Abstract
De novo proteins provide a unique opportunity for investigating the structure-function relationships of metalloproteins in a minimal, well-defined, and controlled scaffold. Herein, we describe the rational programming of function in a de novo designed di-iron carboxylate protein from the due ferri family. Originally created to catalyze O2-dependent, two-electron oxidation of hydroquinones, the protein was reprogrammed to catalyze the selective N-hydroxylation of arylamines by remodeling the substrate access cavity and introducing a critical third His ligand to the metal binding cavity. Additional second-and third-shell modifications were required to stabilize the His ligand in the core of the protein. These changes resulted in at least a 106 –fold increase in the relative rates of the two reactions. This result highlights the potential for using de novo proteins as scaffolds for future investigations of geometric and electronic factors that influence the catalytic tuning of di-iron active sites.
Keywords: de novo design, metalloproteins, di-iron proteins, four-helix bundle, oxidase
Nature uses a limited set of metal ions and amino acids to catalyze a remarkable array of chemical reactions. Not surprisingly, the identity of the metal ion is a relatively poor predictor of functionality. Iron enzymes, for example, function as oxidases,1–4 hydroxylases,5 hydrogenases,6 peroxidases,7–9 and in electron transfer.10,11 Even within the smaller subgroup of non-heme di-iron enzymes, the reactivity can vary from peroxidation to oxidation, and even to NO reduction.12 These vast differences in chemical reactivity are often orchestrated by seemingly subtle changes in the identity or position of first- and second-shell active site amino acid residues. To better understand Nature’s ability to modulate function through structural changes, we have reprogrammed the reactivity of a de novo designed di-iron carboxylate protein from hydroquinone oxidation to N-hydroxylation through a few specific structural alterations.
The due ferri family of de novo designed four-helix bundle di-iron carboxylate proteins were originally designed to mimic the properties of naturally existing dioxygen activating di-iron enzymes such as methane monooxygenase, ribronucleotide reductase, and alternative oxidase.13,14 In these small de novo proteins, the catalytic center (two iron atoms coordinated by two His and four carboxylate residues) is located in the interior of a four-helix bundle as in natural enzymes.15,16 The due ferri proteins bind divalent metal ions and catalyze the oxidation of select hydroquinones to their corresponding quinones.17,18 It is hypothesized that the oxidation reaction mediated by the due ferri proteins proceeds via a mechanistic pathway similar to that proposed for the natural di-iron carboxylate enzymes (Supplementary Fig. S1).19,20
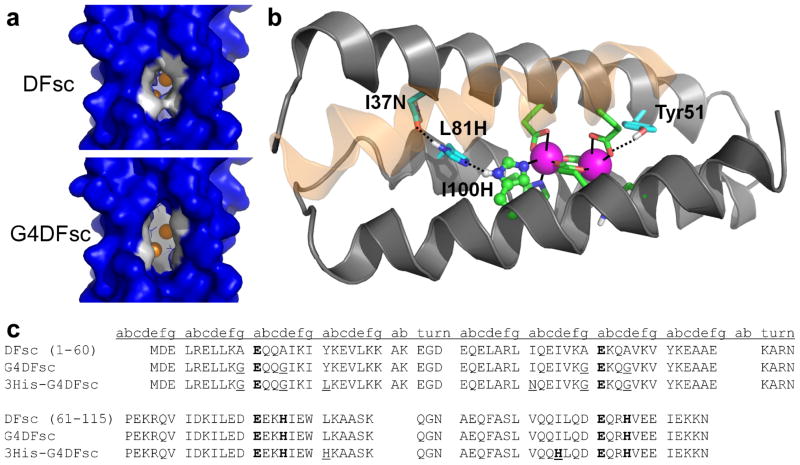
The success of earlier designs suggested that it would be feasible to not only transfer the functional active site of a natural enzyme onto a minimal scaffold, but also to alter its chemical reactivity through specific amino acid substitutions at the active site. The structural simplicity and stability of these de novo proteins make them a logical choice for such structure-function correlation studies. In the current study, we first concentrated on expansion of the substrate-access channel of a single-chain version of the due ferri protein21 to improve binding and oxidation of 4-aminophenol to the corresponding quinone immine (Fig. 1a and 1c). These changes to the substrate access channel also minimized the formation of an off-pathway tyrosinate-iron complex that occurred in previous versions of this protein.22,23
Figure 1.
Important structural features of and amino-acid sequences for the original and redesigned DFsc proteins. (a) Surface models of DFsc (top) and G4DFsc (bottom) based on the initial DFsc computational design.21 The four Ala to Gly substitutions (shown in white) significantly open the substrate access channel. (b) Structure of 3His-G2DFsc variant (PDB 2LFD) highlighting the added active-site His residue (H100) and supporting mutations (I37N and L81H). The helix closest to the viewer is shown as transparent to allow for viewing of the ligands. Note that the structure shown is for a variant with two Ala and two Gly residues along the substrate access channel which proved more stable during the extended data collection times required for the structure determination. This variant still exhibited N-oxygenase activity, but to a lesser extent than 3His-G4DFsc. (c) Amino acid sequences for DFsc, G4DFsc, and 3His-G4DFsc. Metal-binding residues are bolded and mutations introduced are underlined.
We next sought to alter the reactivity of our de novo protein from two-electron oxidation of activated quinols to N-oxygenation of anilines – a class of reactions catalyzed by AurF,24,25 the only structurally-characterized N-oxygenase that contains a di-iron catalytic center. The rarity of this type of chemical reaction in natural enzymes coupled with the potential to expand the range of possible enzymatic reactions catalyzed by de novo designed proteins made AurF an excellent target protein for testing our ability to mimic full length proteins in a minimal scaffold. A third metal-binding His residue was incorporated into the active site analogous to that found in AurF and three additional mutations were ultimately required to stabilize this buried His ligand near the protein core (Fig. 1b and 1c). Remarkably, these simple changes altered the reactivity of the resulting protein from hydroquinone oxidation to selective arylamine N-hydroxylation.
