Abstract
Despite the substantial impact of sleep disturbances on human health and the many years of study dedicated to understanding sleep pathologies, the underlying genetic mechanisms that govern sleep and wake largely remain unknown. Recently, we completed large scale genetic and gene expression analyses in a segregating inbred mouse cross and identified candidate causal genes that regulate the mammalian sleep-wake cycle, across multiple traits including total sleep time, amounts of REM, non-REM, sleep bout duration and sleep fragmentation. Here we describe a novel approach toward validating candidate causal genes, while also identifying potential targets for sleep-related indications. Select small molecule antagonists and agonists were used to interrogate candidate causal gene function in rodent sleep polysomnography assays to determine impact on overall sleep architecture and to evaluate alignment with associated sleep-wake traits. Significant effects on sleep architecture were observed in validation studies using compounds targeting the muscarinic acetylcholine receptor M3 subunit (Chrm3)(wake promotion), nicotinic acetylcholine receptor alpha4 subunit (Chrna4)(wake promotion), dopamine receptor D5 subunit (Drd5)(sleep induction), serotonin 1D receptor (Htr1d)(altered REM fragmentation), glucagon-like peptide-1 receptor (Glp1r)(light sleep promotion and reduction of deep sleep), and Calcium channel, voltage-dependent, T type, alpha 1I subunit (Cacna1i)(increased bout duration slow wave sleep). Taken together, these results show the complexity of genetic components that regulate sleep-wake traits and highlight the importance of evaluating this complex behavior at a systems level. Pharmacological validation of genetically identified putative targets provides a rapid alternative to generating knock out or transgenic animal models, and may ultimately lead towards new therapeutic opportunities.
Keywords: sleep, genetics, rodent, pharmacology, translation
INTRODUCTION
Sleep is a primordial, vital process that is observed across species, and is marked by changes in behavior, neuronal activity, cellular processes, and gene expression. Disordered sleep has been central to a number of human pathological conditions including, but not limited to, metabolic, neurological, psychiatric and cardiovascular disorders (Cohrs, 2008; Compta et al., 2009; Zee & Turek, 2006). The fundamental purpose of sleep is not fully defined and efforts to characterize its molecular basis and underlying genetic regulation are ongoing. While a number of genetic and molecular studies have sought to determine mRNA expression profiles which correlate with sleep/wake state, such approaches have not focused on the determination of the underlying genes that regulate specific sleep-wake states (Thompson et al., 2010; Mackiewicz et al., 2009; Cirelli et al., 2004; Mackiewicz et al., 2007). More recently, genome-wide studies in humans and model organisms have been carried out in order to identify genes which underlie the regulation of sleepwake traits (Millstein et al., 2011; Sehgal & Mignot, 2011; Winrow et al., 2009; Jiang et al., 2011; Miyagawa et al., 2008).
Familial inheritance of sleep disorders, including studies involving monozygotic twins in the 1930’s, have long indicated the genetic basis of sleep (Dauviltiers et al., 2005). Such approaches have led to the discovery of molecular correlates for a spectrum of sleep disorders, including chronic hypersomnia, insomnia, parasomnia, and circadian phase disorders (Sehgal & Mignot, 2011). The hypersomnia disorder, narcolepsy, has well established positive genetic associations that implicate multiple genes in humans (Caylak, 2009; Nishino & Okuro, 2010; Sehgal & Mignot, 2011; Tafti, 2009). While the disorder is multifactorial, predominant genetic factors associated with human narcolepsy include loci for Human Leukocyte Antigen (HLA), T cell receptor variants, and P2RY11 (Chabas et al., 2003; Franken & Tafti, 2003; Hallmayer et al., 2009; Hor et al., 2010; Kornum et al., 2011; Nishino & Okuro, 2010; Sehgal & Mignot, 2011; Tafti, 2009). A seminal discovery was made involving narcolepsy in dogs; a single mutation in the orexin 2 gene results in a fully penetrant, autosomal recessive heritable condition (Lin et al., 1999). Although the human disorder is complex and multifactorial, the findings in dogs identified a genetic target that is leading to potential advances in the treatment of insomnia (Brisbare-Roch et al., 2007; Di Fabio et al., 2011; Winrow et al., 2011).
Significant and categorical changes in gene expression occur between sleep and wake that are consistent with the synaptic homeostasis theory of sleep. Through microarray studies involving different brain regions, signature mRNA expression patterns have emerged for sleep and wake states. For example, in rat cerebral cortex, wakefulness is associated with upregulation of mRNA’s involved in energy metabolism, cellular stress, and synaptic potentiation. In contrast, during sleep, transcripts related to protein anabolism, synthesis of other macromolecules, and synaptic downscaling are upregulated. These changes in expression during sleep and wake support the theory that maintenance of synaptic plasticity is a primary function of sleep (Cirelli, 2009). According to this model, sleep restores baseline synaptic strength, which enables learning, memory and other processes (Tononi & Cirelli, 2003). While the transition that occurs in gene expression between behavioral states provides important insight into processes that are consequential to being awake or asleep, such an approach does not allow for the identification of the genes or genetic networks that are involved, or the factors that ultimately underlie the regulation of sleep/wake states and traits responsible for these expression changes.
Genomic studies involving humans and model organisms, including mice, have begun to identify the genetic basis for sleep and wake. In particular, Quantitative Trait Loci (QTL) analysis, dependant on natural allelic variation or mutagenized alleles (e.g. forward genetic approach), has enabled mapping of sleep-related genes in model organisms (Cirelli, 2009; Winrow et al., 2009). We have previously described an association study in mice that uncovered the genetic landscape for multiple sleep-wake traits, using a forward genetics, phenotype-driven approach to the functional genomics of sleep that enabled identification of QTL. In that study, genetic variation was induced for QTL analysis, by crossing two strains of mice (C57BL/6, and BALB/cByJ ) having substantially different sleep-wake characteristics.The analysis of 20 sleep-wake traits across 269 N2 BALB.B6 hybrid animals revealed 52 significant cQTL representing a minimum of 20 genomic loci (Winrow et al., 2009). Additionally, for each tissue analyzed (cerebral cortex, hypothalamus, thalamus and liver), eQTL’s, defined as genetic loci associated with specific expression changes, were determined for all sleep-wake traits. Causal inference identified candidate genes that govern sleep where cQTL and eQTL regions overlapped, defined as a peak to peak distance no greater than 15 cM and LOD score greater than 3 (Millstein et al., 2011).
Here, we validate a subset of pharmacologically tractable candidate causal genes identified in our previous studies (Winrow et al., 2009)(Millstein et al 2011 sleep). Selected target genes relate specifically to cholinergic (Chrna4 and Chrm3) and monaminergic neuronal signaling (Htr1d and Drd5), as well as insulin synthesis and secretion (Glp1r), and finally mediation of neuronal calcium conductance (Cacna1i). Traditionally, validation of candidate genetic targets has relied on development of genetically engineered animals carrying loss-of-function and/or gain-of-function of candidate genes to determine the impact on a complex trait (i.e. reverse genetic approach). However, time (and cost) needed to develop the animal model, as well as compensatory molecular and developmental mechanisms are potential confounds with this approach and may interfere with rapid phenotype evaluation. To mitigate these challenges, we employed a pharmacological approach to candidate causal sleep gene validation, by using small molecule reagents in rodents that target genetically identified candidates to objectively determine their impact on behavioral state, as defined by electroencephalogram (EEG) and electromyogram (EMG) measurements. Study results indicate significant effects of modulating target gene products (CHRM3, CHRNA4, GLP1r, DRD5, CACNA1I) on rat sleep that are consistent and in alignment with associated sleep/wake traits that were identified in a genetic cross. The results highlight the success of a novel approach to validating genetically identified candidate causal genes, and the exploration of potential pharmaceutical targets.
MATERIALS AND METHODS
All animal studies were performed in accordance with the United States Department of Agriculture (USDA) Guide for the Care and Use of Laboratory Animals and were approved by the Merck Institutional Animal Care and Use Committee.
Animals
Experiments were conducted using adult male Sprague-Dawley rats (450-600g; Taconic Farms, Germantown, NY). Animals were maintained on a reverse, 12 h light/dark cycle (lights on at 16:00) under standard laboratory conditions (22-24°C, ambient humidity), with food and water ad libitum.
Compounds
All drugs were administered at a dose volume of 1 mL per 600 grams body weight. Exendin-4, cytisine, scopolamine hydrochloride, risperidone, and R(+)-SCH-23390 were purchased from Sigma-Aldrich, INC. (St.Louis, MO, USA), dissolved in saline, and administered intraperitoneally (i.p.). Liraglutide (Novo Nordisk A/S, Bagsvaerd, Denmark) and PNU 109291 (Tocris Bioscience, Ellisville, IN) were dissolved in saline, and injected subcutaneously (s.c.). Sumatriptan succinate was purchased from Sigma-Aldrich, INC., dissolved in distilled water and dosed orally. Varenicline tartrate (Pfizer Ireland Labs, Dublin, Ireland) and darifenacin hydrobromide (AmplaChem, Carmal, IN) were dissolved in 0.5% methylcellulose solution, and dosed orally (p.o.). TTA-A2 was synthesized in house as previously described (Shipe et al., 2008), suspended in 0.5% methylcellulose, and dosed orally at 10 mg/kg.
