Abstract
Glutaredoxin (Grx)-catalyzed deglutathionylation of protein–glutathione mixed disulfides (protein-SSG) serves important roles in redox homeostasis and signal transduction, regulating diverse physiological and pathophysiological events. Mammalian cells have two Grx isoforms: Grx1, localized to the cytosol and mitochondrial intermembrane space, and Grx2, localized primarily to the mitochondrial matrix [Pai, H. V., et al. (2007) Antioxid. Redox Signaling 9, 2027–2033]. The catalytic behavior of Grx1 has been characterized extensively, whereas Grx2 catalysis is less well understood. We observed that human Grx1 and Grx2 exhibit key catalytic similarities, including selectivity for protein-SSG substrates and a nucleophilic, double-displacement, monothiol mechanism exhibiting a strong commitment to catalysis. A key distinction between Grx1- and Grx2-mediated deglutathionylation is decreased catalytic efficiency (kcat/KM) of Grx2 for protein deglutathionylation (due primarily to a decreased kcat), reflecting a higher pKa of its catalytic cysteine, as well as a decreased enhancement of nucleophilicity of the second substrate, GSH. As documented previously for hGrx1 [Starke, D. W., et al. (2003) J. Biol. Chem. 278, 14607–14613], hGrx2 catalyzes glutathione-thiyl radical (GS•) scavenging, and it also mediates GS transfer (protein S-glutathionylation) reactions, where GS• serves as a superior glutathionyl donor substrate for formation of GAPDH-SSG, compared to GSNO and GSSG. In contrast to its lower kcat for deglutathionylation reactions, Grx2 promotes GS-transfer to the model protein substrate GAPDH at rates equivalent to those of Grx1. Estimation of Grx1 and Grx2 concentrations within mitochondria predicts comparable deglutathionylation activities within the mitochondrial subcompartments, suggesting localized regulatory functions for both isozymes.
Protein glutathionylation, or mixed disulfide formation between a protein cysteine moiety and the cysteine moiety of glutathione (GSH),1 is a post-translational modification with important roles in cellular signal transduction and sulfhydryl homeostasis within mammalian cells (1–3). During overt oxidative stress, glutathionylation is thought to protect critical sulfhydryl groups from irreversible oxidation to sulfinic and sulfonic acids (2, 4, 5). Under nonstressed conditions, reversible glutathionylation modulates protein activity, through either activation or inactivation, thereby regulating processes such as protein synthesis (6, 7), calcium homeostasis (8), cell growth (9), and transcription factor activity (10, 11). Although mechanisms for protein glutathionylation are not fully understood (3), multiple studies indicate that within mammalian cells, protein deglutathionylation is catalyzed principally by the thiol-disulfide oxidoreductase enzyme glutaredoxin (Grx, also known as thioltransferase) (12–14).
