF 1b early in the infection of cultured cells. Therefore, an RT-PCR assay targeting the N gene may be more sensitive if it is found in higher copy numbers in clinical specimens.
Here, we compare the analytical performance characteristics of an ORF 1b assay first described by Drosten et al. (2) to an N gene assay developed in our laboratory. We also describe the use of a protein-coated RNA transcript as a positive control for RNA extraction, RT, and amplification of the ORF 1b gene from clinical specimens.
MATERIALS AND METHODS
CoV RNA.
A 189-nucleotide (nt) fragment of the ORF 1b region was cloned and transcribed into RNA as previously described (1, 2). This RNA transcript, designated BNI-1, was kindly provided C. Drosten, Bernhard Nocht Institute for Tropical Medicine, National Reference Center for Tropical Infectious Diseases, Hamburg, Germany. The BNI-1 RNA transcript was diluted in RNase-free water to a concentration of 108 copies/ml.
D. Erdman, Respiratory Virus Branch, Centers for Disease Control and Prevention (CDC), Atlanta, Ga., supplied the SARS-CoV genomic RNA through a material transfer agreement. It was provided as a Vero cell lysate in Trizol LS reagent (Life Technologies, Gaithersburg, Md.). RNA was recovered from the lysate according to the manufacturer's instructions.
A 190-nt BNI-1 fragment was assembled with viral coat proteins into a pseudoviral particle for use as an RNase-resistant RNA control for the ORF 1b assay by Ambion RNA Diagnostics as previously described (5). The BNI-1 armored RNA was added to clinical specimens prior to RNA extraction to achieve a 1-μl equivalent of the original armored RNA solution per RT-PCR.
Human CoV OC43 was provided by D. Erdman. Human CoV 229E (VR740) was obtained from the American Type Culture Collection, Rockville, Md. Viral RNA was extracted from these cultures with the QIAamp viral RNA mini kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. All RNA preparations were stored in RNase-free water at −70°C until needed.
RNA extraction.
SARS-CoV armored RNA was added to nasopharyngeal swab, bronchoalveolar lavage (BAL) fluid, plasma, and stool specimens from individuals without SARS, and the RNA was extracted with the QIAamp viral RNA mini kit. An additional clarification step was included for stool specimens, as recommended by the manufacturer. Briefly, the stool specimen was suspended in 1 ml of 0.9% saline and centrifuged for 20 min at 8,000 × g, and then the supernatant was passed through a 0.2-μm-pore-size filter. Nasopharyngeal swab samples were obtained from healthy volunteers and suspended in approximately 5 ml of sterile saline. The others were discarded clinical specimens obtained from the Emory University Hospital clinical microbiology laboratory.
RT.
cDNA was produced from 3 μl of sample RNA with TaqMan RT reagents and random hexamers (Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions in a 10-μl final reaction volume. Cycling parameters were 10 min at 25°C, 30 min at 48°, and 5 min at 95°, followed by a 4°C hold on a Perkin-Elmer 9600 thermal cycler.
Real-time PCR.
The primer (BNITMS1 and BNITMs2) and probe (BNITMP) sequences used for the ORF 1b assay have been previously described (2). The primers amplify a 77-nt sequence within the BNI-1 fragment.
Primers and probe targeting the N gene were based on the Urbani strain SARS-CoV sequence (GenBank accession no. AY278741) as follows: forward primer 5′-GGAGCCTTGAATACACCCAAAG-3′ (nt 28531 to 28550), reverse primer 5′-GCACGGTGGCAGCATTG-3′ (nt 28597 to 28581), and probe 5′-fluorescein-CCACATTGGCACCCGCAATCCTAATA-tetramethylrhodamine-3′ (nt 28554 to 28579). The primers amplify a 66-nt amplicon. The N gene assay primers and probe were designed with Primer Express software, version 1.5 (Applied Biosystems), by following guidelines established for TaqMan assays.