Dose selection
Whenever possible, compound dosages were predetermined from literary references that cite drug-mediated central effects or from available pharmacokinetic data. Doses used in the present study met or exceeded these referenced dosages to ensure CNS exposure. Darifenacin hydrobromide (10 mg/kg, p.o.), a CHRM3 antagonist, and its metabolites have demonstrated brain exposure in Sprague-Dawley rats as quantified in cerebral spinal fluid within 1 to 4 hours of dosing (Devineni et al., 2005). Varenicline tartrate (0.3 to 3 mg/kg, s.c.) increased locomotion in rats, and blocked nicotine mediated hyperlocomotion (Zaniewska et al., 2008). Cytisine (1.5 or 3 mg/kg, s.c.) reduces voluntary ethanol consumption in C57BL/6J mice (Sajja & Rahman, 2011) . SCH-23390 (0.1-0.3 mg/kg, i.p.), the first dopamine D1-like receptor antagonist, demonstrates efficacy in the pilocarpine seizure model in rats (Bourne, 2001). The high affinity D2-like molecule, risperidone (0.33 mg/kg, s.c.) has efficacy in the rat conditioned avoidance response model (Zhang et al., 2011). Sumatriptan, the 5-HT1B/1D receptor antagonist, attenuates rat pain-related behavior at doses as low as 0.1 mg/kg, dosed subcutaneously (Kayser et al., 2002). In a model of guinea pig hypothermia, BRL-15572 (0.1 and 10 mg/kg), a 5-HT1D receptor antagonist, did not have an impact on body temperature, but was found to sufficiently penetrate the CNS (Hagan et al., 1997). The 5-HT1D agonist, PNU-109291 has demonstrated effects in vitro and peripheral effects in vivo, but lacks efficacy in the clinical treatment of migraine (Cutrer et al., 1999; Slassi et al., 2001). PNU-109291 (10 mg/kg and 15 mg/kg, s.c.) was dosed with the understanding that selected doses may not sufficiently cross the blood brain barrier in order to impact arousal. The GLP1R agonists, liraglutide (0.01 mg/kg, i.p.) and exendin-4 (0.003 mg/kg, i.p.) demonstrate anorectic effects in Sprague-Dawley rats within 1 hour of dosing (Kanoski et al., 2011). The potent T-type calcium channel inhibitor, TTA-A2 (3, 10, 30 mg/kg, p.o.) demonstrates central affects on arousal (Uebele et al., 2009).
Rat Sleep Architecture Studies
Electrocorticogram/electroencephalogram (ECoG/EEG) and electromyogram (EMG) were continuously monitored in single housed male Sprague Dawley rats surgically implanted with TL10M3-F50-EEE radio telemetry transmitters and automated sleep stage analysis performed as described previously (Renger et al., 2004; Whitman et al., 2009; Winrow et al., 2011). Current studies employed one of two counterbalanced cross-over designs in which all animals were alternatively treated with drug and vehicle daily. 3 day crossover design: 1-day vehicle-only run-in, a 2-day arm of drug or vehicle treatment, then 3 days of conditional crossover following an appropriate wash-out period (1 to 4 days). 7 day crossover design: 1 day vehicle-only run in, a 7-day arm of drug or vehicle treatment, then 7 days of conditional crossover. Effects of compound relative to vehicle were evaluated following administration at the onset of the active phase (14:30-15:30). Results for all animals were averaged by condition over administration nights and conditional effects statistically compared applying a linear mixed effects model for repeated measures to two-tailed Student’s t-tests for each 30-minute period. Additionally, results for all compounds were analyzed over two intervals, from time of drug administration to 16:30 (active period testing) and also from 16:30 to 18:30 (inactive period testing). For this additional set of analyses, data from each animal were normalized to its vehicle baseline, and plotted as difference of cumulative sleep time, bout duration (mean time in sleep stage divided by bout number) and latency as a percent change (Figures 1 through 5). Difference graphs use one sample t-test with null hypothesis that data are a random sample from a normal distribution with mean of 0.
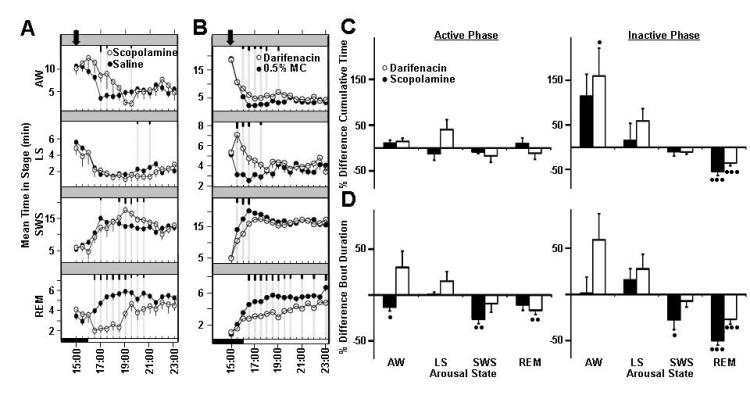
Figure 1.
Effects of modulating muscarinic acetylcholine M3 receptor on rat sleep architecture (associated traits: Wake, Total Sleep, non-REM). Average time spent per 30-minute interval in active wake, light sleep, SWS sleep, and REM sleep in response to vehicle treatment (closed circles) versus treatment with (A) scopolamine and (B) darifenacin (open circles). Dark bars in x -axis represent lights-off period, with time of dosing indicated with an arrowhead on each graph. Values are mean ± SEM (short, medium, and long tic marks represent p ≤ .05, ≤ .01, ≤ .001; linear mixed effects model for repeated measures). Average percent changes in sleep stage (C) cumulative time and (D) bout durations (mean time in sleep stage divided by bout number) following treatment of rats with scopolamine (closed bars) and darifenacin (open bars) for active phase (from time of dosing to ZT 00:30) and inactive phase (ZT 00:30-2:30). Sleep stages are indicated as follows: active wake (Wake), light sleep (LS), slow wave sleep (SWS), and REM sleep (REM). Plotted values represent percent difference from vehicle within independent experiments for each dose, and calculated from raw sleep stage time and bout data. Plotted values are mean ±SEM (• p ≤.05, •• p ≤.01, ••• p ≤.001; two-tailed population t test). Each study run as a balanced crossover design, with 7 sequential days of dosing for scopolamine treatment (n=7 animals, 48 nights/condition total, 0.3 mg/kg, i.p., dosed ZT 23:00) and with 3 days of sequential dosing for darifenacin treatment (n=16 animals, 48 nights/condition total, 50 mg/kg, p.o., dosed ZT 23:00).
Causality Testing Strategy for (B6 × (BALB/C × Sleepless/B6)) Mouse N2 Cross Data Linkage (cQTL) Analysis
The R/qtl package (Broman et al., 2003) was used to compute pseudo markers, defined by the probablilities of the true underlying genotypes given the observed SNP data, at 1 cM intervals in chromosomal regions where typed SNPs were greater than 1 cM apart. A repeated measures framework (Pinheiro et al., 2011) was used where the data consist of four observations per mouse, corresponding to the light and dark periods in each of the two 24 hr periods, according to the model,
where Yij denotes the clinical trait for the jth time point for the ith individual, lij is an indicator for the dark period, and Li represents genotype or genotype probability. Fixed effects were modeled by ωXi to account for mouse age at sleep trait recording, father, scorer, and calendar month of EEG recording. Outliers more than 3 IQRs from the first or third quartiles were removed prior to the analysis. A 2 df likelihood ratio test of β1 and β2 was conducted to test the significance of each marker or pseudomarker. This model allows the QTL to have different effects on sleep in the light and dark periods.
eQTL Analysis
For each tissue, eQTL were identified for all expression traits using the same markers and pseudo-markers used in the cQTL analysis, adjusting for possible important covariates, i.e., father, mouse age at necropsy, and calendar month at necropsy. Robust regression was used on a marker-by-marker basis with M-estimation and Tukey’s biweight initialized by an S-estimator using the R/mass package (Venables & Ripley, 2002). A Wald test was used to compute the significance level for each test position.
Causal Inference for Overlapping cQTL/eQTL Regions
Chromosome regions were identified where a cQTL overlaped with an eQTL as defined by a peak-to-peak distance of less than 15 cM and lod scores were greater than 3. Conditional independence of the cQTL was used to indicate causal mediation of a gene expression trait in an approach related to Schadt et al. (2005) and Millstein et al. (2009) (Millstein et al., 2009; Schadt et al., 2005). The following models,
were used to select between causal, reactive, or independent QTL/expression trait/sleep trait competing scenarios. Here, equation 1 is fitted using ordinary least squares regression and equation 2 is a repeated measures approach in a generalization of the cQTL analysis described above. In equation 1, Yl and Yd denote the sleep trait measured over the light and dark periods, respectively, and R denotes gene expression. A causal scenario is characterized by the expression trait mediating the effect of the cQTL on the sleep trait; in the reactive scenario, the sleep trait mediates the effect of the eQTL on the expression trait; and in the independent scenario, the QTL independently affects the expression and sleep traits. The selection criteria according to estimates generated from 1 and 2 were, causal: (β3,{β4,β5}≠=0)^({β6,β7}=0); reactive: ({β1,β2},{β6,β7}≠0)^(β3=0); and independent: ({β6,β7}≠0)^(β3≠0)
RESULTS
Identification of Pharmacologically Tractable Candidate Causal Genes Impacting Sleep Architecture
Statistical methods employed yielded a comprehensive list of 2,257 causal scenarios (supplemental Table 1), that is, scenarios in which a gene expression feature was statistically consistent with causal mediation of the sleep trait QTL, derived from the previously described cross in mice (Winrow et al., 2009; Millstein et al., 2011). Each scenario describes a trio consisting of the sleep trait QTL, the gene expression feature measured in the respective tissue, and the sleep trait itself. There was a large amount of redundancy, as many identified genes were associated with multiple traits or tissue type gene expression changes. Out of the total 2,257 causal scenarios, there were 1,034 unique candidate causal genes. We explored potential reagents using Ingenuity Pathway Analysis software and internal search methods, and narrowed the candidates to 21 pharmacologically tractable genes. For the purpose of these studies, we chose to focus on a refined set of six target genes for which selective pharmacological tools were readily available (Table 1). Four of these six, Chrm3, Chrna4, Cacna1i, and Glp1r, were identified for a sleep QTL on chromosome one at approximately 76 cM based on associated gene expression pattern changes in the liver, hypothalamus, and frontal cortex, and the remaining two, Htr1d and Drd5 were identified based on associated gene expression profile changes in the thalamus for sleep QTL on chromosomes four and 13, respectively (Table 2).
