Abstract
Burkholderia pseudomallei is the etiologic agent of melioidosis. This multifaceted disease is difficult to treat, resulting in high morbidity and mortality. Treatment of B. pseudomallei infections is lengthy and necessitates an intensive phase (parenteral ceftazidime, amoxicillin–clavulanic acid or meropenem) and an eradication phase (oral trimethoprim–sulfamethoxazole). The main resistance mechanisms affecting these antibiotics include enzymatic inactivation, target deletion and efflux from the cell, and are mediated by chromosomally encoded genes. Overproduction and mutations in the class A PenA β-lactamase cause ceftazidime and amoxicillin–clavulanic acid resistance. Deletion of the penicillin binding protein 3 results in ceftazidime resistance. BpeEF–OprC efflux pump expression causes trimethoprim and trimethoprim–sulfamethoxazole resistance. Although resistance is still relatively rare, therapeutic efficacies may be compromised by resistance emergence due to increased use of antibiotics in endemic regions. Novel agents and therapeutic strategies are being tested and, in some instances, show promise as anti-B. pseudomallei infectives.
Keywords: antibiotics, Burkholderia pseudomallei, melioidosis, resistance, therapy
Burkholderia pseudomallei & melioidosis
Burkholderia pseudomallei is a Gram-negative bacterial pathogen that normally survives as a saprophyte in soil and water, but is also capable of causing serious infections in most mammals (Box 1) [1–8]. B. pseudomallei infection is a major cause of bacterial sepsis and chronic disseminated infections (melioidosis) in humans in Asia and northern Australia [2,5,6,9,10]. Melioidosis exhibits a wide array of clinical symptoms, ranging from acute sepsis to chronic recurrent infections, as well as disease with no clinical symptoms [5,6,8–13]. Even with rapid diagnosis and prompt and aggressive treatment, the fatality rate for melioidosis patients still ranges from 10 to 20% in Australia to over 40% in Thailand. B. pseudomallei is considered an emerging pathogen, and infections have been increasingly reported in many countries in tropical and subtropical regions of the world [14–18]. Melioidosis is also a problem increasingly recognized in travelers who have visited regions of the world where B. pseudomallei is endemic [19–22].
Box 1. Profile of Burkholderia pseudomallei.
-
▪
Gram-negative saprophytic bacterium endemic to tropical and subtropical regions of the world
-
▪
Facultative intracellular pathogen
-
▪
Refractory to antibiotic therapy
-
▪
Adaptable to various environments and hosts
-
▪
Adaptability facilitated by large (>7 Mbp) genome consisting of two chromosomes
-
▪
Genome characterized by plasticity, including genomic islands
-
▪
Other than bacteriophage, mobile genetic elements (plasmids, transposons) are noticeably absent
Melioidosis therapy
Owing to rapid disease progression and the propensity of B. pseudomallei to establish latent infections, melioidosis therapy is biphasic and lengthy [9]. An initial intensive phase involving intravenous administration of antibiotics is followed by an eradication phase to minimize the risk of relapse. Current therapy recommendations are based on the outcome of a number of clinical trials in endemic regions and clinical observations, mostly Thailand and northern Australia, and their evolution is summarized in several recent publications [9,11,23], culminating in the latest guidelines [24]. The intensive phase involves intravenous administration of ceftazidime for 10–14 days or longer. However, this course of therapy may be extended to four or more weeks in cases of more severe disease such as septic shock, deep seated or organ abscesses, extensive lung disease, osteomyelitis or neurological melioidosis. In some instances, for example deep seated infections, parenteral ceftazidime can be supplemented with oral or parenteral administration of trimethoprim–sulfamethoxazole (co-trimoxazole). To preserve the utility of carbapenems, meropenem therapy is only recommended when conditions during ceftazidime therapy worsen or repeat blood cultures remain positive. The agent of choice for the at least 3-month eradication phase is oral co-trimoxazole when dealing with susceptible B. pseudomallei and no documented patient agent allergy. In cases where the organism is resistant or patients are intolerant to co-trimoxazole, the second-line choice is amoxicillin–clavulanic acid (co-amoxiclav). Although the most recent recommendations no longer endorse doxycycline use in clinical melioidosis therapy, the antibiotic was previously recommended for postexposure prophylaxis [11] and is still used as a component of eradication-phase treatment in some parts of the world. In Thailand, chloramphenicol was previously included in a four-drug combination treatment, but a clinical study indicated that this antibiotic did not provide any benefit when included in oral melioidosis therapy [25].
Antibiotic resistance in B. pseudomallei
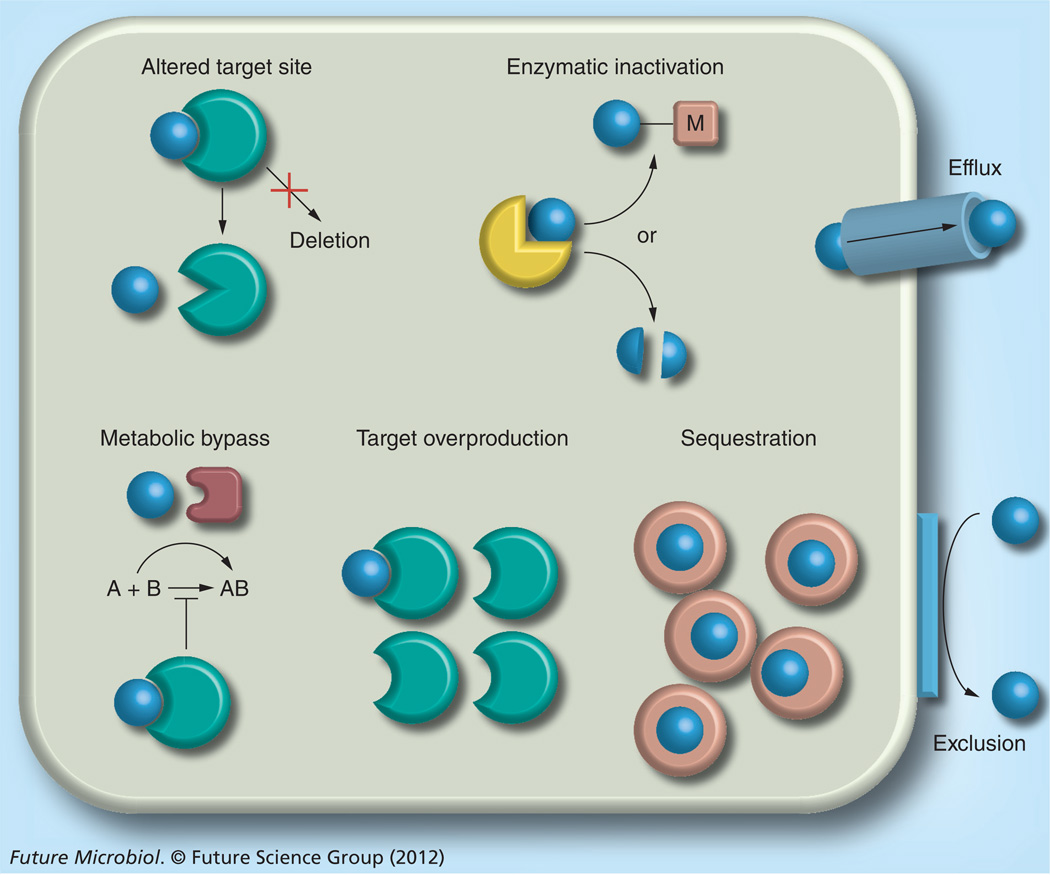
The limited arsenal of antibacterial agents available for melioidosis therapy is mostly due to the inherent resistance of B. pseudomallei to many antibacterial agents. Bacteria possess an impressive antimicrobial resistance armamentarium (Figure 1) [26]. The mechanisms range from exclusion from the cell due to permeability issues bestowed by constituents of the bacterial cell envelope, efflux from the cell, enzymatic inactivation, altered target sites (which in rare instances may include target deletion), metabolic bypass of a susceptible enzyme with a resistant variant, target overproduction and drug sequestration. Of these mechanisms, all except enzymatic modification, target overproduction, metabolic bypass and drug sequestration have been documented in B. pseudomallei (Table 1).