A GenBank basic local alignment search tool (BLAST) search (http://www.ncbi.nlm.nih.gov/BLAST) revealed no homology between the forward primer or probe used in the N gene assay and other human or viral sequences, including those of other CoVs. Only the reverse primer demonstrated some areas of homology with three other sequences: a zebra fish DNA sequence (accession no. AL596024), Nematodirus helvetianus internal transcribed spacer 1 (accession no. AJ251570), and human protein kinase C (accession no. AC090589). Both assays target regions that are conserved among all of the 14 SARS-CoV genomes available in GenBank at the time of the search.
Each 50-μl PCR mixture consisted of 25 μl of TaqMan Universal master mix (Applied Biosystems) and 5 μl of each primer (final concentrations, 200 nM (each) for ORF 1b and 900 nM (each) for N gene) and probe (final concentration, 250 nM) added to the 10-μl RT reaction mixture after cDNA synthesis. Real-time PCR was performed on an ABI Prism 7700 sequence detection system (Applied Biosystems). Thermal cycling conditions were as follows: initial holds for 2 min at 50°C and then 10 min at 95°C, followed by 40 cycles of 15 s at 95° and 1 min at 60°. Amplification plots were recorded and analyzed with the ABI Prism 7700 system software. Real-time RT-PCR data were analyzed with SAS statistical software (Cary, N.C.).
RESULTS
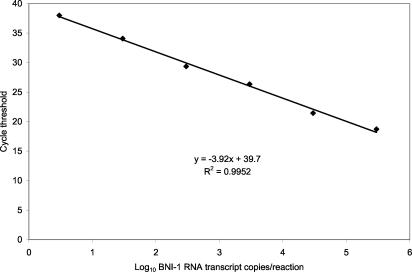
A standard curve for the ORF 1b assay was generated from serial 10-fold dilutions of the BNI-1 RNA transcript (Fig. 1). Each point of the standard curve represents the mean cycle threshold (CT) for duplicates at each concentration determined in two runs. The assay had a linear response over at least a 6-log10 concentration range, from 3 to 300,000 copies/reaction (R2 = 0.995). The ORF 1b assay results were very reproducible with a mean coefficient of variation (CV) in the CT values of only 1.4% (range, 0.7 to 2.4%).
FIG. 1.
Standard curve for the ORF 1b assay generated with the BNI-1 RNA transcript. Each point represents the mean of the results from four determinations.
Eight samples each of nasopharyngeal secretions, BAL fluid, plasma, stool, and water were spiked with the BNI-1 armored RNA transcript prior to RNA extraction to achieve a 1-μl equivalent of the original armored RNA solution per RT-PCR mixture. The armored RNA transcript was reliably recovered and amplified from all of the clinical specimens with the ORF 1b assay. The only detection failure was with one of the water replicates, probably due to a technical error. The mean, range, and standard deviation (SD) of the CT values for the armored RNA in each sample matrix are shown in Table 1. The mean CT values for the armored RNA in the different sample matrices were all well within the detection limit of the assay and ranged from 31.9 to 35.2, with no evidence of a significant sample matrix bias. The overall mean CV of CT values for all samples was 4.5%.
TABLE 1.
Detection of the BNI-1 armored RNA transcript with the ORF 1b assay in different sample matricesa
| Sample matrix | No. detected/no. testedb | Mean CT | Range | SD |
|---|---|---|---|---|
| Nasopharyngeal secretions | 8/8 | 31.9 | 31.2-32.8 | 0.55 |
| BAL fluid | 8/8 | 33 | 29.6-37 | 2.37 |
| Plasma | 8/8 | 31.7 | 30.5-32.8 | 0.9 |
| Stool | 8/8 | 35.2 | 32.4-37.5 | 1.69 |
| Water | 7/8 | 34.4 | 31.6-37.7 | 2.14 |
The BNI-1 armored RNA transcript was added to samples prior to RNA extraction to achieve a 1-μl equivalent of the original armored RNA solution per RT-PCR.
CT < 40.