This successful tuning of chemical reactivity represents a striking example of rational reprogramming of function in a de novo metalloprotein. By comparing the spectroscopic and electronic properties of the structurally similar but functionally distinct proteins, we can begin to delineate the roles (geometric and electronic) of individual amino acids in endowing catalytic activities. These results can then be extrapolated to provide insight into the structural and electronic properties that govern functionality (e.g. oxidation versus N-hydroxylation) in natural di-iron enzymes.
Results
Design
The scaffold for our functional redesign was the single-chain due ferri protein, DFsc (Fig. 1). Unlike other members of the due ferri family (such as DFtet18 and DF317,26), DFsc mimics the asymmetry of natural proteins, allowing for single mutations to the protein sequence.21 DFsc has previously been shown to have native-like folding characteristics and is monomeric, stable, and highly water-soluble.21 Additionally, DFsc tightly binds divalent cations in the desired 2:1 stoichiometry and exhibits modest ferroxidase activity.21
Previous optimizations of other due ferri (DF) proteins established that placing smaller Gly residues along the active site channel improved the efficiency of substrate oxidation.14,17,18,27 To examine this effect in the single-chain scaffold, a quadruple Ala→Gly mutant (G4DFsc) was constructed by incorporating analogous Gly mutations (A10G, A14G, A43G, A47G) into the DFsc sequence. These changes are expected to result in a large increase in the hydration and size of the active site cavity14,17,18,27 (Fig. 1a), the functional consequences of which are explored below and contrasted with a version of the protein containing a third His ligand in the active site.
Inspection of the AurF active site (PDB 2JCD24) revealed a nearly identical structure to that of DFsc (PDB 2HZ828) with the exception of an additional metal-binding His residue in AurF. The corresponding residue in G4DFsc is Ile100 (see Supplementary Fig. S2). Using a recently developed software package (Molecular Software Library29), we investigated various mutations to stabilize the histidine at position 100. First, we built a histidine at position 100 in a similar conformation as seen in AurF and we found, as expected, a favorable ligand interaction with the metal ion cofactor (N-Fe distance, ~2.3 Å). Because AurF and G4DFsc differ significantly in sequence outside the active site, the histidine mutation resulted in a clash with Tyr18 in G4DFsc. To alleviate this steric interaction, the Tyr18 position was computationally scanned with smaller hydrophobic amino acids (Ile, Val, Leu, Phe) (see Supplementary Methods). We experimentally characterized two mutations at position 18: phenylalanine, because it is smaller, yet structurally similar to tyrosine (Y18F/I100H) and leucine, because it is calculated to have the lowest total energy (Y18L/I100H, see Supplementary Table S1). The Y18F/I100H construct exhibited weak metal binding and significant aggregation and was not pursued further. The Y18L/I100H construct did not aggregate, but also had weak metal binding, suggesting that it might be necessary to introduce a second-shell interaction to stabilize His100. Potential second-shell side-chains were examined at the neighboring, buried positions 40, 80, 81, and 96. L81H was found to be the only mutation that could provide both the proper distance and favorable energy for a second-shell interaction to His100 (see Supplementary Table S2). However, this mutation failed to restore stability and metal-binding, possibly because one of its imidazole nitrogens was buried without a compensating hydrogen bond.
To stabilize and terminate the designed polar network (Fe-H100-H81), a third-shell interaction was required to be within hydrogen-bonding distance to H81, yet partially exposed to water. The position I37 met the criteria and was computationally scanned for a potential third-shell interaction using polar amino acids (T, S, Y, H, N, Q). The I37N mutation was selected for experimental validation because it had the lowest total energy (see Supplementary Table S3). The corresponding amino acid in AurF was also an asparagine, which gave us further confidence to proceed with this mutation. The resulting hydrogen-bonded network (Fig. 1b) spans almost the entire length of the DFsc bundle, mimicking an even more elaborate hydrogen-bonding network found in AurF (see Supplementary Fig. S2). All three mutations (Y18L/L81H/I37N) proved necessary to complement the primary ligand, His100, resulting in a stable protein with the desired 2:1 metal/protein stoichiometry.
Protein Folding and Stability
Both G4DFsc and 3His-G4DFsc were well-folded and highly helical in the presence of divalent metal ions at pH 7 as assessed by circular dichroism (CD) spectroscopy (Fig. 2a). The intense negative bands at 208 nm and 222 nm in the spectrum of apo-G4DFsc indicate that the protein is also largely helical in the absence of Zn(II) or Co(II) ions. In contrast, apo-3His-G4DFsc (Fig. 2b) is fully folded only in the presence of divalent metal ions. This finding is not unexpected given that divalent metal ion binding is thermodynamically linked to protein stability in this class of proteins21 and 3His-G4DFsc incorporates numerous strongly helix destabilizing substitutions as well as three apolar-to-polar substitutions within the solvent-inaccessible protein core.
Figure 2.

Folding and metal-binding characterizations of G4DFsc and 3His-G4DFsc. (a) UV-CD spectra of metal-free, Co(II)-bound, and Zn(II)-bound G4DFsc (top) and 3His-G4DFsc (bottom). Each sample contains 20 μM protein and 100 mM metal ions (where applicable). (b) Co(II) titration monitored by visible absorption spectroscopy for 100 μM G4DFsc (top) and 100 μM 3His-G4DFsc (bottom). The red lines are linear regressions of the initial and final linear regions of the curves to determine the stoichiometric ratio of Co(II) to protein. For these individual trials, the lines intersect at 225 μM for G4DFsc and 247 μM for 3His-G4DFsc, versus the theoretical value of 200 μM. The titrations have been performed in triplicate at slightly different protein concentrations, resulting in calculated Co(II):protein stoichiometries of 1.90 ±0.33 and 2.65±0.15. Insets display a representative Abs spectrum collected in the presence of 2 equivalents of Co(II) ions. Extinction coefficients at the peak maxima are 157 M−1 cm−1 for G4DFsc and 94 M−1 cm−1 for 3His-G4DFsc per Co(II) ion. (c) Fe(II) titration monitored by near-IR CD spectroscopy for G4DFsc (top) and 3His-G4DFsc (bottom). For both proteins, the titrations saturate at 2 equivalents of Fe(II).