Table 1.
Candidate Causal Genes and identified small molecule reagents
| Causal Gene |
Reagent |
||
|---|---|---|---|
| Symbol | Name | Name | Description |
| Chrm3 | cholinergic receptor, muscarinic 3 | Scopolamine | Nonselective antagonist |
| Darifenacin | CHRM3 selective antagonist | ||
|
| |||
| Chrna4 | cholinergic receptor, nicotinic, alpha 4 | Varenicline | Partial Agonist |
| Cytisine | Partial Agonist | ||
|
| |||
| Drd5 | dopamine receptor D5 | SCH23390 | High Affinity D1-like antagonist |
| Risperidone | High Affinity D2-like antagonist | ||
|
| |||
| Htr1d | 5-hydroxytryptamine receptor 1D | Sumatriptan | HTR1 specific antagonist |
| PNU-109291 | HTR1D specific agonist | ||
| BRL-15572 | HTR1D specific antagonist | ||
|
| |||
| Cacna1i | Calcium channel, voltage dependent, T-type, alpha1i subunit | TTA-A2 | Selective T-type antagonist |
|
| |||
| Glp1r | glucagon-like peptide 1 receptor | Exendin-4 | Potent GLP1R agonist |
| Liraglutide | Potent GLP1R agonist | ||
Table 2.
Description of causal trios - trait QTL, the gene expression feature measured in the respective tissue, and sleep trait
| Genea | Chromosome | Tissue | Gene Position (cM) |
eQTL LOD |
Associated Traits | Trait Position (cM) |
cQTL LOD |
|---|---|---|---|---|---|---|---|
| Chrm3 | 1 | Frontal Corte | 69 | 4.59 | non-REM Sleep | 76.96 | 3.86 |
| Total Sleep | 76.96 | 3.77 | |||||
| Wake | 62.69 | 3.64 | |||||
|
| |||||||
| Chrna4 | 1 | Frontal Corte | 63.68 | 12.83 | non-REM Sleep | 76.96 | 3.86 |
| Total Sleep | 76.96 | 3.77 | |||||
| Wake | 62.69 | 3.64 | |||||
| Hypothalamus | 68 | 4.14 | Total Sleep | 76.96 | 3.77 | ||
| Wake | 62.69 | 3.64 | |||||
|
| |||||||
| Drd5 | 13 | Thalamus | 3.28 | 3.47 | non-REM Sleep | 0.41 | 3.50 |
|
| |||||||
| Htr1d | 4 | Thalamus | 57.53 | 12.71 | duration bouts between REM Sleep |
44.39 | 3.42 |
|
| |||||||
| Cacna1i | 1 | Hypothalamus | 63 | 3.03 | Duration bouts Total Sleep |
76.96 | 3.62 |
|
| |||||||
| Glp1r | 1 | Liver | 76 | 3.07 | Latency to non-REM sleep | 76.96 | 6.58 |
| non-REM sleep | 76.96 | 3.86 | |||||
| Total Sleep | 76.96 | 3.77 | |||||
| Wake | 62.23 | 3.64 | |||||
Identified candidate genes that govern sleep, according to tissue, where eQTL and cQTL regions overlapped, defined as a peak to peak distance no greater than 15 cM and LOD greater than 3.
Modulation of Cholinergic Neurotransmission Promotes Wakefulness in Rats
Muscarinic Acetylcholine Receptor M3
According to genetic causality testing, the muscarinic acetylcholine receptor M3 subunit is associated with wake, total sleep, and non-REM sleep (Table 2). To probe the effects of muscarinic Acetylcholine receptor modulation, we examined scopolamine, a nonselective muscarinic receptor antagonist, and darifenacin, a selective muscarinic M3 receptor antagonist. Both molecules have overlapping pharmacological profiles and effects on rat electroencephalography. From our studies, both compounds impacted associated sleep parameters.
Scopolamine hydrochloride (0.3 mg/kg, i.p) significantly induced active wake and slow wave sleep (SWS) at the expense of REM sleep. Scopolamine treatment had significant effects on overall sleep architecture for approximately 7 hours post-dose (Figure 1A). Decreased bout duration during the active period indicated increased fragmentation of active wake and SWS. Effects on SWS bout duration persisted into the inactive period (Figure 1D, Table 4). Significant reductions in REM cumulative sleep time (Figure 1C, Table 3) during the inactive period correlated with reduced bout duration (Figure 1D, Table 4).
Table 4.
Percent Difference Bout duration relative to vehicle baseline per Sleep Phase for each Investigated Reagent
| % Difference Bout Durationa | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Causal Gene Symbol |
Reagentb Name |
Active Phase |
Inactive Phase |
||||||
| AW | LS | SWS | REM | AW | LS | SWS | REM | ||
| Chrm3 | Scopolamine | −12.83 ± 4.60 * | 0.57 ± 2.51 | −26.19 ± 5.05 ** | −10.53 ± 6.17 | 1.25 ± 17.51 | 15.83 ± 12.06 | −27.47 ± 10.73 * | −49.93 ± 5.09 *** |
| Darifenacin | 29.92 ± 17.85 | 15.02 ± 10.30 | −9.34 ± 9.36 | −16.61 ± 4.64 ** | 59.19 ± 28.37 | 27.61 ± 15.89 | −7.20 ± 6.37 | −26.90 ± 5.26 *** | |
|
| |||||||||
| Chrna4 | Varenicline | 26.14 ± 15.69 | 6.56 ± 5.77 | −37.56 ± 6.36 *** | −24.24 ± 5.02 ** | −7.43 ± 12.33 | 2.60 ± 7.42 | −26.99 ± 4.33 *** | −17.85 ± 2.59 *** |
| Cytisine | 3.67 ± 9.62 | 14.47 ± 9.96 | −17.06 ± 8.62 | −18.49 ± 3.55 ** | 10.81 ± 16.46 | 0.35 ± 5.72 | −3.87 ± 5.50 | −2.50 ± 4.22 | |
|
| |||||||||
| Drd5 | SCH23390 | −29.87 ± 10.81 * | 20.18 ± 5.82 * | 3.79 ± 16.23 | −22.11 ± 12.15 | −9.96 ± 5.81 | 18.65 ± 5.57 * | 25.67 ± 18.45 | −3.81 ± 6.56 |
| Risperidone | 40.49 ± 23.53 | 40.62 ± 15.60 * | −40.00 ± 8.50 ** | 2.56 ± 9.15 | 51.31 ± 10.57 ** | 56.83 ± 21.38 * | −11.33 ± 18.35 | −15.76 ± 3.19 ** | |
|
| |||||||||
| Htr1d | Sumatriptan | 8.39 ± 7.40 | 16.52 ± 2.93 ** | 9.91 ± 8.59 | −3.00 ± 5.67 | 2.13 ± 12.22 | 6.07 ± 8.10 | 3.86 ± 7.29 | −8.11 ± 3.59 |
| PNU-109291 | −1.60 ± 9.68 | 5.79 ± 5.20 | 25.13 ± 13.05 | 9.58 ± 6.70 | 14.73 ± 12.56 | 5.23 ± 8.75 | 18.17 ± 9.67 | 9.57 ± 8.15 | |
| BRL-15572 | −3.68 ± 5.49 | −1.51 ± 3.55 | 14.59 ± 5.19 * | −13.28 ± 4.47 * | 11.32 ± 8.63 | 5.43 ± 6.52 | 1.93 ± 4.66 | −4.40 ± 3.12 | |
|
| |||||||||
| Cacna1i | TTA-A2 | 6.59 ± 8.82 | −5.79 ± 7.18 | 119.14 ± 20.72 *** | 11.73 ± 11.33 | 11.60 ± 27.54 | −3.55 ± 8.49 | 123.17 ± 26.33 ** | 24.67 ± 16.47 |
|
| |||||||||
| Glp1r | Exendin-4 | −52.88 ± 6.31 ** | 94.97 ± 17.96 ** | −44.70 ± 5.87 *** | −17.67 ± 8.51 | −34.97 ± 9.68 ** | 65.39 ± 24.39 * | −55.34 ± 7.17 *** | −41.87 ± 11.73 * |
| Liraglutide | −22.15 ± 8.56 * | 16.42 ± 7.12 * | 2.38 ± 10.39 | 1.50 ± 9.57 | 16.59 ±12.60 | 20.16 ± 6.15 ** | −28.70 ± 5.99 *** | −30.55 ± 4.28 *** | |
Mean ± SEM percent difference relative to vehicle controls, paired t-test,
Pvalue < * 0.05,
0.01,
0,001.
Doses used: Scopolamine (0.3MPK, i.p.), Darifenacin (50MPK, p.o.), Varenicline (1MPK, p.o.), Cytisine (1.5MPK, i.p.), SCH23390 (0.1MPK, i.p.), Risperidone (3MPK, i.p.), Sumatriptan (10MPK, p.o.), PNU-109291 (15MPK, s.c.), BRL-15572 (10MPK, i.p.), TTA-A2 (10MPK, p.o.), Exendin-4 (0.02MPK, i.p.), Liraglutide (1MPK, s.c.).
Table 3.