Figure 1. Bacterial antibiotic resistance mechanisms.
Bacterial antimicrobial resistance mechanisms are multifaceted, ranging from: exclusion of drug molecules (blue spheres) from the bacterial cell by physicochemical constraints (e.g., porins or lipopolysaccharide); efflux from the cell via active transport mechanisms; enzymatic inactivation, either in the form of substrate cleavage or chemical modification (M = acetylation, adenylation or phosphorylation); target site alteration or, rarely, target deletion; metabolic bypass by substitution of a susceptible enzyme or pathway with a resistant enzyme or pathway; target overproduction by either increased transcription or gene multiplication; and drug sequestration by specific binding proteins akin to immunity proteins. Details about individual resistance mechanisms and specific examples are provided in the text. Bacteria often employ different resistance mechanisms that act synergistically, for instance, efflux and exclusion, to achieve high-level resistance [26].
Table 1.
Burkholderia pseudomallei antibiotic resistance mechanisms.
| Antibiotic or inhibitor class† | Exclusion | Enzymatic inactivation |
Target mutation |
Efflux |
|---|---|---|---|---|
| Aminoglycosides | X | X | ||
| β-lactams | X | X | ||
| Chloramphenicol | X | |||
| Clavulanic acid | X | |||
| Fluoroquinolones | X | X | ||
| Macrolides | X | |||
| Polymyxin B | X | |||
| Tetracyclines | X | |||
| Trimethoprim | X | |||
| Trimethoprim–sulfamethoxazole | X |
Selected antibiotics affected by the indicated resistance mechanisms are: aminoglycosides, including gentamycin, kanamycin, spectinomycin and streptomycin; β-lactams, including ampicillin, amoxicillin, carbenicillin, ceftazidime, imipenem and piperacillin; fluoroquinolones, ciprofloxacin and norfloxacin; macrolides, including clarithromycin, clindamycin and erythromycin; ketolides, cethromycin; tetracyclines, doxycycline and tetracycline.
In many instances, bacterial antibiotic resistance is mediated by mobile elements, such as plasmids, transposons or integrons [26], but all resistance documented to date in B. pseudomallei is mediated by chromosomally encoded genes. The genome of strain K96243 encodes up to seven Ambler class A, B and D β-lactamases (including a cephalosporinase and oxacillinase), up to ten multidrug efflux systems of the resistance nodulation and cell division (RND) family and a putative aminoglycoside acetyl transferase [27]. Most of these resistance mechanisms are putative and, to date, have been found in neither resistant clinical nor laboratory isolates.
Although relatively rare, resistance to clinically significant antibiotics does emerge during treatment. In one study, 7% of patients harbored chloramphenicol-resistant isolates [28] and rates of co-trimoxazole resistance range from 2.5% in Australia [29] to 13–16% in Thailand [30,31]. A recent report that surveyed antimicrobial resistance in clinical B. pseudomallei isolates from northeast Thailand found that over two decades, only 24 out of 4021 (0.6% of isolates) were resistant to ceftazidime, amoxicillin–clavulanic acid or both drugs [32]. As previously discussed, resistance development during the eradication phase may be highly pertinent in those patients that develop relapse with the same strain that may now be recalcitrant to treatment [9]. Resistance may be underdiagnosed, and reports of ceftazidime resistance as a consequence of using this antibiotic regularly for melioidosis therapy are increasingly emerging from endemic areas, for example, Malaysia [33], Australia [34], Thailand [35,36], Singapore [37] and, most recently, India [38]. Resistance to carbapenems has not yet been reported.
B. pseudomallei select agent status & impact on antimicrobial resistance research
Owing to B. pseudomallei’s select agent designation, antibiotic resistance research with this bacterium was, until very recently, severely restricted and limited to clinical isolates. This changed with the construction of B. pseudomallei strain Bp82 [39], which is excluded from select agent regulations. This strain is an attenuated ΔpurM derivative of clinical isolate 1026b and, with Institutional Biosafety Committee approval, can be handled at biosafety level 2 (BSL2) and used for antibiotic resistance research.
B. pseudomallei antibiotic resistance mechanisms
Exclusion from the cell
The exclusionary property of the Gram-negative cell envelope of many nonenteric Gram-negative bacteria is mostly due to reduced outer membrane permeability, which, in species such as Burkholderia cepacia, is only 11% of that observed in Escherichia coli [40]. It is now recognized that this is primarily due to the physicochemical properties of porins [41] and lipopolysaccharide (LPS) [42]. Although outer membrane permeability has not been studied in any detail in B. pseudomallei, there is experimental evidence to suggest that, analogous to what has been observed in other Gram-negative bacteria, it and its key constituents play a key role in intrinsic and, perhaps, acquired resistance. For example, purified Omp38 facilitates permeation of antibiotics, such as ceftazidime and meropenem, in liposome reconstitution assays [43]. LPS contributes to intrinsic high-level polymyxin B resistance in B. pseudomallei, presumably mostly due to lipid A modification by 4-amino-4-deoxy-l-arabinose [44]. When incorporated into the lipid A of the LPS of Pseudomonas aeruginosa, Salmonella enterica serovar Typhimurium and other Gram-negative bacteria, this amino sugar has been deonstrated to cause resistance to cationic antimicrobial peptides, including polymyxin B [42]. Experimental evidence also indicates that LPS O-antigen and outer core components play roles in resistance to cationic antimicrobial peptides [45].
Efflux from the cell
Bacteria are well endowed with efflux systems [46–49]. Efflux transporters are classified into single- or multi-component pumps. In Gram-negative bacteria, single-component pumps transport their substrates across the cytoplasmic membrane and multicomponent pumps span the entire cell envelope and expel their substrates into the external milieu. In some instances, these pumps act in concert to achieve a robust defense against antimicrobials [50]. Owing to the fact that drug efflux is most effective in bacteria with reduced outer membrane permeability [49], it is the efflux pumps of t he RND family that mostly contribute to intrinsic and acquired resistance in Gram-negative bacteria. They consist of three components: the cytoplasmic membrane RND transporter protein, an outer membrane channel protein and a membrane fusion protein, which form an exit channel traversing the entire cell envelope [51]. The synergy of the energy-dependent, active process of extrusion of antibacterial agents to the external cell milieu and the passive process of concentration gradient-dependent diffusion of most such agents results in high-level drug resistance. Owing to the fact that many RND pumps can accommodate multiple chemically and structurally diverse compounds, their expression often results in intrinsic or acquired multidrug resistance [46,49].
Sequenced B. pseudomallei strains encode multiple RND efflux pumps (e.g., ten that are annotated in the K96243 prototype strain) [27]. Efflux pump expression can be detected in many isolates [52], but the clinical relevance, if any, of most of these pumps remains unknown. Only three have been characterized in any detail, AmrAB–OprA [53], BpeAB–OprB [54,55] and BpeEF–OprC [56].