The structure of a 3His-G4DFsc variant containing two Ala and two Gly residues (3His- G2DFsc) along the substrate access channel was determined by NMR spectroscopy with Zn(II) serving as a non-paramagnetic Fe(II) surrogate (PDB 2LFD). These Ala residues were necessary to stabilize the protein during the extended data collection times required for structure determination but resulted in decreased rates of N-oxygenase activity. The expected tertiary structure was confirmed and retention of the overall structure following the mutations speaks to the robust nature of the starting protein (PDB 2lfd, Fig. 1b and Supplementary Fig. S3).
Metal Binding Characterization
Both G4DFsc and 3His-G4DFsc bind divalent metal ions in the expected 2:1 metal/protein ratio, as indicated by UV-visible absorption (Abs) and near-IR CD titrations with Co(II) and Fe(II), respectively (Fig. 2b, 2c and Supplementary Fig. S4). The binding constants for Fe(II) were determined to be 70 ±3 μM and 30 ±2 μM for G4DFsc and 3His-G4DFsc (Supplementary Fig. S5), similar to those observed previously for DFsc.21
While detailed characterization of the spectroscopic properties of the iron cofactors in these proteins is outside the scope of this manuscript, preliminary insight into their coordination environments is obtained from the Abs and CD data. The positions and intensities of features in the Co(II) Abs (Fig. 2b, top) and Fe(II) CD (Fig. 2c, top) spectra for G4DFsc are very similar to those observed for DFsc.21,22 The Fe(II) CD data suggest a distorted 4-coordinate/5-coordinate center while the Co(II) absorption spectrum is similar to that of 5-coordinate Co(II) models.4,29,30 3His-G4DFsc has an altered Abs spectrum (Fig. 2b, bottom) which includes a decrease in the Abs intensity to 94 M−1 cm−1 per Co(II) ion. Based on studies of cobalt complexes containing mixed N/O donor ligands,30 this decrease in intensity may be correlated with an increase in coordination number due to the additional His ligand. The NIR CD spectrum for the Fe(II) bound form of 3His-G4DFsc has significantly changed from that of G4DFsc with its features altered in sign and shifted to higher energy (Fig. 2c, bottom). These data confirm that ligation of the biferrous (and dicobalt(II)) site has been altered in the 3His-G4DFsc relative to G4DFsc.4,30,31
Ferroxidase Activity
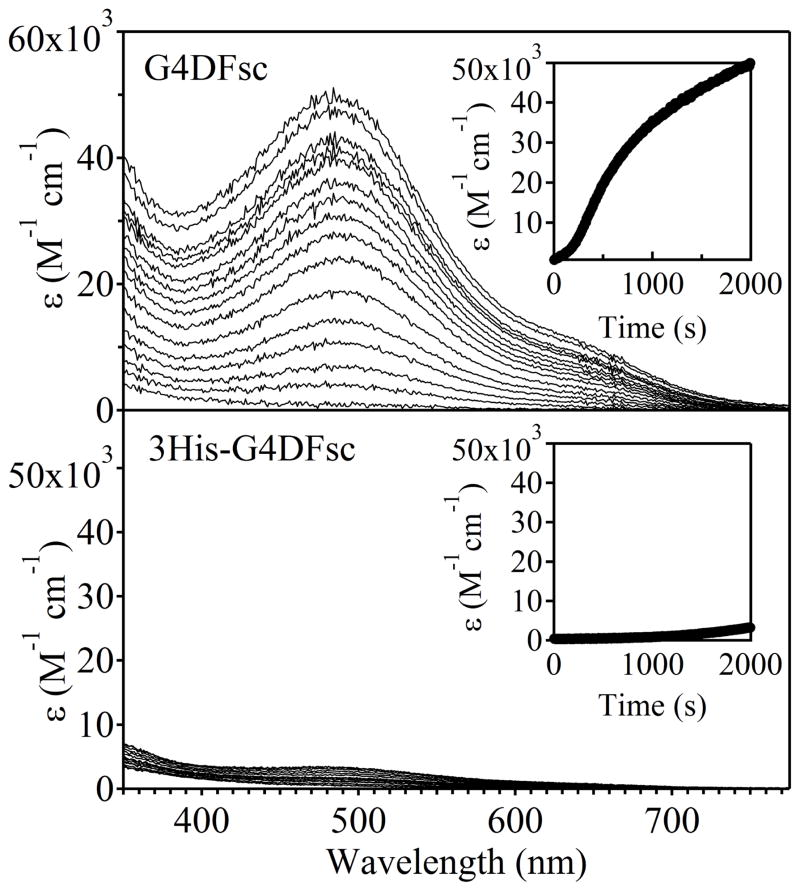
In the presence of ambient O2, both proteins rapidly oxidized Fe(II) to form a species with spectral features consistent with a μ-oxo bridged differic cluster, namely a strong oxo-to-ferric charge transfer band in the absorption spectrum near 360 nm (Fig. 3).32 For both proteins, the formation of the diferric-oxo species occurs significantly faster than in control reactions lacking protein (Supplementary Fig. S6). For G4DFsc, the feature at 360 nm maximizes at a molar absorptivity of ~2300 M−1 cm−1 per di-iron site after 2 minutes. However, a broad absorbance, suggestive of light scattering possibly due to precipitation of the iron atoms, appears over the course of one hour. In the 500–600 nm region, G4DFsc shows only a weak Abs feature (ε of 400 M−1 cm−1 at 520 nm) which implies at most only minimal formation of the previously characterized off-pathway tyrosinate-bound species.22,23
Figure 3.
Ferroxidase activity of G4DFsc (top) and 3His-G4DFsc (bottom) indicated by the formation of a strong oxo-to-ferric charge transfer band near 360 nm. The saturation of this feature occurs much faster for G4DFsc than 3His-G4DFsc (see insets), but is more intense for 3His-G4DFsc (~4600 M-1 cm-1 per di-iron site) than G4DFsc (~2300 M−1 cm−1 per di-iron site). These molar absorptivity values are in range with those observed for natural oxo-bridged di-iron proteins.