Percent Difference Cumulative Sleep time relative to vehicle baseline per Sleep Phase for each Investigated Reagent
| % Difference Cumulative Time* | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Causal Gene Symbol |
Reagentb Name |
Active Phase |
Inactive Phase |
||||||
| AW | LS | SWS | REM | AW | LS | SWS | REM | ||
| Chrm3 | Scopolamine | 10.26 ± 6.62 | −12.41 ± 14.38 | −8.05 ± 3.79 | 9.70 ± 12.42 | 114.43 ± 49.65 | 15.02 ± 38.24 | −9.88 ± 10.25 | −54.49 ± 9.13 *** |
| Darifenacin | 13.92 ± 7.46 | 40.18 ± 21.56 | −17.37 ± 13.70 | −11.34 ± 13.71 | 159.85 ± 62.63 * | 58.85 ± 27.41 | −10.67 ± 5.05 | −35.76 ± 6.04 *** | |
|
| |||||||||
| Chrna4 | Varenicline | 42.12 ± 18.77 | 55.43 ± 22.80 * | −27.12 ± 9.18 * | −10.44 ± 14.07 | 26.26 ± 21.66 | 51.00 ± 21.27 * | −4.21 ± 4.98 | −0.64 ± 7.75 |
| Cytisine | 22.35 ± 11.65 | 36.49 ± 20.28 | −28.02 ± 8.27 * | −39.60 ± 7.69 ** | 34.04 ± 24.00 | −9.33 ± 7.45 | −3.29 ± 5.67 | −5.31 ± 5.71 | |
|
| |||||||||
| Drd5 | SCH23390 | −29.77 ± 27.02 | 42.33 ± 15.58 * | 4.77 ± 6.99 | 4.69 ± 32.77 | −24.24 ±10.95 | 4.76 ± 11.30 | 5.11 ± 3.38 | −9.90 ± 8.58 |
| Risperidone | 4.42 ± 10.22 | 44.04 ± 13.25 * | −36.76 ± 6.08 *** | 54.51 ± 23.00 * | 46.91 ± 22.28 | 109.49 ± 50.71 | −19.29 ± 8.95 | −1.96 ± 11.97 | |
|
| |||||||||
| Htr1d | Sumatriptan | 3.49 ± 6.91 | 22.21 ± 9.48 | −1.47 ± 4.36 | −10.23 ± 4.86 | 2.51 ± 11.63 | 20.09 ± 19.04 | 0.60 ± 3.36 | −7.46 ± 4.68 |
| PNU-109291 | −5.44 ± 7.69 | 2.58 ± 7.98 | 19.29 ± 9.55 | 13.50 ± 11.44 | −11.54 ± 15.91 | −7.58 ± 7.80 | 5.51 ± 4.52 | 6.58 ± 8.42 | |
| BRL-15572 | −1.60 ± 3.44 | −0.80 ± 7.50 | 5.32 ± 3.62 | −28.45 ± 7.05 ** | 9.82 ± 11.62 | 5.92 ± 7.80 | 3.05 ± 3.35 | −8.42 ± 3.88 | |
|
| |||||||||
| Cacna1i | TTA-A2 | −26.63 ± 4.27 *** | −19.36 ± 18.39 | 42.44 ± 11.68 ** | −20.17 ± 12.81 | −28.88 ± 8.98 * | −28.26 ± 16.15 | 24.26 ± 3.10 *** | −28.80 ± 9.90 * |
|
| |||||||||
| Glp1r | Exendin-4 | −52.88 ± 6.31 *** | 191.16 ± 34.13 *** | −41.65 ± 4.55 *** | −4.35 ± 18.98 | −13.44 ± 15.34 | 217.57 ± 57.28 ** | −31.25 ± 4.81 *** | −35.02 ± 19.42 |
| Liraglutide | −20.00 ± 6.25 ** | 110.87 ± 53.76 | 28.64 ± 12.73 * | 40.67 ± 20.18 | 55.30 ± 23.05 * | 126.96 ± 25.87 *** | −13.65 ± 2.99 *** | −22.02 ± 4.10 *** | |
Mean ± SEM percent difference relative to vehicle controls, paired t-test,
Pvalue < 0.05,
0.01,
0,001.
Doses used: Scopolamine (0.3MPK, i.p.), Darifenacin (50MPK, p.o.), Varenicline (1MPK, p.o.), Cytisine (1.5MPK, i.p.), SCH23390 (0.1MPK, i.p.), Risperidone (3MPK, i.p.), Sumatriptan (10MPK, p.o.), PNU-109291 (15MPK, s.c.), BRL-15572 (10MPK, i.p.), TTA-A2 (10MPK, p.o.), Exendin-4 (0.02MPK, i.p.), Liraglutide (1MPK, s.c.).
Darifenacin hydrobromide (50 mg/kg, p.o.) significantly induced arousal, with increases in both wake and light sleep with simultaneous reductions in both SWS and REM sleep (Figure 1B, Supplemental Figure 1). Compound effects on rat polysomnography persisted approximately 9 hours post-dose (Supplemental Figure 1). The effects of darifenacin on rat sleep attenuated over time, and by the last day of dosing only minor effects were observed (supplemental figure 2). Significant fragmentation of REM sleep (reduced bout duration) during the active period occurred (Figure 1D, Table 4). Increased wakefulness during the inactive period were observed, along with compensatory reductions in REM cumulative sleep time (Figure 1C, Table 3) that correlated with reduced REM bout duration (Figure 1D, Table 4).
Darifenacin had more profound effects on wakefulness, especially when accounting for the observed tachyphylaxis. Increases in SWS sleep were observed following administration with scopolamine treatment, but not following darifenacin treatment. This effect may be attributed to another muscarinic receptor subunit or off target interaction.
Nicotinic Receptor α4
The neuronal nicotinic receptor alpha-4 subunit was associated with wake, total sleep, and non-REM sleep, as determined by genetic causality testing (Table 2). To probe the role of nicotinic receptor alpha-4 agonism on sleep architecture, we utilized cytisine and varenicline tartrate, both partial agonists. Varenicline and cytisine have similar pharmacological profiles, but differing potencies and efficacies with regard to neuronal nicotinic receptor subunits (Gonzales et al., 2006).
Varenicline tartrate (1 mg/kg, p.o.), a partial agonist, is a derivative of cytisine with greater potency and efficacy for the alpha4beta2 neuronal nicotinic receptor subunit (Gonzales et al., 2006). Increased wake and light sleep, with concurrent reductions in SWS sleep were observed after treatment, and effects on light sleep persisted for roughly 7 hours (Figure 2A). Significant effects on SWS and REM sleep dissipated over the course of study, with only minor effects observed by the last day of administration (supplemental Figure 3). Cytisine treatment (1.5 mg/kg, i.p.) had only modest and short acting effects on rat electroencephalography. Generally, cytisine treatment had similar effects on rat sleep as did varenicline treatment, but observed effects were relatively brief. Increased wake and light sleep with coincident reduced SWS and REM sleep were observed in the first hour post-dose (Figure 2B). Cytisine treatment altered arousal state during the active phase only, with no significant effects persisting into the inactive period.
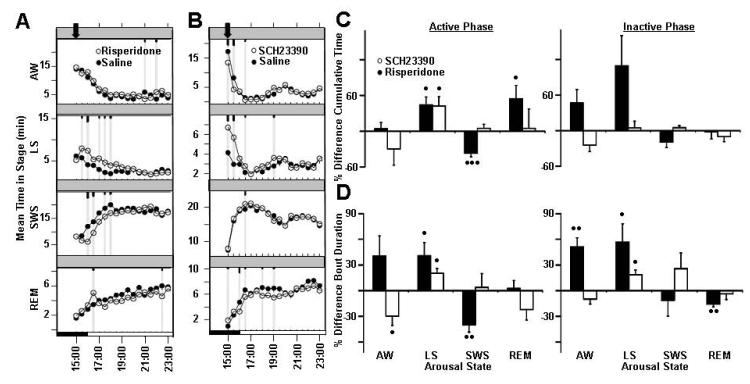
Figure 2.
Effects of Nicotinic acetylcholine receptor α4 on rat sleep architecture (associated traits: Wake, Total Sleep, non-REM). Average time spent per 30-minute interval in active wake, light sleep, SWS sleep, and REM sleep in response to vehicle treatment (closed circles) versus treatment with (A) varenicline and (B) cytisine (open circles). Dark bars in x -axis represent lights-off period, with time of dosing indicated with an arrowhead on each graph. Values are mean ± SEM (short, medium, and long tic marks represent p ≤ .05, ≤ .01, ≤ .001; linear mixed effects model for repeated measures). Average percent changes in sleep stage (C) cumulative time and (D) bout durations (mean time in sleep stage divided by bout number) following treatment of rats with varenicline (closed bars) and cytisine (open bars) for active phase (from time of dosing to ZT 00:30) and inactive phase (ZT 00:30-2:30). Sleep stages are indicated as follows: active wake (AW), light sleep (LS), slow wave sleep (SWS), and REM sleep (REM). Plotted values represent percent difference from vehicle within independent experiments for each dose, and calculated from raw sleep stage time and bout data. Plotted values are mean ±SEM (• p ≤.05, •• p ≤.01, ••• p ≤.001; two-tailed population t test). Each study run as a balanced crossover design, with 7 sequential days of dosing for varenicline treatment (n=8 animals, 56 nights/condition total, 1 mg/kg, p.o., dosed ZT 23:30) and for cytisine treatment (n=8 animals, 56 nights/condition total, 1.5 mg/kg, i.p., dosed ZT 23:00).
More significant effects on rat polysomnography were observed following treatment with varenicline than following treatment with cytisine. Varenicline treatment increased light sleep and reduced SWS during the active period (Figure 2C, Table 3). Treatment with varenicline also significantly increased the number of entries into SWS and REM sleep, which resulted in decreased average bouts of SWS and REM during the active period (Figure 2D, Table 4). During the subsequent inactive phase, light sleep was significantly induced (Figure 2C, Table 3), and SWS and REM sleep consolidation continued, with increased SWS and REM bout duration (Figure 2D, Table 4). Treatment with varenicline increased latency to both SWS (41.9 ± 15.2%, p<0.05) and REM sleep (42.4 ± 14.3%, p<0.05), which is consistent with its wake promoting effects. Acute suppression of REM sleep was the only specific effect that differentiates treatment with cytisine from treatment with varenicline (Figure 2C, Table 3). Both compounds significantly impact each associated trait and promote wake and light sleep. However varenicline has a more profound effect on cumulative time and entries into all stages, which may be attributed to the increased efficacy of varenicline at the alpha4beta2 subunit.