AmrAB–OprA was the first efflux pump characterized in B. pseudomallei [53]. It is responsible for the intrinsic aminoglycoside and macrolide resistance observed in most clinical and environmental strains. AmrAB–OprA also extrudes fluoroquinolones and tetracyclines but the resistance levels are low and probably clinically insignificant [55]. A few aminoglycoside- and macrolide-susceptible B. pseudomallei clinical strains have been reported [57], and this susceptibility was subsequently attributed to deletion or lack of expression of the chromosomal amrAB–oprA operon [58]. Such variants may be more frequent than originally thought, since clinical diagnosis of melioidosis in many instances still relies on use of Ashdown’s agar, whose main selective ingredient is gentamicin [59]. Expression of AmrAB–OprA is the main reason for why aminoglycosides and macrolides are not clinically useful. This efflux pump is also responsible for the reduced efficacy of novel therapeutic agents, such as the ketolide, cethromycin, which was shown to select for mutants overexpressing AmrAB–OprA in response to in vitro cethromycin exposure, resulting in high-level resistance (MIC ≥128 µg/ml) [60].
BpeAB–OprB was originally reported to confer aminoglycoside and macrolide resistance to Singapore strain KHW [54]. However, subsequent molecular genetic studies with strain 1026b, a Thai clinical isolate, revealed that BpeAB–OprB did not bestow significant levels of aminoglycoside resistance in this strain [55]. This pump extrudes macrolides, fluoroquinolones, tetracyclines and, to a lesser extent, chloramphenicol. Despite contributing to intrinsic resistance to these antibiotics, resistance levels are low (with the exception of some macrolides) and, therefore, probably clinically insignificant. Furthermore, although transcripts are detectable in prototype strains, significant BpeAB–OprB expression is only observed in mutants defective in the cognate BpeR repressor [55], but clinical isolates overexpressing this pump have yet to be described.
BpeEF–OprC is still being characterized, but mounting evidence indicates that, in terms of substrate spectrum, it may be the most clinically significant pump identified to date (Box 2). Expression experiments in a drug-susceptible P. aeruginosa strain indicated that this pump effluxes chloramphenicol and trimethoprim [56]. This finding was subsequently corroborated using B. pseudomallei mutants defective in the cognate BpeT activator protein, which constitutively express BpeEF–OprC and exhibit resistance to chloramphenicol, fluoroquinolones, tetracyclines and trimethoprim [Podnecky N et al., Unpublished Data]. A proteomic study with a laboratory-selected chloramphenicol-resistant B. thailandensis mutant confirmed that BpeEF–OprC is responsible for the multidrug-resistance profile observed in the mutant [61]. An analysis of a comprehensive collection of clinical and environmental isolates from Thailand demonstrated that trimethoprim resistance is widespread and attributable to BpeEF–OprC expression. All trimethoprim-resistant isolates remain susceptible to sulfamethoxazole, thus preserving the clinical utility of co-trimoxazole. However, preliminary data indicate that in laboratory-generated Bp82 isolates, co-trimoxazole resistance is due to BpeEF–OprC expression [Podnecky N & Schweizer H, Unpublished Data]. The clinical significance ofBpeEF–OprC is further corroborated by the identification of a pair of sequential isolates, 354b and 354e, from a Thai melioidosis patient, with the secondary isolate, 354e, exhibiting significantly increased resistance to chloramphenicol, ofloxacin and co-trimoxazole. Genomic analyses indicated that a large 800-kb inversion had deleted the last 24 codons of bpeT, which encodes the BpeEF–OprC transcriptional regulator BpeT [62]. As mentioned above, mutations in the C-terminal half of BpeT are known to cause constitutive BpeEF–OprC expression.
Box 2. Burkholderia pseudomallei resistance mechanisms for clinically significant antibiotics.
Ceftazidime resistance
-
▪
Amino acid substitutions in PenA class A β-lactamase
-
▪
Upregulation (overproduction) of PenA β-lactamase
-
▪
Deletion of penicillin binding protein 3 BPSS1219
Resistance to clavulanic acid inhibition
-
▪
Point mutations in PenA β-lactamase
Trimethoprim–sulfamethoxazole resistance
-
▪
BpeEF–OprC efflux pump expression
Doxycycline resistance
-
▪
BpeEF–OprC efflux pump expression
Although much work remains to be done, it is becoming increasingly clear that efflux is one of the dominant antimicrobial resistance mechanisms in B. pseudomallei, presumably owing to the synergy of impermeability and efflux [63].
Enzymatic inactivation
Enzymatic inactivation, either by modification (acetylation, adenylation or phosphorylation) or cleavage, is a widespread bacterial resistance mechanism that affects numerous classes of antibiotics [26]. Surprisingly, to date the only enzymatic inactivation mechanism in B. pseudomallei documented to be of clinical importance is the cleavage of β-lactam antibiotics by a chromosomally encoded class A β-lactamase, PenA. PenA is the first β-lactamase found in a Gram-negative bacterium to be exported via the twin arginine transport system, which exports folded proteins [64].
A report dating back to 1991 documented its involvement in β-lactam and clavulanic acid resistance due to de-repressed production and mutations in the enzyme in response to antibiotic treatment [65], but it was not until relatively recently that PenA was characterized at the molecular level [64,66,67]. In prototype – sometimes referred to as wild-type – B. pseudomallei strains, the enzyme confers resistance to numerous β-lactam antibiotics (e.g., amoxicillin and carbenicillin) and penA deletion strains become susceptible to these antibiotics [64]. Similar analyses indicate that PenA also hydrolyzes ceftazidime to some extent, but this does not impair its clinical use. In these strains, presence of the enzyme has no significant effect on the organism’s carbapenem (imipenem and meropenem) susceptibility, and susceptibilities to other more novel β-lactam antibiotics, such as the sulfactam BAL30072, are similarly not significantly affected [64,68]. Overexpression of PenA in a laboratory strain (exogenous promoter) [64] and clinical isolates (promoter-up mutation) [34] leads to clinically significant ceftazidime resistance. In the clinical strain, the promoter-up mutation arose in response to ceftazidime treatment [34].
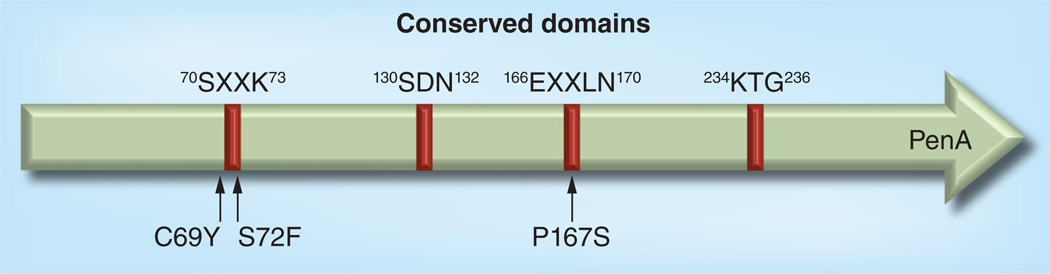
Mutations in PenA have been described that alter critical amino acids affecting the conserved Ambler motifs (Figure 2) [69] and, thus, result in altered substrate specificity, including ceftazidime resistance. A report from Malaysia demonstrated emergence of highly ceftazidime-resistant (MIC ≥256 µg/ml) variants in response to treatment with this antibiotic [33]. The ceftazidime resistance was attributed to a C69Y change near the active site of PenA, a finding that was reproduced by introduction of the same amino acid substitution into laboratory strain Bp82 [64]. Emergence of the same C69Y mutation as a consequence of ceftazidime therapy was subsequently reported in an Australian isolate [34].