The Abs feature associated with the diferric cluster for 3His-G4DFsc is significantly more intense (~4600 M−1 cm−1 per di-iron site), and maximizes more slowly (~15 minutes) than for G4DFsc. The diferric-oxo species is also markedly more stable, with the Abs feature at 360 nm maintaining ~90% of its intensity after 3 days at room temperature. The molar absorbtivity values for both proteins are within the range observed for natural oxo-bridged diferric proteins.33–37
It is interesting to note the lack of an isosbestic point in the 3His-G4DFsc data, suggesting the presence of an intermediate in the oxidation reaction. Also, the time-dependent spectra for this protein are similar to those observed following addition of O2 to diferrous AurF (which gives a diferric-peroxo species that decays in minutes to the final oxo-bridged diferric species38). However, definitive assignment of intermediates in the oxidation of diferrous 3His-G4DFsc will require additional spectroscopic analysis.
Kinetics of Hydroquinone Amine Oxidation
G4DFsc, like other members of the DF family with unencumbered substrate access channels,17,18 successfully catalyzes the 2-electron oxidation of activated 4-aminophenol derivatives to the corresponding quinone immines (Fig. 4, top). The reaction is followed spectrophotometrically by rapid quenching of the oxidation product with m-phenylenediamine to form an aminoindoaniline dye (λmax = 486 nm at pH 7).39,40 The rate of 4-aminophenol conversion for G4DFsc is on the same order of magnitude as observed for DFsc, but reduced to near background autoxidation levels for 3His-G4DFsc (Fig. 4).
Figure 4.
Hydroquinone oxidase activity of G4DFsc (top) and 3His-G4DFsc (bottom). Only G4DFsc reacts to an appreciable extent with 4-aminophenol to form the corresponding quinone (as monitored through a coupling reaction with m-phenylenediamine that produces a species with λmax=486 nm).
Kinetics of Arylamine N-hydroxylation
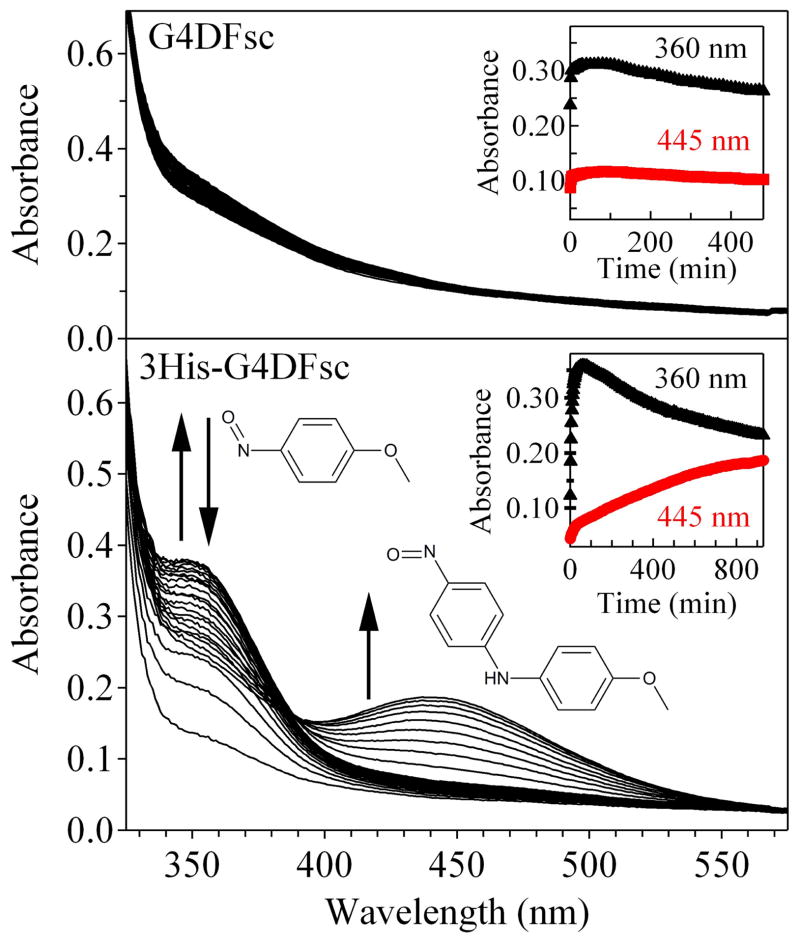
3His-G4DFsc, while not an effective oxidase against activated aminophenols, does successfully catalyze the N-hydroxylation of arylamines. The addition of p-anisidine to 3His-G4DFsc in the presence of two equivalents of Fe(II) led to the formation of a yellow-colored product mixture that exhibited two new features in the Abs spectrum. The first peak near 360 nm saturated after approximately 20 min and then disappeared over the course of several hours concomitant with the appearance of the second feature at 450 nm (Fig. 5, bottom). For the corresponding reaction with G4DFsc, no change in the Abs spectrum was observed (Fig. 5, top) and no product was indicated by HPLC analysis of the reaction mixture.
Figure 5.
The oxygenation of p-anisidine by G4DFsc (top) and 3His-G4DFsc (bottom) in the presence of two equivalents of Fe(II). No reaction is observed in the presence of G4DFsc, but in the 3His-G4DFsc reaction, an absorption feature at 360 nm is observed to grow in and then decay (inset, black triangles) followed by the appearance of a strong absorption feature at 445 nm (inset, red circles) arising from the formation of 4-nitrosodiphenylamine.