Modulation of Dopaminergic Neurotransmission Promotes Light Sleep in Rats
Dopamine receptor D5 was identified as a candidate causal gene associated with non-REM sleep (Table 2). In general, modulation of dopaminergic neurotransmission induced light sleep at the expense of reduced SWS sleep in our studies. Specific day after induction of light sleep was observed following treatment with SCH23390 and coincided with reduction of SWS and REM sleep, which differed from risperidone study results, and may be considered specific DRD5 mediated effects. Additionally, dopamine receptor D1-like antagonism decreased latency time to light sleep.
Overall, risperidone treatment (3 mg/kg, i.p.) resulted in significant induction of Light sleep and reduction of SWS sleep. Drug effects persisted for approximately 3 hours post-dose (Figure 3A). Potentiation to drug treatment was observed, with only minor effects observed on the first day of dosing (Supplemental Figure 4). SCH23390 treatment (0.1 mg/kg, s.c) resulted in an immediate decrease in active wake and an increase in light sleep (Figure 3B). Significant day after effects were also observed; increased light sleep and SWS and reduced REM sleep were recorded during the following active period (Supplemental Figure 5).
Figure 3.
Effects of modulating dopamine 1D receptor on rat sleep architecture (associated trait: non-REM sleep). Average time spent per 30-minute interval in active wake, light sleep, SWS sleep, and REM sleep in response to vehicle treatment (closed circles) versus treatment with (A) risperidone and (B) SCH23390 (open circles). Dark bars in x -axis represent lights-off period, with time of dosing indicated with an arrowhead on each graph. Values are mean ± SEM (short, medium, and long tic marks represent p ≤ .05, ≤ .01, ≤ .001; linear mixed effects model for repeated measures). Average percent changes in sleep stage (C) cumulative time and (D) bout durations (mean time in sleep stage divided by bout number) following treatment of rats with risperidone (closed bars) and SCH23390 (open bars) for active phase (from time of dosing to ZT 00:30) and inactive phase (ZT 00:30-2:30). Sleep stages are indicated as follows: active wake (Wake), light sleep (LS), slow wave sleep (SWS), and REM sleep (REM). Plotted values represent percent difference from vehicle within independent experiments for each dose, and calculated from raw sleep stage time and bout data. Plotted values are mean ±SEM (• p ≤.05, •• p ≤.01, ••• p ≤.001; two-tailed population t test). Each study run as a balanced crossover design, with 7 sequential days of dosing for risperidone treatment (n=8 animals, 56 nights/condition total, 3 mg/kg, i.p., dosed ZT 23:00) and for SCH23390 treatment (n=6 animals, 48 nights/condition total, 0.1 mg/kg, i.p., dosed ZT 23:30).
Both drugs increased light sleep cumulative time immediately post-dose (Figure 3C, Table 3), and light sleep bout duration for the duration of both active and inactive periods (Figure 3D, Table 4). While there were many more observed effects specific to risperidone treatment, only treatment with SCH23390 resulted in reduced active wake mean bout duration during the active period (wake fragmentation) (Figure 3D, Table 4). Also, SCH23390 significantly reduced the latency time to light sleep (50.0 ± 12.7%, p<0.05), which is consistent with its sleep promoting effects.
Modulation of Serotonergic Neurotransmission via the 1D Receptor Subtype Results in Modest Effects on REM Sleep
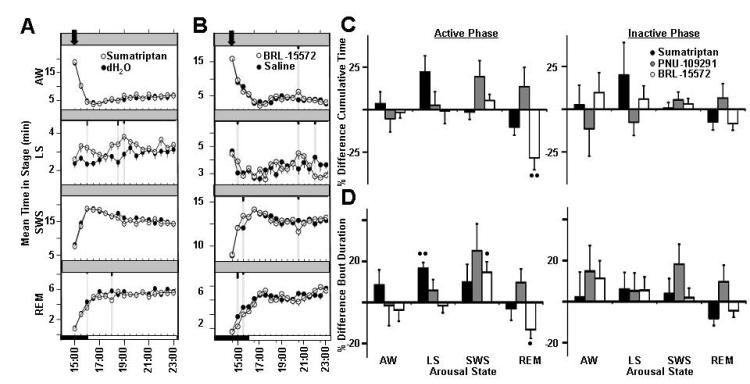
The Htr1d gene is associated with duration between bouts of REM sleep (REM consolidation/fragmentation) (Table 2). Overall, the modulation of serotonergic neurotransmission via the 1D receptor subtype resulted in only modest effects on rat electroencephalography in our studies. Sumatriptan, a 5-HT1B/1D receptor agonist, PNU-109291, a 5-HT1D receptor agonist, and BRL15572, a 5-HT1D receptor antagonist were selected to explore the potential impact of the serotonin 1D receptor on rodent arousal (Kayser et al., 2002; Cutrer et al., 1999; Pauwels & John, 1999). Compound mediated effects on cumulative sleep time and bout duration of REM sleep were specific to BRL 15572, while no significant effects on REM sleep were observed using the nonselective serotonin-1 agonist, sumatriptan. This indicates the effects observed on REM sleep may be mediated by the 5-HT1D receptor.
Sumatriptan, an antimigraine medication, is a nonselective serotonin-1 receptor agonist (Pauwels & John, 1999). Sumatriptan treatment (10 mg/kg, p.o.) resulted in only modest increases in light sleep that persisted for less than 2 hours (Figure 4A). Increases in the average bout duration of light sleep were observed during the active phase (Figure 4D, Table 4).
Figure 4.
Effects of modulating serotonin 1D receptor on rat sleep architecture (associated trait: average duration between bouts of REM sleep). Average time spent per 30-minute interval in active wake, light sleep, SWS sleep, and REM sleep in response to vehicle treatment (closed circles) versus treatment with (A) sumatriptan and (B) BRL-15572 (open circles). Dark bars in x -axis represent lights-off period, with time of dosing indicated with an arrowhead on each graph. Values are mean ± SEM (short, medium, and long tic marks represent p ≤ .05, ≤ .01, ≤ .001; linear mixed effects model for repeated measures). Average percent changes in sleep stage (C) cumulative time and (D) bout durations (mean time in sleep stage divided by bout number) following treatment of rats with sumatriptan (closed bars) and BRL-15572 (open bars) for active phase (from time of dosing to ZT 00:30) and inactive phase (ZT 00:30-2:30). Sleep stages are indicated as follows: active wake (Wake), light sleep (LS), slow wave sleep (SWS), and REM sleep (REM). Plotted values represent percent difference from vehicle within independent experiments for each dose, and calculated from raw sleep stage time and bout data. Plotted values are mean ±SEM (• p ≤.05, •• p ≤.01, ••• p ≤.001; two-tailed population t test). Each study run as a balanced crossover design, with 7 sequential days of dosing for sumatriptan treatment (n=7 animals, 49 nights/condition total, 10 mg/kg, p.o.., dosed ZT 23:00) and 3 days of sequential dosing for BRL-15572 treatment (n=16 animals, 48 nights/condition total, 10 mg/kg, i.p., dosed ZT 22:30).
PNU 109291 is an investigational antimigraine therapy that is a highly selective serotonin-1D receptor agonist (Cutrer et al., 1999). PNU 109291 treatment (10 mg/kg, 15 mg/kg, s.c.) resulted in only minor effects, with no observed trends of treatment on overall rat sleep architecture (Supplemental Figure 6).
Treatment with BRL 15572 (10 mg/kg, i.p.) most notably reduced REM sleep in rats for up to 2 hours (Figure 4B). During the active phase, decreased REM sleep time was observed (Figure 4C, Table 3). Drug treatment also decreased the average bout duration of REM sleep and increased average bout duration of SWS sleep over this same period (Figure 4D, Table 4).
Antagonism of the Voltage-Dependent T-Type Calcium Channel Alpha 1i Subunit (CACNA1I) Promotes Non-REM Sleep
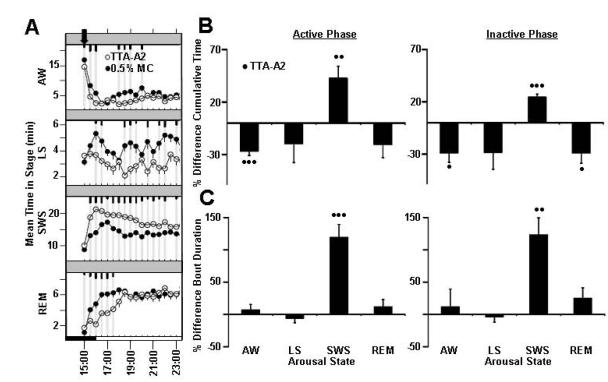
Cacna1i is a candidate gene that was associated with changes in total sleep average bout duration (Table 2). TTA-A2 strongly and preferentially promoted SWS sleep in rats, and also profoundly increased the average bouts of SWS sleep. Treatment with TTA-A2 did not significantly impact light sleep or REM sleep bout duration.
TTA-A2 (10 mg/kg, p.o.) promoted somnolence, specifically increasing SWS sleep at the expense of light and REM sleep (Figure 5A). Suppression of wake and induction of SWS persisted for both active and inactive phases. Decreased REM sleep time was observed during the inactive phase (Figure 5B, Table 3). Significant increases in SWS time coincided with increased bout duration (Figure 5C, Table 4), during both active and inactive phases.
Figure 5.