Figure 2. Burkholderia pseudomallei PenA and locations of mutations leading to clinically significant antibiotic resistance.
Positions of conserved regions and mutations are numbered according to the Ambler scheme [69]. Susceptibilities to clinically significant β-lactam antibiotics range from 1.5 to 3 µg/ml (wild-type) [33,34,36,64], ≥256 µg/ml (C69Y) [33,34,64] and 24 to 64 µg/ml (P167S) [36,64] for ceftazidime, and 3 to 8 µg/ml (wild-type) and 16 to 32 µg/ml (P72F) for amoxicillin–clavulanic acid [64,67]. By comparison, the meropenem MICs in all isolates are not significantly affected and range from 0.75 to 1.5 µg/ml [36,64]. Current susceptibility breakpoints are ≤8 µg/ml for ceftazidime, ≤8 µg/ml for amoxicillin and ≤4 µg/ml for clavulanic acid.
Following an earlier report describing resistance due to mutations in penA as a consequence of antibiotic therapy, a later study attributed ceftazidime and amoxicillin–clavulanic acid resistance to P167S and S72F substitutions in PenA, respectively [67]. These attributions were subsequently confirmed by allelic exchange in laboratory strain Bp82 [64]. The P167S mutation was also found to be the cause for ceftazidime resistance in another laboratory-selected B. pseudomallei mutant [70] and in a Thai melioidosis patient, where it arose as a consequence of ceftazidime therapy of an acute infection [36].
The PenA enzyme is conserved amongst Burkholderia species, including Burkholderia thailandensis, which is closely related to B. pseudomallei [71]. In vitro selection of a ceftazidime-resistant B. thailandensis isolate revealed a three-base deletion of the codon for glutamate 168 (Glu168del), which affects the so-called omega loop in PenA [72]. This mutation has not yet been found in any ceftazidime-resistant B. pseudomallei isolate.
PenA mutants carrying either C69Y P167S or Glu168del are sensitized to other β-lactams, for example amoxicillin, thus possibly providing an avenue for alternative therapeutic strategies [64,72]. As noted above, judged by MIC, PenA is not significantly active against imipenem and inactive against meropenem, and none of these mutations extend the activity spectrum to these carbapenems [36,64,72].
Two studies described the cloning and characterization of B. pseudomallei class D β-lactamases (oxacillinases named OXA-42, -43 and -57) from various B. pseudomallei strains [73,74]. Expression of OXA-43 was significantly increased in laboratory-selected ceftazidime-resistant mutants, when compared with the parental strain from which they were derived [73]. However, no oxacillinase has yet been shown to cause ceftazidime resistance in clinical B. pseudomallei isolates.
Altered target sites
Investigations of six strains from Thai patients that failed ceftazidime therapy revealed that they contained large (145–309 kb) chromosome 2 deletions [35]. These deletions affected a common 49 genes and resulted in high-level ceftazidime resistance (MIC ≥256 µg/ml). This resistance was accompanied by filamentation and a severe growth defect that prevented growth on commonly used laboratory culture media. The deleted genomic region contained two penicillin-binding proteins (PBPs) of the PBP3 family and a PBP belonging to the PBP5/6 family. The experimentation pinpointed PBP3 BPSS1219 as the ceftazidime target and its deletion as the sole cause of high-level ceftazidime resistance. Since most antibiotics target essential cellular functions, target deletion is an unusual resistance mechanism because of the undesired consequences on bacterial fitness. Nonetheless, it seems to be a rather common ceftazidime-resistance mechanism in B. pseudomallei.
Other resistance mechanisms
This review focuses on traditional antibiotic resistance mechanisms but numerous other factors have been demonstrated to affect the antimicrobial susceptibility of B. pseudomallei. These include: the biofilm mode of growth known to substantially lower antimicrobial susceptibility [75–77]; the chronic or latent infection state in which bacteria presumably reside intracellularly in a nonreplicating altered metabolic state (e.g., anaerobiosis) where they are less susceptible to conventional antibiotics [78]; small colony variants arising in response to treatment with diverse antibiotics giving rise to crossresistance to chemically unrelated agents [79]; and growth under stress conditions that may induce antimicrobial resistance mechanisms, for example growth of B. pseudomallei under salt stress resulted in induction of a β-lactamase-like protein that was accompanied by increased ceftazidime resistance [80]. Some of these mechanisms may not be regarded as true resistance, but rather tolerance mechanisms. Nonetheless, some of these mechanisms may have clinical implications. However, lack of clinical evidence and of studies in suitable animal models, make it rather difficult to assess the contributions of these resistance – or tolerance – mechanisms to clinically significant resistance.
Future perspective
Although, thus far, resistance to clinically significant antibiotics in B. pseudomallei is relatively rare, there is mounting evidence that resistance is more prevalent than previously thought. Not surprisingly, resistance is emerging in response to antimicrobial treatment, both during the intensive and eradication phase, and may become a more prevalent issue with more widespread use of antibiotics throughout regions with endemic melioidosis. Given the paucity of antimicrobial agents useful for melioidosis therapy, resistance to any of the currently used key antibiotics severely undermines the ability to successfully treat the disease. Resistance to carbapenems has not yet been observed and, apart from clinical manifestations that warrant their use, carbapenems should therefore remain drugs of last resort. As mentioned previously, mobile genetic elements, such as plasmids, seem notably absent from B. pseudomallei. However, many strains are naturally transformable [81], and conjugative plasmids can be introduced and, with selective pressure, stably maintained in B. pseudomallei [82]. Some conjugative multiresistance determinants, especially those containing carbapenemases, including NDM-1 [83] are disseminating rapidly worldwide [84] and challenge the treatment of Gram-negative infections. Since many of the regions where carbapenemases and other resistance determinants are emerging overlap with those that are endemic for B. pseudomallei, it will be wise to monitor drug-resistant B. pseudomallei for resistance determinants known to be associated with mobile elements.
Understanding resistance mechanisms provides tangible benefits, such as the development of PCR-based assays for rapid detection of known resistance alleles. For example, a SYBR® Green-based mismatch amplification mutation assay was developed for the detection of single nucleotide polymorphisms in B. pseudomallei penA that result in ceftazidime resistance [34,36]. Ceftazidime-resistant isolates carrying the PBP3 deletion do not grow on common laboratory media unless they are supplemented with glycerol [35]. The ceftazidime-resistant Thai patient isolates were detected because they were fortuitously plated on Ashdown’s agar, which contains glycerol [59]. However, this is not common practice and one of the lessons learned from this study is that, following ceftazidime therapy, patient isolates should be routinely plated onto Ashdown’s agar so that the growth-deficient resistant variants can be detected early.