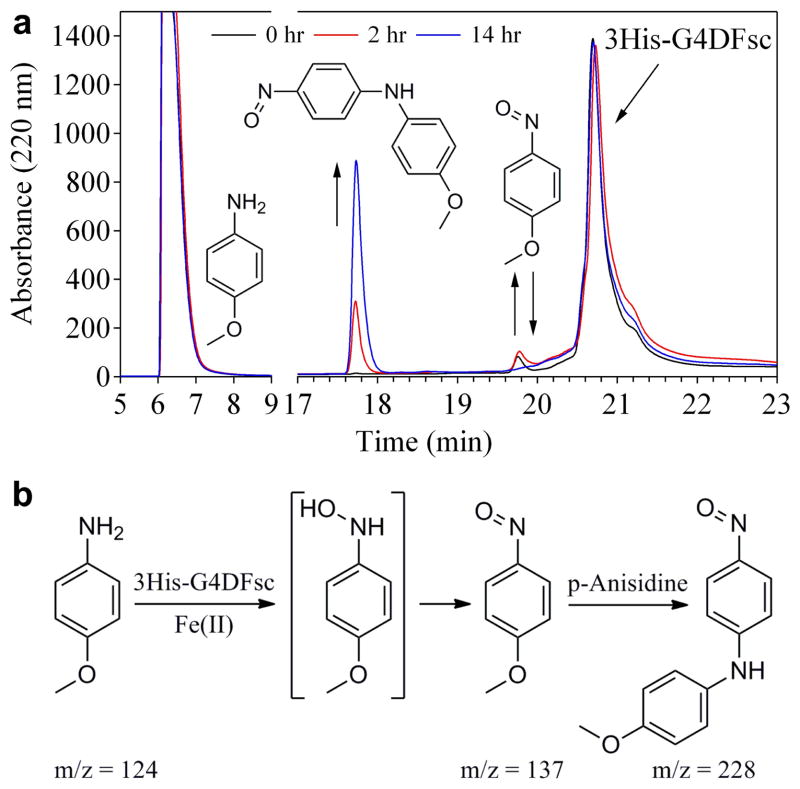
To identify the compounds being formed in the 3His-G4DFsc/p-anisidine reaction, aliquots of the mixture were analyzed via analytical HPLC at 0, 2, and 14 hours after mixing (Fig. 6a). Species giving rise to each of the observed peaks in the chromatograms were identified based on their Abs spectra, mass-to-charge ratios via LC-MS/MS, and comparison with authentic samples (obtained commercially or synthetically; see Supplementary Methods). p-anisidine, which is used in large excess and not fully consumed in the reaction, elutes at 6.2 min, and the 3His-G4DFsc protein elutes at 20.7 min. A weak feature corresponding to a p-nitrosoanisole intermediate at 19.8 minutes is observed to increase and then decrease over time, fully consistent with the Abs results described above. The peak at 17.7 minutes gains intensity over time and is identified as 4-nitrosodiphenylamine, a diaryl product which likely arises from a nucleophilic aromatic substitution reaction between the p-nitrosoanisole intermediate and unreacted p-anisidine (Fig. 6b).
Figure 6.
N-hydroxylation of p-anisidine by 3His-G4DFsc. (a) HPLC chromatograms (monitoring Abs at 220 nm) for the reaction mixture of 0.25 mM 3His-G4DFsc with 5 mM p-anisidine at 0 (black), 2 (red), and 14 (blue) hours after mixing. Products were identified by LC-MS/MS (m/z values indicated in panel b) and comparison with authentic samples. (b) Proposed reaction scheme depicting the oxidation of p-anisidine to p-nitrosoanisole and subsequent nucleophilic aromatic substitution with unreacted p-anisidine to form 4-nitrosodiphenylamine that gives rise to a visible absorption features at 445 nm.
We postulate that the p-nitrosoanisole intermediate arises from the disproportionation of N-hydroxyamino-anisole (Fig. 6b), which is the expected product of the initial hydroxylation reaction in the proposed AurF mechanism, but is not stable under these conditions.25,38,41
To support this hypothesis, a second substrate, p-aminobenzonitrile, was incubated with apo-3His-G4DFsc and two equivalents of Fe(II). Importantly, p-aminobenzonitrile lacks the electrophilic site that is responsible for the secondary product formation observed with p-anisidine. While Abs spectra collected over several hours were complicated by overlapping absorption features of the reactant and products, analysis of the product mixture by analytical HPLC revealed the formation and decay of a p-N-hydroxybenzonitrile intermediate (see Supplementary Fig. S7). This intermediate proved to be highly reactive and resulted in the formation of a wide variety of products which could not be definitively identified. A corresponding reaction mixture with G4DFsc showed no appreciable formation of any products or intermediates.
Additional substituted anilines, including o-anisidine, m-anisidine, 2,4-dimethoxyaniline, p-chloroaniline, p-toluidine, and p-aminobenzoic acid were also assayed for their reactivity with 3His-G4DFsc, with only p-chloroaniline and p-toluidine exhibiting any appreciable product formation. The inherent chemical reactivities vary widely for the reactive small molecule substrates, raising the possibility that specific substrate-protein interactions are playing a role in directing substrate recognition and reactivity. Detailed kinetic and mechanistic studies on these substrates, along with p-anisidine and p-aminobenzonitrile, are currently underway. These studies should yield insight into the factors that tune the specificity of 3His-G4DFsc.
It is important to note that no appreciable levels of substrate oxidation were observed by Fe(II) in the absence of 3His-DFsc. Similarly, no products were formed upon the addition of substrates to 3His-DFsc in the absence of Fe(II) or upon substitution of Mn(II) for Fe(II) in the reaction mixtures. In all cases, reaction rates were maximized when Fe(II) was added last to the reaction mixtures. Control reactions indicated that this rate enhancement did not result from direct Fe(II)/substrate interactions, which were equivalent to background levels. Additional control reactions were performed, including the addition of superoxide dismutase, catalase, and a radical trap (1-hydroxy-2,2,5,5-tetramethyl-3-imidazoline-3-oxide) to rule out the contribution of superoxide, peroxide, or diffusible radicals in the catalytic mechanisms (data not shown). These results imply that the substrates must undergo oxidation only when bound to the active site of 3His-G4DFsc.
Discussion
Advances in protein design research have made it possible to create functional proteins from first principles, and to explore how nature tunes active sites for specific chemical reactivities. With the goal of rationally redesigning the functionality of DFsc from hydroquinone oxidation to N-oxygenation, two new members of the due ferri family were produced: G4DFsc and 3His-G4DFsc.
