Abstract
We developed a group B streptococcus multiplex PCR assay which allows, by direct analysis of the amplicon size, determination of the surface protein antigen genes of alpha-C protein, epsilon protein, Rib, Alp2, Alp3, and Alp4. The multiplex PCR assay offers a rapid and simple method of subtyping Streptococcus agalactiae based on surface protein genes.
Streptococcus agalactiae (group B streptococcus [GBS]) is the leading pathogen in newborn infants in both developing and industrialized countries throughout the world, causing sepsis and meningitis. It also causes disease in parturient women and immunocompromised adults (18).
Epidemiological studies on S. agalactiae infections are mainly based on capsule serotyping. Currently, nine different S. agalactiae serotypes have been described (serotypes Ia, Ib, and II to VIII). Serotype distribution varies with geographical region and ethnic origin, and the virulence of clinical isolates with similar capsular composition can vary widely, suggesting that other bacterial virulence factors, in addition to capsule, are involved in the pathogenesis of GBS (19).
The major surface-localized protein antigens of group B streptococci belong to a family of surface proteins named the alpha-C protein, Rib, Alp2, Alp3, Alp4, and the epsilon protein (alpha-protein-like proteins). They contain large internal tandem repeats and are potential virulence factors (2, 5, 10, 12, 15, 16, 20). All these proteins are encoded by stable mosaic genes, generated by a recombination of modules at the same chromosomal locus (4, 12). The proteins exhibit size variation between strains depending on the number of repeats in the corresponding gene. Moreover, it has been demonstrated that, in the course of infection, the number of repeats inside the alpha-C proteins can undergo internal deletions as a means for evading the host immune response (14).
Protein typing of GBS is determined by the use of C and R antisera. Indeed, antisera are not protein specific (10, 12). While the alpha-C and Rib proteins are immunologically distinct from each other, the alpha-C, Alp2 and Alp3 (Alp2/3), and the epsilon proteins cross-react during Western blotting (11).
C antiserum also recognizes beta antigen, a GBS immunoglobulin A binding protein that was not included in this study because its structure is unrelated to that of the family of alpha-protein-like proteins (7, 8).
Protocols to identify serotypes and surface protein antigen genes by molecular methods have been recently published (3, 9, 10). For the protein subtyping, parallel PCRs using different primer pairs for each gene were set up; detection of epsilon gene had not previously been considered. We developed a multiplex PCR that allows determination of the following GBS surface protein genes directly by the analysis of the amplicon size: the alpha-C protein, the epsilon protein, Rib, Alp2/3, and Alp4.
While the repeats present extensive homology between alpha-protein-like proteins, the N-terminal portion is distinctive. The analysis of a ClustalW amino acid multisequence alignment of the N-terminal portions of the alpha-C and epsilon proteins, Rib, Alp2, Alp3, and Alp4 identified distinctive strings for each protein (Fig. 1), except for the Alp2 and Alp3 proteins, which are identical over the first half of their length.
FIG. 1.
Amino acid multisequence alignment of the N-terminal portions of S. agalactiae alpha-C protein, Alp2, Alp3, Rib, and the epsilon protein. The nucleotide sequence corresponding to the string from positions 11 to 17, indicated in grey, was used as the forward universal primer. The reverse primers sequences, specific for each protein gene (except for Alp2/3), are also indicated in grey.
Primer nucleotide sequences corresponding to the distinctive strings were used in a multiplex assay as the reverse primers, while a nucleotide string, common to all the surface protein genes, was used as the forward primer (Table 1). Briefly, total DNA preparations were prepared from GBS cultures, according to the instructions provided with a DNeasy tissue kit (QIAGEN), and 50 ng was used as a template in a final volume of 25 μl of PCR mixture containing the following: 1× PCR buffer; 2 mM MgCl2; 200 μM concentrations of dATP, dCTP, dGTP, and dTTP; 400 nM concentrations of each of the six primers; and 0.25 U of TaqDNA polymerase (Life Technologies).
TABLE 1.