Effects of TTA-A2, a T-type calcium channel antagonist on rat sleep architecture (associated traits: duration of the bouts of total sleep). Average time spent per 30-minute interval in active wake, light sleep, SWS sleep, and REM sleep in response to vehicle treatment (closed circles) versus treatment with (A) TTA-A2 (open circles). Dark bars in x -axis represent lights-off period, with time of dosing indicated with an arrowhead on each graph. Values are mean ± SEM (short, medium, and long tic marks represent p ≤ .05, ≤ .01, ≤ .001; linear mixed effects model for repeated measures). Average percent changes in sleep stage (B) cumulative time and (C) bout durations (mean time in sleep stage divided by bout number) following treatment of rats with TTA-A2 (closed bars) for active phase (from time of dosing to ZT 00:30) and inactive phase (ZT 00:30-2:30). Sleep stages are indicated as follows: active wake (Wake), light sleep (LS), slow wave sleep (SWS), and REM sleep (REM). Plotted values represent percent difference from vehicle within independent experiments for each dose, and calculated from raw sleep stage time and bout data. Plotted values are mean ±SEM (• p ≤.05, •• p ≤.01, ••• p ≤.001; two-tailed population t test). Study run as a balanced crossover design, with 7 sequential days of dosing for TTA-A2 treatment (n=8 animals, 56 nights/condition total, 10 mg/kg, p.o., dosed ZT 23:00).
Activation of Glucagon-like Peptide 1 Receptor (GLP1R) has Profound Effects on Rat Sleep Architecture
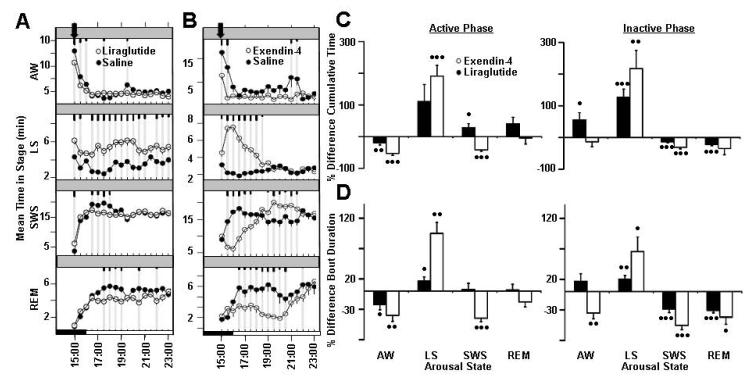
As determined by causality testing, the Glp1r gene was associated with wake and non-REM sleep (Table 2). Liraglutide and exendin-4 are specific and highly potent agonists of glucagon-like peptide receptor, and each had similar impact on rat electroencephalography. Both compounds preferentially induced light sleep at the expense of deep sleep and wake.
Liraglutide is a high affinity GLP1R agonist, developed with a prolonged half-life, and intended for the treatment of type-2 diabetes (Hayes et al., 2011). Liraglutide treatment (1 mg/kg, s.c.) significantly induced light sleep for nearly all time points, at the expense of wake, SWS and REM sleep (Figure 6A). Additionally, day after rebound of both SWS and REM sleep was observed at the expense of active wake (Supplemental Figure 7). Exendin-4 is another potent GLP-1 receptor agonist and pancreatic secretagogue (Yin et al., 2005). Overall, significant induction of light sleep was observed, with compensatory reductions in wake, REM sleep, and SWS in response to exendin-4 treatment (0.02 mg/kg, i.p.) in the late active period. Interestingly, compound treatment had a mixed effect on SWS time. For approximately 4 hours post-dose, exendin-4 reduced SWS significantly, followed by increased SWS during the subsequent 2 hours as compared to vehicle treatment group (Figure 6B).
Figure 6.
Incretin mimetics, exendin-4 and liraglutide, effects on rat sleep architecture (associated traits: Wake, Total Sleep, non-REM, latency to non-REM sleep). Average time spent per 30-minute interval in active wake, light sleep, SWS sleep, and REM sleep in response to vehicle treatment (closed circles) versus treatment with (A) liraglutide and (B) exendin-4 (open circles). Dark bars in x -axis represent lights-off period, with time of dosing indicated with an arrowhead on each graph. Values are mean ± SEM (short, medium, and long tic marks represent p ≤ .05, ≤ .01, ≤ .001; linear mixed effects model for repeated measures). Average percent changes in sleep stage (C) cumulative time and (D) bout durations (mean time in sleep stage divided by bout number) following treatment of rats with liraglutide (closed bars) and exendin-4 (open bars) for active phase (from time of dosing to ZT 00:30) and inactive phase (ZT 00:30-2:30). Sleep stages are indicated as follows: active wake (Wake), light sleep (LS), slow wave sleep (SWS), and REM sleep (REM). Plotted values represent percent difference from vehicle within independent experiments for each dose, and calculated from raw sleep stage time and bout data. Plotted values are mean ±SEM (• p ≤.05, •• p ≤.01, ••• p ≤.001; two-tailed population t test). Each study run as a balanced crossover design, with 3 sequential days of dosing for liraglutide treatment (n=14 animals, 42 nights/condition total, 1 mg/kg, s.c., dosed ZT 23:00) and with 7 days of sequential dosing for exendin-4 treatment (n=7 animals, 49 nights/condition total, 0.02 mg/kg, i.p., dosed ZT 23:00).
Both drugs reduced wake during the active period, and increased light sleep at the expense of SWS during the inactive period. Effects of liraglutide on light sleep and SWS were more acute and were observed immediately post-dose. In contrast, exendin-4 treatment increased SWS during the active period, and increased active wake during the inactive period (Figure 6C, Table 3). Treatment with either drug resulted in reduced active wake and increased light sleep average bout duration immediately post-dose, with effects on light sleep bout duration continuing into the inactive period. Decreased SWS and REM bout duration during the inactive period were coupled with increased light sleep average bout duration in both treatment groups. Liraglutide treatment had more acute effects on SWS bout duration, and more prolonged effects on active wake bout duration (Figure 6D, Table 4).
DISCUSSION
The identification and validation of candidate causal genes regulating sleep and wake may provide new therapeutic targets for insomnia as well as other diseases in which sleep is dysregulated. The approach used in our studies is direct; causal inference generated a comprehensive list of candidate genes, which we used to identify pharmacological reagents specific to these targets. Such an approach allowed for the alignment of genetic causality with pharmacodynamics using small molecule tools in preclinical sleep studies. The objective of confirming associated sleep/wake traits identified from a genetically segregating population is met with certain challenges. In our studies, causal inference identified candidate genes that govern sleep in an N2 cross involving mice, while validation studies were conducted on Sprague-Dawley rats. For each genetic target, select molecules had significant effects on rat sleep architecture that were in alignment with associated sleep traits discovered in mice. Validating genetic targets in rats that were originally identified in mice is a testament to the strength and potential of these targets as being impactful on sleep and arousal across species. Further work may determine if these results translate to humans.
Behavioral state as determined by EEG provides a complex readout on the overall electrical activity in the brain that includes information on many complex and inter-related traits including sleep state, total time spent in each state, and fragmentation of sleep. Identified candidate genes may primarily impact a single sleep-wake trait, which at the same time may affect sleep architecture due to compensation; a loss or gain in a single sleep state affects total time spent in other sleep states. Therefore, alignment of associated sleep-wake traits with effects of target engagement on rat electroencephalography is not straightforward. Our criterion for alignment is therefore either positive (e.g. gain of time spent) or negative (e.g. loss of time spent) effects on the associated sleep-wake trait, with the expectation of consequential compensatory effects on other parameters.
Reagent polypharmacology and pharmacokinetics are other considerations and potential challenges to this approach. Even molecules with high specificity for target gene products have the potential for low affinity off-target interactions that produce effects on behavioral state. Selection of reagents that have complementary or contrasting pharmacology offers the best approach to differentiating target and off-target effects. Additionally, certain small molecules may not sufficiently penetrate the central nervous system to produce significant effects on behavioral state. Whenever possible, in the present study we utilized doses that had demonstrated central effects on other behaviors. A negative finding, however, is inconclusive in the absence of in depth characterization, such as receptor occupancy, and may indicate the compound did not have effects at selected doses due to poor CNS bioavailability. Our choice of reagents targeting CHRM3, CHRNA4, HTR1D, DRD5, GLP1R and CACNA1I satisfied the above selection requirements, and demonstrated effects on rat sleep architecture that aligned with associated sleep-wake traits as elaborated on below.
The cholinergic receptors, nicotinic acetycholine receptor alpha4 subunit and the muscarinic acetycholine receptor M3 subunit, were both associated with total sleep, wake and non-REM sleep traits. The selective pharmacological tools had the same overall impact on rat sleep architecture. Scopolamine, darifenacin, cytisine, and varenicline significantly promote wake at the expense of SWS and REM sleep. The selective muscarinic M3 antagonist, darifenacin, and more potent nicotinic alpha4beta2 agonist, varenicline, has more substantial, quantifiable, and prolonged impact on wake, total sleep and non-REM sleep. These findings are consistent with the observation that cholinergic neurotransmission provides input for the Ascending Arousal System, neuronal circuitry that induces cortical activity and modulate behavioral state. Major cholinergic contributors to this pathway originate in the mesopontine area and activate thalamic relay neurons, which in turn promote cortical activity and wake. Secondary contribution bypasses thalamic control of wakefulness, and originates in the basal forebrain (Saper et al., 2005). Muscarinic M3 receptor containing neurons are localized to the mesopontine region, in the pedunculopontine and laterodorsal tegmental nuclei and may contribute to both branches of the Ascending Arousal System (Vilaro et al., 1994). These regions of the mesopontine area fire rapidly during wake and REM sleep (Saper et al., 2005).
In addition to promoting wake through the Ascending Arousal System, cholinergic signaling also inhibits key neurons in the ventrolateral preoptic nucleus that promote sleep (Saper et al., 2005). The nicotinic acetylcholine receptor, alpha 4 subunit may contribute to this sleep effect (Dani & Bertrand, 2007). In this way, modulating cholinergic neurotransmission may primarily inhibit sleep and induce a compensatory shift in sleep architecture favoring light sleep and wake.