In G4DFsc, four glycine residues were incorporated along the substrate access channel facilitating access of oxygen and substrates to the di-iron center. These changes also inhibited the formation of a non-productive tyrosinate-bound Fe(III) complex. Crystallographic studies of a dimeric version of the DF protein (DF3) showed that mutations equivalent to the A14G and A47G mutations introduced into DFsc led to small but significant changes in helix-helix packing that improved the solvation and binding of the dimetal cofactor.27 Similar effects might help stabilize the folding of G4DFsc into the desired structure and minimize off-pathway formation of the undesired tyrosinate ligation.
There are, however, trade-offs for these improvements. Unlike DFsc, the apo-form of G4DFsc is slightly less alpha-helical than the holo-form. Such a change is predicted as Ala residues have a greater propensity for helix formation and would contribute to stability of the helices via small increases in hydrophobic packing of the bundle. Additionally, greater active site accessibility increases exposure of the iron atoms to the aqueous solvent, rendering them prone to hydrolysis. This reduces the stability of the Fe-protein complex and leads to the formation of iron oxide precipitates.
In designing 3His-G4DFsc, the goal was to alter the functionality of a designed protein through minimal structural perturbations. Through incorporation of four mutations (one first-shell, two second-shell, and one third-shell), the due ferri protein was successfully transformed from a hydroquinone oxidase to an N-hydroxylase. These mutations resulted in the creation of a hydrogen-bonding chain through the helix bundle similar to that found in AurF (see supplementary Fig. S2). Although the second- and third-shell mutations were necessary to reduce side-chain clashes and improve the metal-binding properties of 3His-G4DFsc, they each contributed to a disruption of the hydrophobic packing of the four-helix bundle interior, significantly destabilizing the apo-protein. Fortunately, the addition of divalent cations provided enough driving force to promote folding (Fig. 2a) and once folded in the holo-form, the protein proved sufficiently robust for characterization and reactivity studies. It may even be argued that this decrease in the stability of the protein fold could facilitate substrate access to the active site. This stabilization of the apo-protein by the metal ions highlights the delicate balance between function and stability that nature manipulates so elegantly and that further studies of de novo proteins can help us to understand.
The altered chemical reactivity of 3His-G4DFsc compared to its predecessor (G4DFsc) was dramatic. 3His-G4DFsc successfully catalyzes the first of two successive two-electron oxidations of an aniline substrate, similar to the reactivity observed for AurF.42 Importantly, this reaction was selective as hydroquinone substrates readily oxidized by G4DFsc were unaltered by 3His-G4DFsc and arylamine substrates with no apparent reactivity with G4DFsc were efficiently oxidized by 3His-G4DFsc. This change in reactivity has precedent in natural di-iron proteins, with the additional His ligand in AurF leading to amine hydroxylase activity.25,38,42 The mechanism by which the reactivity switching occurs in the natural proteins has not been fully established and is hard to understand from protein structures alone given the very large number of changes in the tertiary structure and active site configuration between AurF and other di-iron carboxylate enzymes. However, our studies with a protein that is only a small fraction of the size of AurF (Supplementary Fig. S2) show that the reactivity change is likely due to the additional His ligand. It has been proposed that the additional His residue stabilizes a diferric peroxo intermediate, which reacts directly with anilines rather than breaking down to higher valent iron species as in some di-iron proteins.38,42,43 In conclusion, the differences in active site ligation between 3His-G4DFsc and G4DFsc indicate a direct relationship between active site structure and these differing reactivities. The parallels between the reactivities of these designed proteins and their natural counterparts are striking and imply that simplified, model proteins can be effective tools for studying and understanding the structure/function relationships in natural enzymes.
Methods
Design
G4DFsc was designed based on previous studies in which four Ala residues located along the active site access channel were substituted with smaller Gly residues.18 Starting from the DFsc design sequence,21,28 the following residues were selected for mutagenesis: A10G, A14G, A43G, and A47G. 3His-G4DFsc was designed based on comparisons between the original DFsc design (2HZ8)21,28 and the four-helix bundle of AurF containing the active site (2JCD)24 (see Supplementary Fig. S2), along with geometric and energetic calculations of potential second and third shell mutations (see Supplementary Methods).
Materials
All reagents were purchased commercially and used without further purification with the exception of of p-N-hydroxybenzonitrile, p-nitrosoanisole, p-nitrosobenzonitrile, and p-nitrosodiphenylamine, whose syntheses are described in the Supplementary Methods. 10 mM stock solutions of ferrous ammonium sulfate were made by dissolving the solid in 0.1% sulfuric acid and diluting to a final concentration of 10 mM as measured by a spectrophotometric assay using ferrozine. Stock solutions of the arylamines were made by dissolving the solids in neat DMSO to concentrations of 1 M.
Mutagenesis and Expression
Mutations were introduced into the pET-28a (Novagen) vector containing the DFsc gene using the quickchange mutagenesis protocol from Stratagene (Agilent) and HPLC purified primers from either MWG biotech or Applied Biosystems. Genes were sequenced (UPenn Cell Center) following TAQ amplification to confirm the incorporation of the correct mutations and then transformed and expressed in E. coli BL21(DE3) cells (Novagen). Cells were lysed using the freeze-thaw method43 and the crude lysate was collected via centrifugation at 15,000 rpm for 20 minutes. The protein was purified and reconstituted as described in the Supplementary Methods.
Spectroscopic Analyses
CD and UV-visible absorption spectra were collected according to standard procedures and are described in the Supplementary Methods.
Ferroxidase Assay
50 μM protein samples in 150 mM MOPS, 150 mM NaCl pH 7 buffer were rapidly mixed with 100 μM of ferrous ammonium sulfate in 0.1% sulfuric acid under ambient oxygen conditions. Within 30 seconds of mixing, the collection of UV-Visible absorption spectra was initialized and continued every 10 seconds for up to 30 minutes using an HP 8243 diode array spectrophotometer. Spectra were collected following centrifugation at 13k rpm after an additional hour and after 72 hours to analyze the stability of the diferric-oxo bridged cofactor.