Nucleotide primer sequences and amplicon size expected for each S. agalactiae surface protein gene considered in this study
| Primer | Sequence (5′-3′) | GenBank accession no. | Position from start codon (nt)a | Amplicon size (bp) |
|---|---|---|---|---|
| Universal forward | TGATACTTCACAGACGAAACAACG | 30 | ||
| Alpha-C reverse | TACATGTGGTAGTCCATCTTCACC | M97256 | 428 | 398 |
| Rib reverse | CATACTGAGCTTTTAAATCAGGTGA | U583333 | 325 | 295 |
| Epsilon reverse | CCAGATACATTTTTTACTAAAGCGG | U33554 | 230 | 200 |
| Alp2/3 reverse | CACTCGGATTACTATAATATTTAGCAC | AF208158 | 364 | 334 |
| Alp4 reverse | TTAATTTGCACCGGATTAACACCAC | AJ488912 | 140 | 110 |
nt, nucleotides.
The samples were amplified on a DNA thermal cycler (MJ Research, Inc.) by heating for 3 min at 96°C, followed by 30 cycles of 95°C for 60 s, 58°C for 45 s, and 72°C for 45 s and concluding with a cycle of 72°C for 10 min.
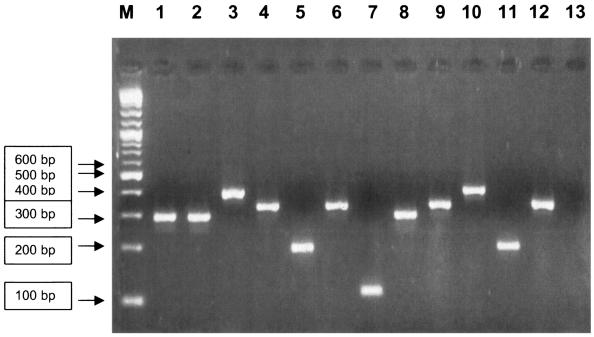
The PCR products were analyzed by electrophoresis in a 2% (wt/vol) agarose-1000 gel (Life Technologies). UV transillumination of the bands on the agarose gel showed different sizes of amplicons, which allowed direct identification of the surface protein gene possessed by each GBS strain tested (Fig. 2).
FIG. 2.
Gel electrophoresis of multiplex PCR amplification products. Direct analysis of amplicon size allowed the determination of the genes encoding the surface protein of each GBS strain as follows: Rib (lanes 1, 2, and 8), alpha-C protein (lanes 3 and 10), Alp2/3 (lanes 4, 6, 9, and 12), Alp4 (lane 7), and the epsilon protein (lanes 5 and 1l). Lane 13 is the negative control. M, 2-log DNA ladder (0.1 to 10.0 kb; New England Biolabs, Inc.).
A serologically typed collection of GBS reference strains (10 strains), selected clinical isolates (55 strains), and bovine strains (9 strains) was analyzed by multiplex PCR, and results derived from the direct analysis of amplicon size were correlated with the gene sequence of the corresponding amplified product for each individual strain (Table 2). As noted above, C and R antisera cross-reacted with different protein genes. Moreover, as previously reported (4, 10), GBS strains can possess an alpha-protein-like protein gene even if the protein is not expressed on the surface. In two cases (reference strain Prague 10/84 and a bovine strain), no surface protein gene was detected. The absence of a surface protein gene in GBS isolates, although rare, has previously been reported (10).
TABLE 2.
Characteristics of GBS strains used in this study
| Strain or isolate | Serotype | Alpha-protein-like proteina | Strain or isolate | Serotype | Alpha-protein-like proteina | |
|---|---|---|---|---|---|---|
| Reference strains | ||||||
| NCTC 11078 (A909) | Ia/C | Alpha | ||||
| 090 | Ia | Alp2b | ||||
| NCTC 8187 (H36B) | Ib/C | Alpha | ||||
| M 781 | III/R | Rib | ||||
| Prague 1/82 | IV | Epsilon | ||||
| Prague 10/84 | V | Negative | ||||
| Prague 118754 | VI | Epsilon | ||||
| Prague 7271 | VII | Epsilon | ||||
| JM9-130013 | VIII/R | Alp3b | ||||
| NCTC 9828 | NT/R | Alp4 | ||||
| Clinical isolates | ||||||
| 1319 | Ia | Epsilon | ||||
| 1325 | Ia | Epsilon | ||||
| 2129 | Ib | Alpha | ||||
| 5518 | Ib | Alpha | ||||
| 2088 | Ia/C | Epsilon | ||||
| 5551 | Ia/C | Alpha | ||||
| 1599 | Ib/C | Alpha | ||||
| 1977 | Ib/C | Alpha | ||||
| 5401 | II | Alpha | ||||
| 3050 | II | Rib | ||||
| 2452 | II/C | Epsilon | ||||
| 5405 | II/C | Alpha | ||||
| 2448 | II/R | Rib | ||||
| 2141 | II/R | Rib | ||||