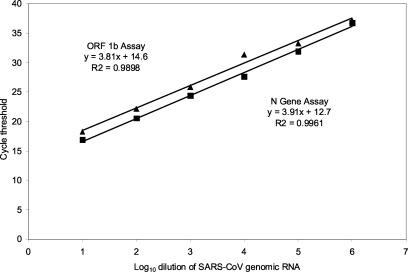
Serial 10-fold dilutions of the SARS-CoV RNA were prepared in water, and each dilution was tested in duplicate in two separate runs of the ORF 1b and N gene assays (Fig. 2). The limit of detection was the 10−6 dilution for both assays. Both assays also demonstrated a linear response over a 6-log10 concentration range (R2 > 0.99). The efficiencies of the two amplification reactions, as indicated by the slopes (m) of the regression lines, were very similar (ORF 1b, m = 3.8; N gene, m = 3.9).
FIG. 2.
Serial log10 dilutions of SARS-CoV genomic RNA tested with the ORF 1b (▴) and N gene (▪) assays. Each point represents the mean of the results from four determinations.
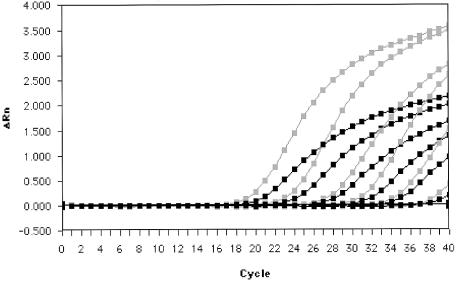
However, lower CT values were obtained at all dilutions with the N gene assay. The mean CT values, SD, and CV for replicates at each dilution in both assays are compared in Table 2. The mean change in CT (ΔCT) (ORF 1b CT − N gene CT) between the assays over the dilution series was 1.6 (P = 0.001, analysis of variance). The amplification plots revealed that the N gene assay had a higher change in fluorescence (ΔRn) than the ORF 1b assay at all dilutions tested (Fig. 3). The reproducibilities of the ORF 1b and N gene assays were similar, with average CV of CT values of replicates of 2.6 and 2.4%, respectively.
TABLE 2.
Comparison of the ORF 1b and N gene assays with SARS-CoV RNAa
| Log RNA dilution | ORF 1b assay
|
N gene assay
|
ΔCTb | ||||
|---|---|---|---|---|---|---|---|
| Mean CT | SD | % CV | Mean CT | SD | % CV | ||
| 1 | 18.2 | 0.17 | 0.93 | 16.9 | 0.17 | 0.99 | 1.3 |
| 2 | 22.1 | 0.75 | 3.38 | 20.5 | 0.31 | 1.52 | 1.6 |
| 3 | 25.8 | 0.18 | 0.7 | 24.4 | 0.66 | 2.69 | 1.4 |
| 4 | 31.4 | 2.57 | 8.21 | 27.6 | 2.18 | 7.87 | 3.8 |
| 5 | 33.3 | 0.7 | 2.1 | 31.9 | 0.19 | 0.58 | 1.4 |
| 6 | 37.1 | 0.1 | 0.27 | 36.8 | 0.23 | 0.63 | 0.3 |
Each dilution of SARS-CoV RNA was tested in duplicate in two runs of each assay (n = 4).
ΔCT = ORF 1b CT − N gene CT.
FIG. 3.
Amplification plots of serial log10 dilutions of SARS-CoV genomic RNA in the ORF 1b (black) and N gene (gray) assays.
A limiting twofold dilution series of SARS-CoV RNA was prepared, and 8 replicates of each dilution were tested with both assays to document any small differences in analytical sensitivity (Table 3). Both assays detected 100% of the samples at an estimated concentration of 1,500 genome equivalents (GE)/ml. Both assays also detected approximately half of the replicates at 750 GE/ml. Below 750 GE/ml detection occurred sporadically with both tests.
TABLE 3.