UV-Visible Absorption Kinetic Assays
Kinetic measurements of the rates of oxidation of 4-aminophenol (4AP) by G4DFsc and 3His-G4DFsc were conducted in the presence of m-phenylenediamine (MPD), which led to the formation of an aminoindoaniline dye allowing for spectrophotometric detection at the absorption maximum of 486 nm.39,40 Specific reaction conditions can be found in the Supplementary Methods. The kinetics of arylamine N-hydroxylation by G4DFsc and 3His-G4DFsc were monitored by UV-Visible absorption spectroscopy of the products. Additional experimental conditions can be found in the Supplementary Methods.
HPLC Kinetic Assays
Pure samples of the expected products for retention time comparisons were either obtained commercially or synthesized according to literature procedures as described in the Supplementary Methods. Descriptions of the reaction mixtures and HPLC assay conditions can be found in the Supplementary Methods. Following HPLC analysis, the products were extracted from buffer with methanol and then analyzed by HPLC coupled to an electrospray ionization (ESI) mass spectrometer (Applied Biosystems QTrap 3200) in negative-ion mode under gradient conditions identical to those described in the Supplementary Methods.
Supplementary Material
Acknowledgments
This work was supported by the NIH (F32-GM808852 to A.J.R., F32-GM095242 to M.M.P, and U54 GM094597 to T.S.) and the NSF (MCB-0919027 to E.I.S.).
Footnotes
Author Contributions
J.R.C. mutated, expressed, and purified the initial G4DFsc construct. J.R.C. and A.J.R. performed characterization and kinetics studies for G4DFsc. D.W.K. performed computational modeling studies to determine feasibility of 3His-G4DFsc mutations. A.J.R. mutated, expressed, produced, and purified the 3His-G4DFsc construct, characterized the protein, and designed, conducted, and analyzed kinetic assays. S.E.B. expressed, produced, and purified the 3His-G4DFsc construct and performed Co(II) binding titrations. M.M.P. conducted HPLC analyses of the arylamine oxidation reaction products and produced labeled protein for the NMR studies. H.J. synthesized, purified, and characterized possible reaction products for retention time comparisons. Y.W. collected and, with T.G.S., analyzed the NMR structural data. R.S. performed and, with E.I.S., analyzed Fe(II) CD titrations. A.J.R. and W.F.D. wrote the manuscript in consultation with the other contributors.
References
- 1.Kolberg M, Strand KR, Graff P, Andersson KK. Structure, function, and mechanism of ribonucleotide reductases. BBA-Proteins Proteom. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Shanklin J, Guy JE, Mishra G, Lindqvist Y. Desaturases: Emerging models for understanding functional diversification of diiron-containing enzymes. J Biol Chem. 2009;284:18559–18563. doi: 10.1074/jbc.R900009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthold DA, Stenmark P. Membrane-bound diiron carboxylate proteins. Annu Rev Plant Biol. 2003;54:497–517. doi: 10.1146/annurev.arplant.54.031902.134915. [DOI] [PubMed] [Google Scholar]
- 4.Solomon EI, et al. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem Rev. 2000;100:235–350. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 5.Lippard SJ. Hydroxylation of C–H bonds at carboxylate-bridged diiron centres. Philos T R Soc A. 2005;363:861–877. doi: 10.1098/rsta.2004.1532. [DOI] [PubMed] [Google Scholar]
- 6.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 7.Iyer RB, Silaghi-Dumitrescu R, Kurtz DM, Lanzilotta WN. High-resolution crystal structures of Desulfovibrio vulgaris (Hildenborough) nigerythrin: facile, redox-dependent iron movement, domain interface variability, and peroxidase activity in the rubrerythrins. J Biol Inorg Chem. 2005;10:407–416. doi: 10.1007/s00775-005-0650-8. [DOI] [PubMed] [Google Scholar]
- 8.Jin S, Kurtz DM, Liu ZJ, Rose J, Wang BC. X-ray crystal structures of reduced rubrerythrin and its azide adduct: a structure-based mechanism for a non-heme diiron peroxidase. J Am Chem Soc. 2002;124:9845–9855. doi: 10.1021/ja026587u. [DOI] [PubMed] [Google Scholar]
- 9.Tempel W, et al. Structural genomics of Pyrococcus furiosus: X-ray crystallography reveals 3D domain swapping in rubrerythrin. Proteins. 2004;57:878–882. doi: 10.1002/prot.20280. [DOI] [PubMed] [Google Scholar]
- 10.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 11.Yu CA, Wen X, Xiao K, Xia D, Yu L. Inter- and intra-molecular electron transfer in the cytochrome bc(1) complex. Biochim Biophys Acta. 2002;1555:65–70. doi: 10.1016/s0005-2728(02)00256-6. [DOI] [PubMed] [Google Scholar]
- 12.Silaghi-Dumitrescu R, Kurtz DM, Ljungdahl LG, Lanzilotta WN. X-ray crystal structures of moorella thermoacetica FprA. Novel diiron site structure and mechanistic insights into a scavenging nitric oxide reductase. Biochemistry. 2005;44:6492–6501. doi: 10.1021/bi0473049. [DOI] [PubMed] [Google Scholar]
- 13.Calhoun JR, et al. Artificial diiron proteins: From structure to function. Peptide Sci. 2005;80:264–278. doi: 10.1002/bip.20230. [DOI] [PubMed] [Google Scholar]
- 14.Maglio O, et al. Diiron-containing metalloproteins: Developing functional models. CR Chim. 2007;10:703–720. [Google Scholar]
- 15.Andreini C, Bertini I, Cavallaro G, Najmanovich RJ, Thornton JM. Structural analysis of metal sites in proteins: Non-heme iron sites as a case study. J Mol Biol. 2009;388:356–380. doi: 10.1016/j.jmb.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 16.Lombardi A, et al. Retrostructural analysis of metalloproteins: application to the design of a minimal model for diiron proteins. Proc Natl Acad Sci USA. 2000;97:6298–6305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faiella M, et al. An artificial di-iron oxo-protein with phenol oxidase activity. Nat Chem Biol. 2009;5:882–884. doi: 10.1038/nchembio.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc Natl Acad Sci USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D, Lippard SJ. Nonheme di-iron enzymes. Comprehensive Coordination Chemistry II. 2003:309–342. [Google Scholar]