Owing to B. pseudomallei’s biothreat potential and with no viable vaccine in the pipeline [85,86], there is continued interest in developing alternative therapeutic agents that are not (yet) subject to existing resistance mechanisms. Potential novel therapeutic approaches, such as immunoantimicrobial therapy [87] and use of anti-inflammatories, such as glyburide [88], have been described, but the perceived patient benefits are, to date, largely observational and will need to be clinically proven. Other, mostly experimental, strategies (e.g., isocitrate lyase inhibitors and silver carbine compounds, among others) have been reviewed [89]. Most promising are novel compounds in various stages of preclinical or clinical development with activity against B. pseudomallei. Basilea Pharmaceutica’s (Basel, Switzerland) sulfactam BAL30072 exhibits stellar in vitro efficacy against B. pseudomallei [68]; however, the compound’s in vivo efficacy in animal models is yet unknown. Several other pharmaceutical companies presented data at diverse scientific meetings on novel developmental compounds with in vitro and in vivo efficacy against B. pseudomallei. These include novel tetracyclines from Tetraphase Pharmaceuticals (MA, USA) [90], novel GyrB/ParE inhibitors from Trius Therapeutics (CA, USA) [91] and EV-035 from Evolva SA (Switzerland) [92]. Aside from the perceived need for new therapeutics for biodefense purposes, melioidosis is an emerging infectious disease and further development of novel antimicrobial agents with efficacy against a pathogen for which there is a paucity of efficacious anti-infectives is warranted.
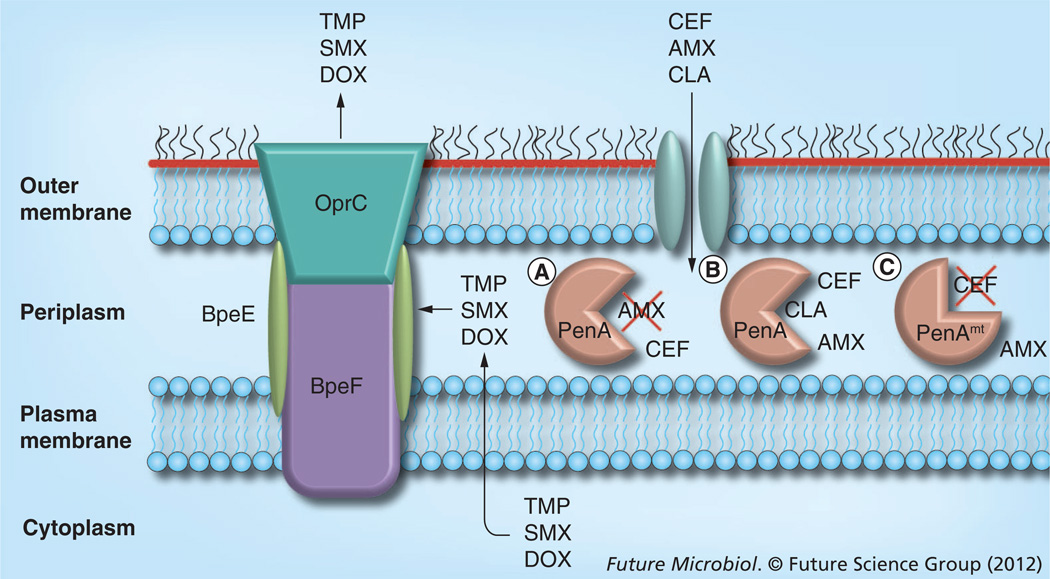
With the increased attention that B. pseudomallei received since its listing as a category B select agent [93], our knowledge about its antimicrobial resistance mechanisms and their implications for treatment of melioidosis has significantly increased. At the time of writing of this review, several clinically significant chromosomally mediated resistance mechanisms have been described, including that of PenA β-lactamase, deletion of PBP3 and efflux (the roles of PenA and efflux are summarized in Figure 3). However, the picture is still rather incomplete and more research is needed to fill the remaining gaps in our knowledge.
Figure 3. Summary of Burkholderia pseudomallei resistance mechanisms compromising therapy with clinically significant antibiotics.
Enzymatic inactivation and efflux from the cell are mechanisms that compromise the use of antibiotics employed in intensive and eradication phase therapy. The CEF and AMX targets are located in the periplasm. There is experimental evidence that CEF and other β-lactams permeate the outer membrane through porins. Periplasmic β-lactams, such as AMX, are (A) inactivated by the wild-type PenA class A β-lactamase, (B) unless its activity is inhibited by CLA. Although wild-type PenA hydrolyzes CEF to some extent, it does not confer clinically significant resistance to CEF. (C) Some mutant PenA (PenAmt) derivatives catalyze CEF hydrolysis and others (not illustrated) are refractory to CLA inhibition. Antibiotics such as DOX, TMP and TMP–SMX have cytoplasmic targets and are extruded to the extracellular milieu by the multicomponent BpeEF–OprC resistance nodulation and cell division efflux pump. As resistance nodulation and cell division pumps extrude substrates acquired from the periplasmic space, the listed antibiotics are most likely extruded either during their transit into the cell or after extrusion to the periplasm from the cytoplasmic space via an unknown mechanism.
AMX: Amoxicillin; CEF: Ceftazidime; CLA: Clavulanic acid; DOX: Doxycycline; SMX: Sulfamethoxazole; TMP: Trimethoprim.
Executive summary.
Burkholderia pseudomallei
-
▪
B. pseudomallei is an emerging pathogen that resides in soil and water.
-
▪
The bacterium is found around the globe in tropical and subtropical regions, with hotspots in northern Australia and southeast Asia, especially in northeastern Thailand.
-
▪
B. pseudomallei readily adapts to adverse conditions and this adaptation is facilitated by a large and plastic genome consisting of two chromosomes.
Melioidosis
-
▪
B. pseudomallei infections in animals and humans cause melioidosis.
-
▪
Melioidosis is a multifaceted disease and a major cause of bacterial sepsis and chronic disseminated infections in humans in northern Australia and southeast Asia.
-
▪
Melioidosis exhibits a wide array of clinical symptoms, ranging from acute sepsis to chronic recurrent infections, as well as disease with no clinical symptoms.
-
▪
Even with rapid diagnosis and prompt and aggressive treatment, the fatality rate for melioidosis patients still ranges from 10 to 20% in Australia to over 40% in Thailand.
Resistance in clinical isolates impacting therapy
-
▪
The development of antibiotic resistance in response to therapy is a cause for concern.
-
▪
Upregulation of penA transcription causes ceftazidime resistance.
-
▪
Amino acid substitutions in class A β-lactamase cause ceftazidime and amoxicillin–clavulanic acid resistance.
-
▪
Deletion of the penicillin binding protein 3 results in high-level ceftazidime resistance.
B. pseudomallei resistance mechanisms in clinical strains & resistant laboratory isolates affecting clinically significant antibiotics
-
▪
Efflux mediated by the BpeEF–OprC pump affects doxycycline, trimethoprim and trimethoprim–sulfamethoxazole.
-
▪
Doxycycline resistance in Australian isolates.
Future melioidosis therapies
-
▪
The development of methods for rapid detection of resistance mechanisms allows for the initiation or redirection of therapeutic interventions.
-
▪
Preclinical and clinical development of novel therapeutics not prone to existing resistance mechanisms.
-
▪
Immunoantimicrobial therapies that demonstrate efficacy in vitro and in murine melioidosis models.
-
▪
Adjunct therapies preventing or lessening the effects of septic shock.
Acknowledgments
Burkholderia research in the H Schweizer laboratory is supported by NIH funding: U54 AI065357 (Rocky Mountain Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research).
Footnotes
Financial & competing interests disclosure
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Ashdown LR, Duffy VA, Douglas RA. Melioidosis. Med. J. Aust. 1980;1:314–316. [PubMed] [Google Scholar]
- 2.White NJ. Melioidosis. Lancet. 2003;361(9370):1715–1722. doi: 10.1016/s0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- 3.Leelarasamee A. Recent development in melioidosis. Curr. Opin. Infect. Dis. 2004;17:131–136. doi: 10.1097/00001432-200404000-00011. [DOI] [PubMed] [Google Scholar]
- 4. Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 2005;18(2):383–416. doi: 10.1128/CMR.18.2.383-416.2005. ▪▪ Excellent review on the various aspects of melioidosis.