We studied effects of modulating monoaminergic neurotransmission on rat sleep. Genes that encode serotonin 1D receptor and dopamine D5 receptor were both identified in our causality study (associated with duration between REM bouts, and non-REM sleep respectively). Monoaminergic signaling pathways feed into thalamic relay neurons that ultimately stimulate cortical activity and arousal, including those that originate from the ventral tegmental area (dopaminergic), and the raphe nuclei (serotonergic) (Saper et al., 2005). Certain psychiatric and neurological pathology are attributed to dysfunction in the serotinergic and dopaminergic pathways that also display co-morbidity with disordered sleep. Depression, Parkinson’s disease, and schizophrenia are all conditions that exhibit altered monoaminergic signaling, and are also associated with disruptions in sleep architecture (Peterson & Benca, 2006; Cohrs, 2008; Compta et al., 2009).
Dopamine receptor D5 was identified as a candidate gene associated with non-REM sleep. Subtypes D1 and D5, are classified D1-like receptors, while dopamine receptors D2, D3, and D4 are considered D2-like. The two subfamilies of dopamine receptors have differing pharmacology, while, within subfamilies, receptor subtypes are nearly indistinct (Missale et al., 1998). SCH23390 is a potent dopamine D1 and D5 (D1-like) receptor antagonist (Bourne, 2001). In contrast, risperidone, an atypical antipsychotic, was examined for its high affinity for the dopamine D2 (D2-like) receptor. In our studies, both risperidone and SCH23390 impact non-REM sleep. However, SCH23390, the D1-like specific antagonist impacted latency to non-REM sleep, and had profound day after effects on non-REM and other sleep parameters as compared to the D2-like compound, risperidone. This day after effect is paradoxical, since the compound has a short half life of 25 minutes in rats, and may result from rapid neuronal expression changes or another compensatory or anticipatory mechanism(Bourne, 2001).
The Htr1d gene was associated with duration between bouts of REM sleep (REM consolidation/fragmentation). Studies involving 5-HT1D receptor antagonism detected acute effects on cumulative time spent and average bout duration of REM sleep. Within 30 minutes of dosing, mean number of entries into REM sleep decreased. REM effects were specific to BRL-15572, a 5-HT1D receptor antagonist, and indicate reduction of the number of cycles and the average duration of REM sleep. And while the magnitude was relatively small but significant, and brief in duration, causality does not necessarily discriminate effect size. Dosing BRL-15572 early in the active period may reveal more prolonged effects on REM sleep.
We have previously reported effects of T-type calcium channel (Cacna1i) antagonism on rat electroencephalography (Uebele et al., 2009). The Cacna1i gene was identified as a candidate causal gene for average bout duration of total sleep and SWS. TTA-A2 promotes SWS at the expense of active wake, light and REM sleep. TTA-A2 not only increases total sleep, but it specifically increases bout duration of SWS sleep, demonstrating remarkable alignment with the associated trait.
Glucagon-like peptide-1 (GLP1) functions as an incretin, to stimulate insulin and inhibit glucagon (Kanoski et al., 2011). Both liraglutide and exendin-4 are incretin mimetics that also demonstrate central effects on appetite control when administered peripherally (Kanoski et al., 2011). The Glp1r gene was associated with wake, total sleep, non-REM sleep and latency to non-REM sleep. Both GLP1r agonists are sleep promoting compounds that specifically induce light sleep over SWS and REM sleep. Glucagon-like peptide-1 receptors are found throughout the hypothalamus, in regions that impact arousal, and when taken together with the observed pharmacology of liraglutide and exendin-4, GLP-1 receptors may play a role in regulating sleep and wake homeostasis.
In summary, pharmacological validation of candidate genetic targets is a direct approach that offers advantages over characterization of target genes in knockout or transgenic models. Interrogating gene function using small molecule tools provides an accelerated route to evaluate potential new targets for sleep disorders as well as for psychiatric diseases. In addition, small molecules test the targets in an organism that developed normally, without any genetic lesion, that could have led to abnormal physiology via compensatory gene regulation during development. This approach many not only validate or invalidate candidate genes as targets for drug discovery, but may also provide insight into the potential causal or reactive nature of specific genes and networks for regulating behavior. Taken together, the results from the present study demonstrate robust agreement between genetic causality in mice and pharmacologically induced sleep effects in rats, providing novel insights into the underlying physiology of sleep and wake regulation conserved between these species as well as the identification of new avenues for therapeutic investigation.
Supplementary Material
Supplemental Figure 1. Effects of Darifenacin (50 mg/kg, p.o.) on rat sleep architecture.
Supplemental Figure 2. Effects of Darifenacin (50 mg/kg, p.o.) on rat sleep architecture on first and last day of treatment.
Supplemental Figure 3. Effects of Varenicline (1mg/kg, p.o.) on rat sleep architecture on first and last day of treatment.
Supplemental Figure 4. Effects of Risperidone (3 mg/kg, i.p.) on rat sleep architecture on first and last day of treatment.
Supplemental Figure 5. Effects of SCH23390 (0.1 mg/kg, s.c.) on rat sleep architecture.
Supplemental Figure 6. Effects of PNU-109291 (15, 10 mg/kg, s.c.) on rat sleep architecture.
Supplemental Figure 7. Effects of Liraglutide (1 mg/kg, s.c.) on rat sleep architecture.
Supplemental Table 1 - Causal Gene List
Acknowledgements
Assistance from Merck’s Laboratory Animal Resources Staff (Central Pharmacology, Merck Research Laboratories) for expert animal handling and treatments are much appreciated. This work was supported by a research grant from Merck and Co. (FWT) and by the Army Research Office (ARO), award number DAAD19-02-1-0038 from the Defense Advance Research Projects Agency. Investigators from the sponsoring institution (Merck/Rosetta) were involved in data analysis (genotyping, QTL identification, Baeysian network construction and statistics) and interpretation of the results. J.I.B, A.L.G., J.M., S.G., J.B., S.M.D, S.V.F., A.K., J.J.R., and C.J.W. are or have been employed by Merck & Co., Inc. (USA) and potentially own stock and/or stock options in the company.
Footnotes
Authors contributions Participated in research design: Brunner, Gotter, Kasarskis, Renger, Turek, Webber, Winrow Conducted experiments: Brunner, Garson, Savitz, Yang, Fitzpatrick, Zhou, Owens, Vitaterna Performed data analysis: Brunner, Garson, Fox, Millstein Wrote or contributed to the writing of the manuscript: Brunner, Gotter, Kasarskis, Millstein, Renger, Turek, Uebele, Webber, Winrow.
REFERENCES
- Bourne JA. SCH 23390: The first selective dopamine D-1-like receptor antagonist. CNS Drug Reviews. 2001;7:399–414. doi: 10.1111/j.1527-3458.2001.tb00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisbare-Roch C, Dingemanse J, Koberstein R, Hoever P, Aissaoui H, Flores S, et al. Promotion of sleep by targeting the orexin system in rats, dogs and humans. Nature Medicine. 2007;13:150–155. doi: 10.1038/nm1544. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Caylak E. The genetics of sleep disorders in humans: narcolepsy, restless legs syndrome, and obstructive sleep apnea syndrome. Am.J Med.Genet.A. 2009;149A:2612–2626. doi: 10.1002/ajmg.a.33087. [DOI] [PubMed] [Google Scholar]
- Chabas D, Taheri S, Renier C, Mignot E. The genetics of narcolepsy. Annual Review of Genomics and Human Genetics. 2003;4:459–483. doi: 10.1146/annurev.genom.4.070802.110432. [DOI] [PubMed] [Google Scholar]
- Cirelli C. The genetic and molecular regulation of sleep: from fruit flies to humans. Nature Reviews Neuroscience. 2009;10:549–560. doi: 10.1038/nrn2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Cohrs S. Sleep Disturbances in Patients with Schizophrenia Impact and Effect of Antipsychotics. Cns Drugs. 2008;22:939–962. doi: 10.2165/00023210-200822110-00004. [DOI] [PubMed] [Google Scholar]
- Compta Y, Santamaria J, Ratti L, Tolosa E, Iranzo A, Munoz E, et al. Cerebrospinal hypocretin, daytime sleepiness and sleep architecture in Parkinson’s disease dementia. Brain. 2009;132:3308–3317. doi: 10.1093/brain/awp263. [DOI] [PubMed] [Google Scholar]
- Cutrer FM, Yu XJ, Ayata G, Moskowitz MA, Waeber C. Effects of PNU-109,291, a selective 5-HT1D receptor agonist, on electrically induced dural plasma extravasation and capsaicin-evoked c-fos immunoreactivity within trigeminal nucleus caudalis. Neuropharmacology. 1999;38:1043–1053. doi: 10.1016/s0028-3908(99)00032-5. [DOI] [PubMed] [Google Scholar]
- Dani JA, Bertrand D. Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annual Review of Pharmacology and Toxicology. 2007;47:699–729. doi: 10.1146/annurev.pharmtox.47.120505.105214. [DOI] [PubMed] [Google Scholar]
- Dauviltiers Y, Maret S, Tafti M. Genetics of normal and pathological sleep in humans. Sleep Medicine Reviews. 2005;9:91–100. doi: 10.1016/j.smrv.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Devineni D, Skerjanec A, Woodworth TG. Low Central Nervous System (CNS) Penetration by Darifenacin, a Muscarinic M3 Selective Receptor Antagonist, in Rats. pA2 Online E-journal of the British Pharmacological Society. 2005 [Google Scholar]