| 5368 | III | Rib | ||||
| 5435 | III | Rib | ||||
| 5400 | III | Alp2b | ||||
| 1056 | III/C | Epsilon | ||||
| 1496 | III/C | Epsilon | ||||
| 2601 | III/C | Alpha | ||||
| 4383 | III/R | Rib | ||||
| 1998 | III/R | Rib | ||||
| 1999 | IV | Epsilon | ||||
| 2274 | IV | Alpha | ||||
| 1613 | IV | Epsilon | ||||
| 2273 | IV | Alpha | ||||
| 2620 | IV | Epsilon | ||||
| 2102 | V | Epsilon | ||||
| 2361 | VII/R | Alp3b | ||||
| 2362 | VII/R | Alp3b | ||||
| 5408 | VIII | Alp2b | ||||
| 2179 | VIII/C | Epsilon | ||||
| 1715 | VIII/R | Rib | ||||
| 2198 | VIII/R | Rib | ||||
| 2442 | VIII/R | Alp2b | ||||
| 2925 | NT | Alpha | ||||
| 2933 | NT | Alpha | ||||
| 2450 | NT | Alpha | ||||
| 2927 | NT | Alpha | ||||
| 3002 | NT | Rib | ||||
| 2277 | NT/C | Alpha | ||||
| 2914 | NT/C | Epsilon | ||||
| 2116 | NT/R | Rib | ||||
| 5366 | NT/R | Rib | ||||
| Bovine strains | ||||||
| 3492 | III | Epsilon | ||||
| 3491 | IV | Epsilon | ||||
| 3476 | IV | Epsilon | ||||
| 3477 | IV | Epsilon | ||||
| 3479 | NT | Epsilon | ||||
| 3485 | NT | Epsilon | ||||
| 3487 | NT/C | Alpha | ||||
| 3490 | NT | Epsilon | ||||
| 3488 | NT | Negative | ||||
| 2628 | V | Rib | ||||
| 2068 | V | Alp3b | ||||
| 2110 | V/C | Epsilon | ||||
| 2929 | V/C | Alpha | ||||
| 5403 | V/R | Rib | ||||
| 1989 | VI | Epsilon | ||||
| 1992 | VI | Epsilon | ||||
| 1990 | VI/C | Epsilon | ||||
| 1994 | VI/C | Epsilon | ||||
| 1272 | VII/C | Alpha | ||||
| 2928 | VII/C |
The designation of the alpha-protein-like protein encoded by the gene is based both on PCR results and sequencing of the PCR product.
The differentiation between the proteins encoded by the alp2 and alp3 genes was performed by sequencing entire genes as described in reference 10.
Relationships between serotypes and surface protein genes were noted but to a lesser degree than previously reported (10, 12). The association of serotypes Ia, Ib, and II with the alpha protein, of serotype III with Rib, and of serotypes V and VIII with Alp3 was found, but these associations were not absolute. In particular, the alpha protein was present in most serotypes, Rib was detected in serotypes II, V, and VIII as well as in nontypeable strains, and Alp3 was also present in serotype VII. The Alp2 protein, first detected in serotype V (12) and subsequently detected in serotypes Ia and III (10), was also associated with type VIII in our study. No epidemiological studies surveyed the distribution of the epsilon protein that, in our study, was evenly distributed among GBS serotypes and prevalent in bovine strains.
The possibility of looking at the protein gene profile increases the potential of GBS subtyping by limiting the use of C and R antisera to the most appropriate strains after PCR has been used to ascertain protein expression.
A glycoconjugate vaccine to prevent GBS disease is being tested in clinical trials (1, 17). Promising data on using GBS surface proteins as protein carriers instead of tetanus toxoid have been published (6, 13). In this context, it is extremely important to have a rapid and reliable molecular test capable of determining the GBS surface protein genes by direct evaluation of amplicon size, which would permit extensive epidemiological studies on the profiles of protein subtypes of circulating GBS strains without consuming additional resources.
Acknowledgments
We thank M. Pataracchia for serotyping GBS isolates.
This work was supported by the Italian Ministry of Health Project 1%, grant 0AD-F (C.V.H.), and by ISS grant 1024/RI (C.V.H.).