- 5. Wiersinga WJ, Van Der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 2006;4:272–282. doi: 10.1038/nrmicro1385. ▪▪ Excellent review of Burkholderia pseudomallei and melioidosis.
- 6.Peacock SJ. Melioidosis. Curr. Opinion Infect. Dis. 2006;19:421–428. doi: 10.1097/01.qco.0000244046.31135.b3. [DOI] [PubMed] [Google Scholar]
- 7.Adler NR, Govan B, Cullinane M, Adler B, Boyce JD. The molecular and cellular basis of pathogenesis in melioidosis: how does Burkholderia pseudomallei cause disease? FEMS Microbiol. Rev. 2009;33:1079–1099. doi: 10.1111/j.1574-6976.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- 8.Limmathurotsakul D, Peacock SJ. Melioidosis: a clinical overview. Br. Med. Bull. 2011;99:125–139. doi: 10.1093/bmb/ldr007. [DOI] [PubMed] [Google Scholar]
- 9. Wuthiekanun V, Peacock SJ. Management of melioidosis. Expert Rev. Anti Infect. Ther. 2006;4:445–455. doi: 10.1586/14787210.4.3.445. ▪▪ Comprehensive review of the clinical management of melioidosis.
- 10.Inglis TJ, Rolim DB, Rodriguez JL. Clinical guideline for diagnosis and management of melioidosis. Rev. Inst. Med. Trop. Sao Paulo. 2006;48:1–4. doi: 10.1590/s0036-46652006000100001. [DOI] [PubMed] [Google Scholar]
- 11.Peacock SJ, Schweizer HP, Dance DAB, et al. Management of accidental laboratory exposure to Burkholderia pseudomallei and B. mallei. Emerg. Infect. Dis. 2008;14(7):e2. doi: 10.3201/eid1407.071501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiersinga WJ, van’t Weer C, van den Pangaart PS, et al. Immunosuppression associated with interleukin-1R-assictated kinase M upregulation predicts mortality in Gram-negative sepsis (melioidosis) Crit. Care Med. 2009;37:569–576. doi: 10.1097/CCM.0b013e318194b1bf. [DOI] [PubMed] [Google Scholar]
- 13.Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl. Trop. Dis. 2010;4(11):e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Currie BJ, Dance DAB, Cheng AC. The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans. R. Soc. Trop. Med. Hyg. 2008;102/S1:S1–S4. doi: 10.1016/S0035-9203(08)70002-6. [DOI] [PubMed] [Google Scholar]
- 15.Kanungo R, Padhan P, Bhattacharya S, Srimannarayana J, Jayanthi S, Swaminathan RP. Melioidosis – a report from Pondicherry, South India. J. Assoc. Physicians India. 2002;50:1438–1439. [PubMed] [Google Scholar]
- 16.Rolim DB, Vilar DCFL, Sousa AQ, et al. Melioidosis, northeastern Brazil. Emerg. Infect. Dis. 2005;11:1458–1460. doi: 10.3201/eid1109.050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rolim DB, Rocha MF, Brilhante RS, et al. Environmental isolates of Burkholderia pseudomallei in Ceara State, northeastern Brazil. Appl. Env. Microbiol. 2009;75:1215–1218. doi: 10.1128/AEM.01953-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhopadhyay C, Kaestli M, Vandana KE, et al. Molecular characterizaion of clinical Burkholderia pseudomallei isolates from India. Am. J. Trop. Med. Hyg. 2011;85:121–123. doi: 10.4269/ajtmh.2011.11-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visca P, Cazzola G, Petrucca A, Braggion C. Travel-associated Burkholderia pseudomallei infection (melioidosis) in a patient with cystic fibrosis: a case report. Clin. Infect. Dis. 2001;32(1):E15–E16. doi: 10.1086/317528. [DOI] [PubMed] [Google Scholar]
- 20.Kite-Powell A, Livengood JR, Suarez J, Hopkins R, Clark TA, Chertow D. Imported melioidosis – south Florida, 2005. Morb. Mortal. Wkly Rep. 2006;55:873–876. [PubMed] [Google Scholar]
- 21.Ciervo A, Mattei R, Cassone A. Melioidosis in an Italian tourist injured by the tsunami in Thailand. J. Chemother. 2006;18(4):443–444. doi: 10.1179/joc.2006.18.4.443. [DOI] [PubMed] [Google Scholar]
- 22.Svensson E, Welinder-Olsson C, Claesson BA, Studahl M. Cutaneous melioidosis in a Swedish tourist after the tsunami in 2004. Scand. J. Infect. Dis. 2006;38(1):71–74. doi: 10.1080/00365540500264738. [DOI] [PubMed] [Google Scholar]
- 23.Cheng AC. Melioidosis: advances in diagnosis and treatment. Curr. Opin. Infect. Dis. 2010;23(6):554–559. doi: 10.1097/QCO.0b013e32833fb88c. [DOI] [PubMed] [Google Scholar]
- 24. Lipsitz R, Garges S, Aurigemma R, et al. Workshop on treatment of and prophylaxis for Burkholderia spp. infections, Australia, 2010. Emerg. Infect. Dis. 2012 doi: 10.3201/eid1812.120638. (In press). ▪ Provides updated consensus guidelines for melioidosis therapy.
- 25.Chaowagul W, Chierakul W, Simpson AJ, et al. Open-label randomized trial of oral trimethoprim–sulfamethoxazole, doxycycline, and chloramphenicol compared with trimethoprim–sulfamethoxazole and doxycycline for maintenance therapy of melioidosis. Antimicrob. Agents Chemother. 2005;49(10):4020–4025. doi: 10.1128/AAC.49.10.4020-4025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh C. Antibiotic Resistance. Washington, DC, USA: American Society for Microbiology Press; [Google Scholar]
- 27. Holden MTG, Titball RW, Peacock SJ, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl Acad. Sci. USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. ▪ Describes the elucidation of the first B. pseudomallei genome sequence.
- 28.Dance DA, Wuthiekanun V, Chaowagul W, White NJ. The antimicrobial susceptibility of Pseudomonas pseudomallei. Emergence of resistance in vitro and during treatment. J. Antimicrob. Chemother. 1989;24:295–309. doi: 10.1093/jac/24.3.295. [DOI] [PubMed] [Google Scholar]
- 29.Piliouras P, Ulett GC, Ashurst-Smith C, Hirst RG, Norton RE. A comparison of antibiotic suscebptibility testing methods for cotrimoxazole with Burkholderia pseudomallei. Int. J. Antimicrob. Agents. 2002;19:427–429. doi: 10.1016/s0924-8579(02)00016-x. [DOI] [PubMed] [Google Scholar]
- 30.Lumbiganon P, Tattawasatra U, Chetchotisakd P, Wongratanacheewin S, Thinkamrop B. Comparison between the antimicrobial susceptibility of Burkholderia pseudomallei to trimethoprim–sulfamethoxazole by standard disk diffusion method and by minimal inhibitory concentration determination. J. Med. Assoc. Thai. 2000;83:856–860. [PubMed] [Google Scholar]
- 31.Wuthiekanun V, Cheng AC, Chierakul W, et al. Trimethoprim/sulfamethoxazole resistance in clinical isolates of Burkholderia pseudomallei. J. Antimicrob. Chemother. 2005;55(6):1029–1031. doi: 10.1093/jac/dki151. [DOI] [PubMed] [Google Scholar]
- 32.Wuthiekanun V, Amornchai P, Saiprom N, et al. Survey of antimicrobial resistance in clinical Burkholderia pseudomallei isolates over two decades in northeast Thailand. Antimicrob. Agents Chemother. 2011;55(11):5388–5391. doi: 10.1128/AAC.05517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sam IC, See KH, Puthucheary SD. Variations in ceftazidime and amoxicillin-clavulanate susceptibilities within a clonal infection of Burkholderia pseudomallei. J. Clin. Microbiol. 2009;47(5):1556–1558. doi: 10.1128/JCM.01657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sarovich DS, Price EP, Von Schulze AT, et al. Characterization of ceftazidime resistance mechanisms in clinical isolates of Burkholderia pseudomallei from Australia. PLoS One. 2012;7(2):e30789. doi: 10.1371/journal.pone.0030789. ▪▪ Along with [36], describes a real-time PCR-based SYBR® Green-mismatch amplification assay for rapid detection of penA alleles causing ceftazidime resistance.