- Di Fabio R, Pellacani A, Faedo S, Roth A, Piccoli L, Gerrard P, et al. Discovery process and pharmacological characterization of a novel dual orexin 1 and orexin 2 receptor antagonist useful for treatment of sleep disorders. Bioorganic & Medicinal Chemistry Letters. 2011;21:5562–5567. doi: 10.1016/j.bmcl.2011.06.086. [DOI] [PubMed] [Google Scholar]
- Franken P, Tafti M. Genetics of sleep and sleep disorders. Frontiers in Bioscience. 2003;8:E381–E397. doi: 10.2741/1084. [DOI] [PubMed] [Google Scholar]
- Gonzales D, Rennard SI, Nides M, Oncken C, Azoulay S, Billing CB, et al. Varenicline, an alpha 4 beta 2 nicotinic acetylcholine receptor partial agonist, vs sustained-release bupropion and placebo for smoking cessation - A randomized controlled trial. Jama-Journal of the American Medical Association. 2006;296:47–55. doi: 10.1001/jama.296.1.47. [DOI] [PubMed] [Google Scholar]
- Hagan JJ, Slade PD, Gaster L, Jeffrey P, Hatcher JP, Middlemiss DN. Stimulation of 5-HT1B receptors causes hypothermia in the guinea pig. European Journal of Pharmacology. 1997;331:169–174. doi: 10.1016/s0014-2999(97)01055-8. [DOI] [PubMed] [Google Scholar]
- Hallmayer J, Faraco J, Lin L, Hesselson S, Winkelmann J, Kawashima M, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nature Genetics. 2009;41:708–711. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MR, Kanoski SE, Alhadeff AL, Grill HJ. Comparative Effects of the Long-Acting GLP-1 Receptor Ligands, Liraglutide and Exendin-4, on Food Intake and Body Weight Suppression in Rats. Obesity. 2011;19:1342–1349. doi: 10.1038/oby.2011.50. [DOI] [PubMed] [Google Scholar]
- Hor H, Kutalik Z, Dauvilliers Y, Valsesia A, Lammers GJ, Donjacour CEHM, et al. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nature Genetics. 2010;42:786–U80. doi: 10.1038/ng.647. [DOI] [PubMed] [Google Scholar]
- Jiang P, Striz M, Wisor JP, O’Hara BF. Behavioral and genetic dissection of a mouse model for advanced sleep phase syndrome. SLEEP. 2011;34:39–48. doi: 10.1093/sleep/34.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoski SE, Fortin SM, Arnold M, Grill HJ, Hayes MR. Peripheral and Central GLP-1 Receptor Populations Mediate the Anorectic Effects of Peripherally Administered GLP-1 Receptor Agonists, Liraglutide and Exendin-4. Endocrinology. 2011;152:3103–3112. doi: 10.1210/en.2011-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser V, Aubel B, Hamon M, Bourgoin S. The antimigraine 5-HT1B/1D receptor agonists, sumatriptan, zolmitriptan and dihydroergotamine, attenuate pain-related behaviour in a rat model of trigeminal neuropathic pain. British Journal of Pharmacology. 2002;137:1287–1297. doi: 10.1038/sj.bjp.0704979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornum BR, Kawashima M, Faraco J, Lin L, Rico TJ, Hesselson S, et al. Common variants in P2RY11 are associated with narcolepsy. Nature Genetics. 2011;43:66–U90. doi: 10.1038/ng.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin XY, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, et al. Macromolecule biosynthesis: a key function of sleep. Physiol Genomics. 2007;31:441–457. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- Mackiewicz M, Zimmerman JE, Shockley KR, Churchill GA, Pack AI. What are microarrays teaching us about sleep? Trends Mol.Med. 2009;15:79–87. doi: 10.1016/j.molmed.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millstein J, Winrow CJ, Kasarskis A, Owens JR, Zhou L, Summa KC, et al. Identification of Causal Genes, Networks and Transcriptional Regulators of REM Sleep and Wake. SLEEP. doi: 10.5665/sleep.1378. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. Bmc Genetics. 2009;10 doi: 10.1186/1471-2156-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine receptors: From structure to function. Physiological Reviews. 1998;78:189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- Miyagawa T, Kawashima M, Nishida N, Ohashi J, Kimura R, Fujimoto A, et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat.Genet. 2008;40:1324–1328. doi: 10.1038/ng.231. [DOI] [PubMed] [Google Scholar]
- Nishino S, Okuro M. Emerging treatments for narcolepsy and its related disorders. Expert Opin.Emerg.Drugs. 2010;15:139–158. doi: 10.1517/14728210903559852. [DOI] [PubMed] [Google Scholar]
- Pauwels PJ, John GW. Present and future of 5-HT receptor agonists as antimigraine drugs. Clinical Neuropharmacology. 1999;22:123–136. [PubMed] [Google Scholar]
- Peterson MJ, Benca RM. Sleep in mood disorders. Psychiatric Clinics of North America. 2006;29:1009. doi: 10.1016/j.psc.2006.09.003. + [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, the R Development Core Team nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-98. 2011. [Computer software]
- Renger JJ, Dunn SL, Motzel SL, Johnson C, Koblan KS. Sub-chronic administration of zolpidem affects modifications to rat sleep architecture. Brain Research. 2004;1010:45–54. doi: 10.1016/j.brainres.2004.02.067. [DOI] [PubMed] [Google Scholar]
- Sajja RK, Rahman S. Lobeline and cytisine reduce voluntary ethanol drinking behavior in male C57BL/6J mice. Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2011;35:257–264. doi: 10.1016/j.pnpbp.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, GuhaThakurta D, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nature Genetics. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Mignot E. Genetics of Sleep and Sleep Disorders. Cell. 2011;146:194–207. doi: 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipe WD, Barrow JC, Yang ZQ, Lindsley CW, Yang FV, Schlegel KA, et al. Design, Synthesis, and Evaluation of a Novel 4-Aminomethyl-4-fluoropiperidine as a T-Type Ca(2+) Channel Antagonist. J Med Chem. 2008;51:3692–3695. doi: 10.1021/jm800419w. [DOI] [PubMed] [Google Scholar]
- Slassi A, Isaac M, Arora J. Novel serotonergic and non-serotonergic migraine headache therapies. Expert Opinion on Therapeutic Patents. 2001;11:625–649. [Google Scholar]
- Tafti M. Genetic aspects of normal and disturbed sleep. Sleep Med. 2009;10(Suppl 1):S17–S21. doi: 10.1016/j.sleep.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Wisor JP, Lee CK, Pathak SD, Gerashchenko D, Smith KA, et al. Molecular and anatomical signatures of sleep deprivation in the mouse brain. Front Neurosci. 2010;4:165. doi: 10.3389/fnins.2010.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Cirelli C. Sleep and synaptic homeostasis: a hypothesis. Brain Research Bulletin. 2003;62:143–150. doi: 10.1016/j.brainresbull.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Uebele VN, Gotter AL, Nuss CE, Kraus RL, Doran SM, Garson SL, et al. Antagonism of T-type calcium channels inhibits high-fat diet-induced weight gain in mice. Journal of Clinical Investigation. 2009;119:1659–1667. doi: 10.1172/JCI36954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern Applied Statistics with S. Fourth ed Springer; New York: 2002. [Google Scholar]
- Vilaro MT, Palacios JM, Mengod G. Multiplicity of Muscarinic Autoreceptor Subtypes - Comparison of the Distribution of Cholinergic Cells and Cells Containing Messenger-Rna for 5 Subtypes of Muscarinic Receptors in the Rat-Brain. Molecular Brain Research. 1994;21:30–46. doi: 10.1016/0169-328x(94)90375-1. [DOI] [PubMed] [Google Scholar]
- Whitman DB, Cox CD, Breslin MJ, Brashear KM, Schreier JD, Bogusky MJ, et al. Discovery of a Potent, CNS-Penetrant Orexin Receptor Antagonist Based on an N,N-Disubstituted-1,4-diazepane Scaffold that Promotes Sleep in Rats. Chemmedchem. 2009;4:1069–1074. doi: 10.1002/cmdc.200900069. [DOI] [PubMed] [Google Scholar]
- Winrow CJ, Gotter AL, Cox CD, Doran SM, Tannenbaum PL, Breslin MJ, et al. Promotion of Sleep by Suvorexant--A Novel Dual Orexin Receptor Antagonist. Journal of Neurogenetics. 2011;25:52–61. doi: 10.3109/01677063.2011.566953. [DOI] [PubMed] [Google Scholar]
- Winrow CJ, Williams DL, Kasarskis A, Millstein J, Laposky AD, Yang HS, et al. Uncovering the Genetic Landscape for Multiple Sleep-Wake Traits. Plos One. 2009;4 doi: 10.1371/journal.pone.0005161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XP, Wei DZ, Yi LN, Tao XY, Ma YS. Expression and purification of exendin-4, a GLP-1 receptor agonist, in Escherichia coli. Protein Expression and Purification. 2005;41:259–265. doi: 10.1016/j.pep.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Zaniewska M, McCreary AC, Stefanski R, Przegalinski E, Filip M. Effect of Varenicline on the Acute and Repeated Locomotor Responses to Nicotine in Rats. Synapse. 2008;62:935–939. doi: 10.1002/syn.20564. [DOI] [PubMed] [Google Scholar]
- Zee PC, Turek FW. Sleep and health - Everywhere and in both directions. Archives of Internal Medicine. 2006;166:1686–1688. doi: 10.1001/archinte.166.16.1686. [DOI] [PubMed] [Google Scholar]
- Zhang C, Fang YR, Li M. Olanzapine and risperidone disrupt conditioned avoidance responding by selectively weakening motivational salience of conditioned stimulus: Further evidence. Pharmacology Biochemistry and Behavior. 2011;98:155–160. doi: 10.1016/j.pbb.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Effects of Darifenacin (50 mg/kg, p.o.) on rat sleep architecture.
Supplemental Figure 2. Effects of Darifenacin (50 mg/kg, p.o.) on rat sleep architecture on first and last day of treatment.
Supplemental Figure 3. Effects of Varenicline (1mg/kg, p.o.) on rat sleep architecture on first and last day of treatment.
Supplemental Figure 4. Effects of Risperidone (3 mg/kg, i.p.) on rat sleep architecture on first and last day of treatment.
Supplemental Figure 5. Effects of SCH23390 (0.1 mg/kg, s.c.) on rat sleep architecture.
Supplemental Figure 6. Effects of PNU-109291 (15, 10 mg/kg, s.c.) on rat sleep architecture.
Supplemental Figure 7. Effects of Liraglutide (1 mg/kg, s.c.) on rat sleep architecture.
Supplemental Table 1 - Causal Gene List