- 20.Bou-Abdallah F. The iron redox and hydrolysis chemistry of the ferritins. BBA-Gen Subjects. 2010;1800:719–731. doi: 10.1016/j.bbagen.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Calhoun JR, et al. Computational design and characterization of a monomeric helical dinuclear metalloprotein. J Mol Biol. 2003;334:1101–1115. doi: 10.1016/j.jmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Bell CB, et al. Spectroscopic definition of the biferrous and biferric sites in de novo designed four- helix bundle DFsc peptides: implications for O2 reactivity of binuclear non-heme iron enzymes. Biochemistry. 2009;48:59–73. doi: 10.1021/bi8016087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calhoun JR, et al. Oxygen reactivity of the biferrous site in the de novo designed four helix bundle peptide DFsc: Nature of the “intermediate” and reaction mechanism. J Am Chem Soc. 2008;130:9188–9189. doi: 10.1021/ja801657y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zocher G, Winkler R, Hertweck C, Schulz GE. Structure and action of the N-oxygenase AurF from Streptomyces thioluteus. J Mol Biol. 2007;373:65–74. doi: 10.1016/j.jmb.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Choi YS, Zhang H, Brunzelle JS, Nair SK, Zhao H. In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis. Proc Natl Acad Sci USA. 2008;105:6858–6863. doi: 10.1073/pnas.0712073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeGrado WF, Summa CM, Pavone V, Nastri F, Lombardi A. De novo design and structural characterization of proteins and metalloproteins. Annu Rev Biochem. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 27.Di Costanzo L, et al. Toward the de novo design of a catalytically active helix bundle: A substrate-accessible carboxylate-bridged dinuclear metal center. J Am Chem Soc. 2001;123:12749–12757. doi: 10.1021/ja010506x. [DOI] [PubMed] [Google Scholar]
- 28.Calhoun JR, et al. Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement. Structure. 2008;16:210–215. doi: 10.1016/j.str.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulp DW, et al. Structural informatics, modeling and design using a new open-source Molecular Software Library (MSL) J Comp Chem. 2012;33:1645–1661. doi: 10.1002/jcc.22968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertini L. C High spin cobalt(II) as a probe for the investigation of metalloproteins. Advances in Inorganic Biochemistry. 1984:71–111. [PubMed] [Google Scholar]
- 31.Solomon EI, Pavel EG, Loeb KE, Campochiaro C. Magnetic circular dichroism spectroscopy as a probe of the geometric and electronic structure of non-heme ferrous enzymes. Coordin Chem Rev. 1995;144:369–460. [Google Scholar]
- 32.Brown CA, Remar GJ, Musselman RL, Solomon EI. Spectroscopic and electronic structure studies of met-hemerythrin model complexes: A description of the ferric-oxo dimer bond. Inorg Chem. 1995;34:688–717. [Google Scholar]
- 33.Yang X, Chen-Barrett Y, Arosio P, Chasteen ND. Reaction paths of iron oxidation and hydrolysis in horse spleen and recombinant human ferritins. Biochemistry. 1998;37:9743–9750. doi: 10.1021/bi973128a. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Le Brun NE, Thomson AJ, Moore GR, Chasteen ND. The iron oxidation and hydrolysis chemistry of Escherichia coli bacterioferritin. Biochemistry. 2000;39:4915–4923. doi: 10.1021/bi992631f. [DOI] [PubMed] [Google Scholar]
- 35.Fox BG, Shanklin J, Ai J, Loehr TM, Sanders-Loehr J. Resonance raman evidence for an Fe-O-Fe center in stearoyl-ACP desaturase. Primary sequence identity with other diiron-oxo proteins. Biochemistry. 1994;33:12776–12786. doi: 10.1021/bi00209a008. [DOI] [PubMed] [Google Scholar]
- 36.Bollinger J, et al. Mechanism of assembly of the tyrosyl radical-dinuclear iron cluster cofactor of ribonucleotide reductase. Science. 1991;253:292–298. doi: 10.1126/science.1650033. [DOI] [PubMed] [Google Scholar]
- 37.Vincent JB, Olivier-Lilley GL, Averill BA. Proteins containing oxo-bridged dinuclear iron centers: a bioinorganic perspective. Chem Rev. 1990;90:1447–1467. [Google Scholar]
- 38.Korboukh VK, Li N, Barr EW, Bollinger JM, Krebs C. A long-lived, substrate-hydroxylating peroxodiiron(III/III) intermediate in the amine oxygenase, AurF, from Streptomyces thioluteus. J Am Chem Soc. 2009;131:13608–13609. doi: 10.1021/ja9064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corbett JF. Benzoquinone imines. Part V. Mechanism and kinetics of the reaction of p-benzoquinone monoimines with m-phenylenediamines. J Chem Soc, B. 1969:823–826. [Google Scholar]
- 40.Corbett JF, Gamson EP. Benzoquinone imines. Part XI. Mechanism and kinetics of the reaction of p-benzoquinone di-imines with aniline and its derivatives. J Chem Soc, Perkin Trans. 1972;2:1531–1537. [Google Scholar]
- 41.Simurdiak M, Lee J, Zhao H. A new class of arylamine oxygenases: evidence that p-aminobenzoate N-oxygenase (AurF) is a di-iron enzyme and further mechanistic studies. Chembiochem. 2006;7:1169–1172. doi: 10.1002/cbic.200600136. [DOI] [PubMed] [Google Scholar]
- 42.Li N, Korboukh VK, Krebs C, Bollinger JM. Four-electron oxidation of p-hydroxylaminobenzoate to p-nitrobenzoate by a peroxodiferric complex in AurF from Streptomyces thioluteus. Proceedings of the National Academy of Sciences. 2010;107:15722–15727. doi: 10.1073/pnas.1002785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fries A, Bretschneider T, Winkler R, Hertweck C. A ribonucleotide reductase-like electron transfer system in the nitroaryl-forming N-oxygenase AurF. ChemBioChem. 2011;12:1832–1835. doi: 10.1002/cbic.201100138. [DOI] [PubMed] [Google Scholar]
- 44.Johnson BH, Hecht MH. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Nat Biotech. 1994;12:1357–1360. doi: 10.1038/nbt1294-1357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.