- 35. Chantratita N, Rholl DA, Sim B, et al. Antimicrobial resistance to ceftazidime involving loss of penicillin-binding protein 3 in Burkholderia pseudomallei. Proc. Natl Acad. Sci. USA. 2011;108:17165–17170. doi: 10.1073/pnas.1111020108. ▪▪ Describes large-scale loss of the genomic region containing the drug target as a unique ceftazidime resistance mechanism.
- 36. Sarovich DS, Price EP, Limmathurotsakul D, et al. Development of ceftazidime resistance in an acute Burkholderia pseudomallei infection. Infect. Drug Resist. 2012;5:129–132. doi: 10.2147/IDR.S35529. ▪▪ Along with [34], describes a real-time PCR-based SYBR Green-mismatch amplification assay for rapid detection of penA alleles causing ceftazidime resistance.
- 37.Kung CT, Lee CH, Li CJ, Lu HI, Ko SF, Liu JW. Development of ceftazidime resistance in Burkhoderia pseudomallei in a patient experiencing melioidosis with mediastinal lymphadenitis. Ann. Acad. Med. Singapore. 2010;39(12):945–947. [PubMed] [Google Scholar]
- 38.Behera B, Prasad Babu TL, Kamalesh A, Reddy G. Ceftazidime resistance in Burkholderia pseudomallei: first report from India. Asian Pac. J. Trop. Med. 2012;5(4):329–330. doi: 10.1016/S1995-7645(12)60050-9. [DOI] [PubMed] [Google Scholar]
- 39.Propst KL, Mima T, Choi KH, Dow SW, Schweizer HP. A Burkholderia pseudomallei ΔpurM mutant is avirulent in immunocompetent and immunodeficient animals: candidate strain for exclusion from select-agent lists. Infect. Immun. 2010;78:3136–3143. doi: 10.1128/IAI.01313-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hancock REW. Resistance mechanisms in Pseudomonas aeruginosa and other non-fermentative bacteria. Clin. Infect. Dis. 1998;27(Suppl. 1):S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 41.Pages JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008;6(12):893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 42.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in Gramnegative bacteria. Annu. Rev. Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siritapetawee J, Prinz H, Samosornsuk W, Ashley RH, Suginta W. Functional reconstitution, gene isolation and topology modelling of porins from Burkholderia pseudomallei and Burkholderia thailandensis. Biochem. J. 2004;377(Pt 3):579–587. doi: 10.1042/BJ20031118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novem V, Shui G, Wang D, et al. Structural and biological diversity of lipopolysaccharides from Burkholderia pseudomallei and Burkholderia thailandensis. Clin. Vaccine Immunol. 2009;16(10):1420–1428. doi: 10.1128/CVI.00472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burtnick MN, Woods DE. Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob. Agents Chemother. 1999;43(11):2648–2656. doi: 10.1128/aac.43.11.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poole K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 47.Kumar A, Schweizer HP. Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv. Drug Deliv. Rev. 2005;57:1486–1513. doi: 10.1016/j.addr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 48.Langton KP, Henderson PJ, Herbert RB. Antibiotic resistance: multidrug efflux proteins, a common transport mechanism? Nat. Prod. Rep. 2005;22(4):439–451. doi: 10.1039/b413734p. [DOI] [PubMed] [Google Scholar]
- 49.Nikaido H, Pages JM. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2012;36(2):340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc. Natl Acad. Sci. USA. 2009;106(22):9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Three’s company: component structures bring a closer view of tripartite drug efflux pumps. Curr. Opin. Struct. Biol. 2004;14:741–747. doi: 10.1016/j.sbi.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Kumar A, Mayo M, Trunck LA, Cheng AC, Currie BJ, Schweizer HP. Expression of resistance-nodulation-cell division efflux pumps in commonly used Burkholderia pseudomallei strains and clinical isolates from Northern Australia. Trans. R. Soc. Trop. Med. Hyg. 2008;102(Suppl. 1):S145–S151. doi: 10.1016/S0035-9203(08)70032-4. [DOI] [PubMed] [Google Scholar]
- 53.Moore RA, Deshazer D, Reckseidler S, Weissman A, Woods DE. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan YY, Tan TMC, Ong YM, Chua KL. BpeAB–OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2004;48(4):1128–1135. doi: 10.1128/AAC.48.4.1128-1135.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mima T, Schweizer HP. The BpeAB–OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob. Agents Chemother. 2010;54(8):3113–3120. doi: 10.1128/AAC.01803-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar A, Chua K-L, Schweizer HP. Method for regulated expression of single-copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF–OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob. Agents Chemother. 2006;50:3460–3463. doi: 10.1128/AAC.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simpson AJ, White NJ, Wuthiekanun V. Aminoglycoside and macrolide resistance in Bukholderia pseudomallei. Antimicrob. Agents Chemother. 1999;43:2332. doi: 10.1128/aac.43.9.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trunck LA, Propst KL, Wuthiekanun V, et al. Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical isolates from Thailand. PLoS Negl. Trop. Dis. 2009;3 doi: 10.1371/journal.pntd.0000519. e0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashdown LR. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology. 1979;11:293–297. doi: 10.3109/00313027909061954. [DOI] [PubMed] [Google Scholar]
- 60.Mima T, Schweizer HP, Xu Z-Q. In vitro activity of cethromycin against Burkholderia pseudomallei and investigation of mechanism of resistance. J. Antimicrob. Chemother. 2011;66:73–78. doi: 10.1093/jac/dkq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biot FV, Valade E, Garnotel E, et al. Involvement of the efflux pumps in chloramphenicol selected strains of Burkholderia thailandensis: proteomic and mechanistic evidence. PLoS One. 2011;6:e16892. doi: 10.1371/journal.pone.0016892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hayden HS, Lim R, Brittnacher MJ, et al. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One. 2012;7(5):e36507. doi: 10.1371/journal.pone.0036507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schweizer HP. When it comes to drug discovery not all Gram-negative bacterial biodefence pathogens are created equal: Burkholderia pseudomallei is different. Microb. Biotechnol. 2012;5:581–583. doi: 10.1111/j.1751-7915.2012.00334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rholl DA, Papp-Wallace KM, Tomaras AP, Vasil ML, Bonomo RA, Schweizer HP. Molecular Investigations of PenA-mediated beta-lactam Resistance in Burkholderia pseudomallei. Front. Microbiol. 2011;2:139. doi: 10.3389/fmicb.2011.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Godfrey AJ, Wong S, Dance DA, Chaowagul W, Bryan LE, et al. Pseudomonas pseudomallei resistance to beta-lactam antibiotics due to alterations in the chromosomally encoded beta-lactamase. Antimicrob. Agents Chemother. 1991;35(8):1635–1640. doi: 10.1128/aac.35.8.1635. ▪ First paper attributing β-lactam resistance in clinical isolates to the presence of a chromosomal β-lactamase.