Limits of detection of the ORF 1b and N gene assays as determined with a terminal dilution series of SARS-CoV RNA
| Dilution | Estimated GE/mla | No. detected/no. tested (%)b for assay
|
|
|---|---|---|---|
| ORF 1b | N gene | ||
| Neat | 1,500 | 8/8 (100) | 8/8 (100) |
| 1:2 | 750 | 4/8 (50) | 3/8 (38) |
| 1:4 | 380 | 0/8 (0) | 2/8 (25) |
| 1:8 | 190 | 1/8 (13) | 1/8 (13) |
| 1:16 | 90 | 0/8 (0) | 2/8 (25) |
GE were estimated by using the BNI-1 standard curve (Fig. 1).
CT < 40.
The specificity of each assay was tested with 32 clinical specimens from patients without SARS and with RNA purified from the two other known human CoVs, 229E and OC43. None of these samples were positive with either test.
DISCUSSION
The ORF 1b assay described here is a modification of the real-time RT-PCR assay first described by Drosten et al. (2). Their assay was used to evaluate respiratory specimens from 5 patients with probable SARS, 13 patients with suspected SARS, and 21 asymptomatic contacts of patients with SARS. All patients with probable SARS, 3 patients with suspected SARS, and none of the case contacts were positive for SARS-CoV RNA. Commercially available conventional (Qiagen) and real-time (Artus GmbH, Hamburg, Germany) RT-PCR tests are also based on this assay.
Our modifications of the original ORF 1b assay included use of the standard reagents and thermal cycle parameters recommended for real-time RT-PCR on the ABI Prism platform. Also, the PCR master mix contained dUTP and uracil-N-glycosylase to protect against false-positive test results due to amplicon cross-contamination. We used random hexamers rather than specific primers to initiate cDNA synthesis because the same RT reaction could be used for assays designed to detect other RNA viruses. We verified that the ORF 1b assay could consistently detect as few as 10 copies of the BNI-1 RNA transcript per reaction mixture, as originally reported.
We also demonstrated that a commercially available armored RNA transcript could serve as an effective positive control for the ORF 1b assay. Unlike naked RNA transcripts or purified genomic RNA, armored RNA can be added to the specimen prior to processing as a control for all of the steps in the RT-PCR: RNA extraction, RT, and amplification of cDNA. The armored RNA was reliably recovered and amplified from nasopharyngeal secretions, BAL fluid, serum, and stool sampels. The virus has been detected in all of these specimen types in patients with SARS, and testing of multiple body sites has been shown to increase the diagnostic sensitivity of RT-PCR (6). The armored RNA can be added to a separate aliquot of each specimen to ensure that samples are free of inhibitors and that all assay components are functioning properly. Although this approach is more labor intensive than incorporating an internal control in each reaction, it is technically simpler to accomplish and avoids concern about reduced sensitivity due to competition when two or more products are simultaneously amplified in the same reaction.
The protocol described here for extraction of viral RNA worked well with a variety of clinical specimens. Pretreatment of specimens with 1% acetylcysteine has been used by others to liquefy highly viscous specimens (2).
CoVs use a unique strategy to synthesize a set of eight subgenomic RNAs. ORF 1b is found on only one of them, whereas the N gene is present in all of the seven remaining subgenomic RNAs. Thus, by targeting the N gene, the sensitivity of nucleic acid tests for the SARS-CoV may be substantially improved. To test this hypothesis, we developed an N gene assay and compared its analytical performance characteristics with the ORF 1b assay by using a dilution series of SARS genomic RNA harvested at a late phase of viral replication.
We found that the amplification efficiency, dynamic range, and reproducibility of the two assays were similar but that the N gene assay was more robust, with consistently lower CT and higher ΔRn values at all concentrations tested. Using equal primer concentrations in the two assays had little effect on the ΔCT and ΔRn values (data not shown). Thus, the differences probably result from a slightly higher copy number of the N gene target and inherent differences in kinetics of binding of the probes to the different target regions.