REFERENCES
- 1.Baker, C. J., and M. S. Edwards. 2003. Group B streptococcal conjugate vaccines. Arch. Dis. Child. 88:375-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolduc, G. R., M. J. Baron, C. Gravekamp, C. S. Lachenauer, and L. C. Madoff. 2002. The alpha C protein mediates internalization of group B streptococcus within human cervical epithelial cells. Cell Microbiol. 4:751-758. [DOI] [PubMed] [Google Scholar]
- 3.Borchardt, S. M., B. Foxman, D. Chaffin, C. Rubens, P. A. Tallman, S. D. Manning, C. J. Baker, and C. F. Marrs. 2003. Molecular serotyping of group B streptococci (GBS) by PCR and DNA dot blot hybridization. Ann. Epidemiol. 13:581. [Google Scholar]
- 4.Creti, R., J. L. Michel, and G. Orefici. 2000. Genetic variability of the locus encoding alpha-C like proteins in Streptococcus agalactiae, p. 397-399. In D. R. Martin and John Tagg (ed.), Streptococci and streptococcal diseases: entering the new millenium. Proceedings of the XIV Lancefield International Symposium on Streptococci and Streptococcal Diseases. Securacopy, Porirua, New Zealand.
- 5.Ferrieri, P. 1988. Surface-localized protein antigens of group B streptococci. Rev. Infect. Dis. 10(Suppl. 2):S363-S366. [DOI] [PubMed] [Google Scholar]
- 6.Gravekamp, C., D. L. Kasper, L. C. Paoletti, and L. C. Madoff. 1999. Alpha C protein as a carrier for type III capsular polysaccharide and as protective protein in group B streptococcal vaccines. Infect. Immun. 67:2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heden, L. O., E. Fritz, and G. Lindahl. 1991. Molecular characterization of an IgA receptor from group B streptococci: sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA-binding capacity. Eur. J. Immunol. 21:1481-1490. [DOI] [PubMed] [Google Scholar]
- 8.Jerlstrom, P. G., G. S. Chhatwal, and K. N. Timmis. 1991. The IgA-binding beta antigen of the C protein complex of group B streptococci: sequence determination of its gene and detection of two binding regions. Mol. Microbiol. 5:843-849. [DOI] [PubMed] [Google Scholar]
- 9.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Molecular profiles of group B streptococcal surface protein antigen genes: relationship to molecular serotypes. J. Clin. Microbiol. 40:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lachenauer, C. S., and L. C. Madoff. 1996. A protective surface protein from type V group B streptococci shares N-terminal sequence homology with the alpha C protein. Infect. Immun. 64:4255-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lachenauer, C. S., R. Creti, J. L. Michel, and L. C. Madoff. 2000. Mosaicism in the alpha-like protein genes of group B streptococci. Proc. Natl. Acad. Sci. USA 97:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson, C., M. Stalhammar-Carlemalm, and G. Lindahl. 1996. Experimental vaccination against group B streptococcus, an encapsulated bacterium, with highly purified preparations of cell surface proteins Rib and α. Infect. Immun. 64:3518-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madoff, L. C., J. L. Michel, E. W. Gong, D. E. Kling, and D. L. Kasper. 1996. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc. Natl. Acad. Sci. USA 93:4131-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel, J. L., L. C. Madoff, D. E. Kling, D. L. Kasper, and F. M. Ausubel. 1991. C proteins of group B streptococci, p. 214-218. In G. M. Dunny, P. P. Cleary, and L. L. McKay (ed.), Genetics and molecular biology of streptococci, lactococci, and enterococci. American Society for Microbiology, Washington, D.C.
- 16.Michel, J. L., L. C. Madoff, K. Olson, D. E. Kling, D. L. Kasper, and F. M. Ausubel. 1992. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc. Natl. Acad. Sci. USA 89:10060-10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paoletti, L. C., and D. L. Kasper. 2003. Glycoconjugate vaccines to prevent group B streptococcal infections. Expert Opin. Biol. Ther. 3:975-984. [DOI] [PubMed] [Google Scholar]
- 18.Schuchat, A. 1999. Group B streptococcus. Lancet 353:51-56. [DOI] [PubMed] [Google Scholar]
- 19.Spelleberg, B. 2000. Pathogenesis of neonatal S. agalactiae infections. Microbes Infect. 2:1733-1742. [DOI] [PubMed] [Google Scholar]
- 20.Wastfelt, M., M. Stalhammar-Carlemalm, A. M. Delisse, T. Cabezon, and G. Lindhal. 1996. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J. Biol: Chem. 271:18892-18897. [DOI] [PubMed] [Google Scholar]