- 66.Cheung TK, Ho PL, Woo PC, Yuen KY, Chau PY. Cloning and expression of class A beta-lactamase gene blaA(BPS) in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2002;46(4):1132–1135. doi: 10.1128/AAC.46.4.1132-1135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tribuddharat C, Moore RA, Baker P, Woods DE, et al. Burkholderia pseudomallei class A beta-lactamase mutations that confer selective resistance against ceftazidime or clavulanic acid inhibition. Antimicrob. Agents Chemother. 2003;47:2082–2087. doi: 10.1128/AAC.47.7.2082-2087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mima T, Kvitko BH, Rholl DA, Page MGP, Desarbre E, Schweizer HP. In vitro activity of BAL30072 against Burkholderia pseudomallei. Int. J. Antimicrob. Agents. 2011;38:157–159. doi: 10.1016/j.ijantimicag.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ambler RP, Coulson AF, Frere J-M, et al. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho PL, Cheung TK, Yam WC, Yuen KY. Characterization of a laboratory-generated variant of BPS beta-lactamase from Burkholderia pseudomallei that hydrolyses ceftazidime. J. Antimicrob. Chemother. 2002;50(5):723–726. doi: 10.1093/jac/dkf208. [DOI] [PubMed] [Google Scholar]
- 71.Brett PJ, Deshazer D, Woods DE. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 72.Yi H, Kim K, Cho KH, Jung O, Kim HS. Substrate apectrum extension of PenA in Burkholderia thailandensis with a single amino acid deletion, Glu168del. Antimicrob. Agents Chemother. 2012;56(7):4005–4008. doi: 10.1128/AAC.00598-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niumsup P, Wuthiekanun V. Cloning of the class D beta-lactamase gene from Burkholderia pseudomallei and studies on its expression in ceftazidime-susceptible and -resistant strains. J. Antimicrob. Chemother. 2002;50(4):445–455. doi: 10.1093/jac/dkf165. [DOI] [PubMed] [Google Scholar]
- 74.Keith KE, Oyston PC, Crossett B, et al. Functional characterization of OXA-57, a class D beta-lactamase from Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2005;49(4):1639–1641. doi: 10.1128/AAC.49.4.1639-1641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vorachit M, Lam K, Jayanetra P, Costerton JW. Resistance of Pseudomonas pseudomallei growing as a biofilm on silastic discs to ceftazidime and co-trimoxazole. Antimicrob. Agents Chemother. 1993;37:2000–2002. doi: 10.1128/aac.37.9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sawasdidoln C, Taweechaisupapong S, Sermswan RW, Tattawasart U, Tungpradabkul S, Wongratanacheewin S. Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS One. 2010;5(2):e9196. doi: 10.1371/journal.pone.0009196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pibalpakdee P, Wongratanacheewin S, Taweechaisupapong S, Niumsup PR. Diffusion and activity of antibiotics against Burkholderia pseudomallei biofilms. Int. J. Antimicrob. Agents. 2012;39(4):356–359. doi: 10.1016/j.ijantimicag.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 78.Hamad MA, Austin CR, Stewart AL, Higgins M, Vazquez-Torres A, Voskuil MI. Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2011;55(7):3313–3323. doi: 10.1128/AAC.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haussler S, Rohde M, Steinmetz I. Highly resistant Burkholderia pseudomallei small colony variants isolated in vitro and in experimental melioidosis. Med. Microbiol. Immunol. 1999;188:91–97. doi: 10.1007/s004300050110. [DOI] [PubMed] [Google Scholar]
- 80.Pumirat P, Saetun P, Sinchaikul S, Chen ST, Korbsrisate S, Thongboonkerd V. Altered secretome of Burkholderia pseudomallei induced by salt stress. Biochim. Biophys. Acta. 2009;1794(6):898–904. doi: 10.1016/j.bbapap.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 81.Thongdee M, Gallagher LA, Schell M, Dharakul T, Songsivilai S, Manoil C. Targeted mutagenesis of Burkholderia thailandensis and Burkholderia pseudomallei through natural transformation of PCR fragments. Appl. Environ. Microbiol. 2008;74(10):2985–2999. doi: 10.1128/AEM.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi K-H, Mima T, Casart Y, et al. Genetic tools for select agent compliant manipulation of Burkholderia pseudomallei. Appl. Env. Microbiol. 2008;74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009;53(12):5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarkar-Tyson M, Titball RW. Progress toward development of vaccines against melioidosis: a review. Clin. Ther. 2010;32(8):1437–1445. doi: 10.1016/j.clinthera.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 86.Peacock SJ, Limmathurotsakul D, Lubell Y, et al. Melioidosis vaccines: a systematic review and appraisal of the potential to exploit biodefense vaccines for public health purposes. PLoS Negl. Trop. Dis. 2012;6(1):e1488. doi: 10.1371/journal.pntd.0001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Propst KL, Troyer RM, Kellihan LM, Schweizer HP, Dow SW. Immunotherapy markedly increases the effectiveness of antimicrobial therapy for treatment of Burkholderia pseudomallei infection. Antimicrob. Agents Chemother. 2010;54(5):1785–1792. doi: 10.1128/AAC.01513-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koh GC, Maude RR, Schreiber MF, et al. Glyburide is anti-inflammatory and associated with reduced mortality in melioidosis. Clin. Infect. Dis. 2011;52(6):717–725. doi: 10.1093/cid/ciq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Estes DM, Dow SW, Schweizer HP, Torres AG. Present and future therapeutic strategies for melioidosis and glanders. Expert Rev. Anti Infect. Ther. 2010;8:325–338. doi: 10.1586/eri.10.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grossman T, O’Brien W, Fyfe C, Sutcliffe J. The novel isoindoline-containing pentacycline TP-834 is active against community and biothreat respiratory pathogens; Presented at: 22nd European Congress of Clinical Microbiology and Infectious Diseases; 31 March–3 April 2012; London, UK. [Google Scholar]
- 91.Brown-Driver V, Podnecky NL, Marlenee N, et al. Antimicrobial activity and in vivo efficacy of novel GyrB/ParE inhibitors versus Burkholderia pseudomallei; Presented at: ASM Interscience Conference on Antimicrobial Agents and Chemotherapy; 9–12 September 2012; San Francisco, CA, USA. [Google Scholar]
- 92.Salerno G, Goldman S, Milligan D, et al. EV-035: potent antibiotic series against biothreat agents, in particular Burkholderia pseudomallei; Presented at: ASM Interscience Conference on Antimicrobial Agents and Chemotherapy; 9–12 September 2012; San Francisco, CA, USA. 2012. [Google Scholar]
- 93.Dance DAB. Melioidosis and glanders as possible biological weapons. In: Fong W, Alibek K, editors. Bioterrorism and Infectious Agents. A New Dilemma for the 21st Century. >NY, USA: Springer Science and Business Media; 2005. pp. 99–145. [Google Scholar]