The average ΔCT for the two assays was 1.6, which implied that the N gene assay should be approximately threefold (21.6) more sensitive than the ORF 1b assay. In an attempt to document the predicted small difference in sensitivity, we used a twofold terminal dilution series of SARS-CoV genomic RNA and tested replicates at each dilution with both assays. We found no measurable difference in analytical sensitivity of the two assays. Both assays detected 100% of the replicates at a concentration of 1,500 GE/ml and approximately 50% of the replicates at 750 GE/ml. It is unlikely that further optimization of either assay would substantially improve the analytical sensitivity, since both reliably detected as few as 5 GE per reaction.
Whether the N gene represents a better target for SARS diagnostics depends on the nature of the predominant viral RNA species in clinical specimens. Since subgenomic RNAs are short-lived and restricted to the intracellular environment, it is unlikely that they would be found in large numbers in respiratory secretions, plasma, or stool specimens. If genomic RNA is the most abundant species in clinical specimens, then assays targeting the ORF 1b and N gene should have equal sensitivity for detection of SARS-CoV in clinical specimens. Our data demonstrate that the N gene assay was no more sensitive than the ORF 1b assay for detection of SARS-CoV RNA isolated from infected Vero cells and suggest that genomic RNA predominates in late-stage infection in cell culture. Comparative data from assays targeting different genomic regions to test clinical specimens from patients with documented SARS are lacking.
Current WHO and CDC guidelines for SARS-CoV nucleic acid diagnostics recommend that a positive PCR should be confirmed by testing a second sample, repeating the test on the same sample, or amplifying a second target region. Amplifying a second target region should provide the best specificity. The N gene assay described here has performance characteristics similar to the well-characterized ORF 1b assay and could serve as confirmatory test. The use of the armored RNA as a positive control for the ORF 1b assay provides an increased level of confidence in the negative results.
Although the CDC has developed SARS-CoV RT-PCR assays for the public health laboratory sector, the performance characteristics of these tests have not been published. It is unlikely that these laboratories would have the surge capacity to provide the results in a clinically relevant time frame if the United States were to experience a large SARS outbreak. The assays described here employ standard reagents and a platform already in use in many hospital-based clinical laboratories and positive-control material that is readily available.
The lack of appropriate clinical material has hampered development and verification of SARS diagnostics. Our study is also limited by the lack of clinical specimens from documented cases of SARS. To expedite the development of better diagnostic tests, the WHO announced the establishment of a SARS specimen bank in June 2003 to hold specimens representing all stages of disease from different body sites and to make them available to interested laboratories. Although this was a welcomed development, the administrative procedures for distribution of these specimens had not been established by the WHO as of November 2003.
Sensitive real-time RT-PCR assays will play an important role in the early diagnosis of SARS should it return. Prompt identification of SARS-CoV in clinical specimens early in the disease should lead to better control of its spread and improved clinical management. It is hoped that the information provided here can help clinical laboratories prepare for this newly recognized threat to public health.
REFERENCES
- 1.Drosten, C., S. Gottig, S. Schilling, M. Asper, M. Panning, H. Schmitz, and S. Gunther. 2002. Rapid detection and quantification of RNA or Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 40:2323-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and S. W. Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 4.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 5.Pasloske, B. L., C. R. Walkerpeach, R. D. Obermoeller, M. Winkler, and D. B. DuBois. 1998. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J. Clin. Microbiol. 36:3590-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, K. Y. Yuen, and H. U. S. S. Group. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, K. Y. Yuen, and S. S. group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poon, L. L., O. K. Wong, K. H. Chan, W. Luk, K. Y. Yuen, J. S. Peiris, and Y. Guan. 2003. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS). Clin. Chem. 49:953-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, A. J. McGeer, and the Canadian Severe Acute Respiratory Syndrome Study Team. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 10.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 11.Thiel, V., K. A. Ivanov, Á. Putics, T. Hertzig, B. Schelle, S. Bayer, B. WeiBbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 12.Yam, W. C., K. H. Chan, L. L. Poon, Y. Guan, K. Y. Yuen, W. H. Seto, and J. S. M. Peiris. 2003. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 41:4521-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]