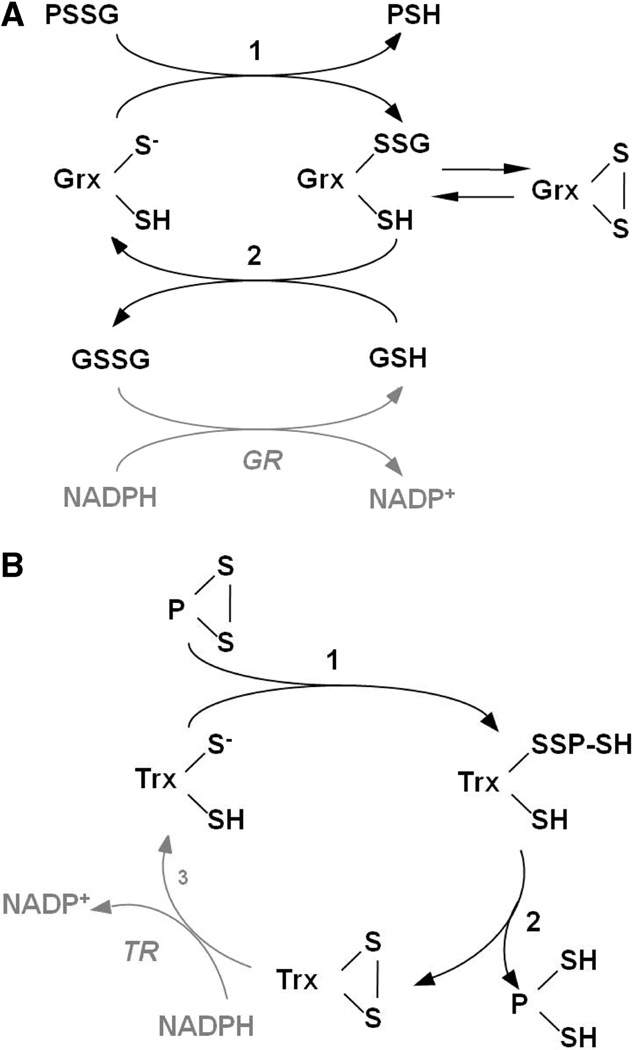
Mammalian glutaredoxin (Grx1), localized to the cytosol and mitochondrial intermembrane space (15), is well-characterized as a specific and efficient catalyst of deglutathionylation of protein–SSG, proceeding via a nucleophilic, double-displacement mechanism in which the N-terminal Cys (Cys22) of the CPY(F)C active site attacks the glutathionyl sulfur of the protein–SSG disulfide bond, releasing protein–SH and forming a Grx1–SSG covalent intermediate [Scheme 1 (16)]. In the second, rate-determining step of the reaction, GSH serves as the nucleophile in the thiol–disulfide exchange with Grx1–SSG, forming Grx1–S− and GSSG (16,17). Human Grx1 is distinguished from other thiol–disulfide oxidoreductase enzymes, such as thioredoxin (Trx), in this monothiol mechanism; i.e., only the N-terminal active site Cys is required for catalysis (18). In fact, the presence of the C-terminal active site Cys limits kcat by promoting a side reaction whereby the Grx1–SSG intermediate is converted to an intramolecular disulfide (Scheme 1) that can be recruited back into the catalytic cycle by reaction with GSH. Rate enhancement of the Grx1-catalyzed reaction over the nonenzymatic reaction is attributed primarily to the superior ability of the active site thiolate to act as a leaving group in the second step of the reaction (17), due to its unusually low pKa of 3.5 (19, 20). In addition, Grx1 appears to enhance the nucleophilicity of GSH for the second step of the reaction (17) (Scheme 1).
Scheme 1.
(A) Catalytic Mechanism of Protein Deglutathionylation by Mammalian Grx1 (16, 18) and Grx2 and (B) Catalytic Mechanism of Intramolecular Disulfide Reduction by Mammalian Trx (5, 57)a
a In the first step of panel A, the catalytic cysteine thiolate of Grx attacks the glutathionyl sulfur of the protein–SSG mixed disulfide, releasing the first product, protein–SH. In the second step of the reaction, GSH attacks the enzyme–SSG intermediate, restoring the reduced enzyme and releasing GSSG. Alternatively, the cysteine adjacent to the active site may attack the Grx–SSG intermediate, forming an intramolecular disulfide and releasing GSH. The intramolecular disulfide may be recruited back into the catalytic cycle by reaction with GSH, re-forming Grx–SSG, and proceeding with step 2. Reduction of GSSG is catalyzed by GSSG reductase, coupled to oxidation of NADPH. PSSG, protein–GSH mixed disulfide. In the first step of panel B, the acidic cysteine thiolate of Trx attacks an intramolecular protein disulfide, forming a transient HS–Trx–SS–protein–SH intermediate in which the second active site cysteine of Trx attacks the intermolecular disulfide and displaces protein–(SH)2, forming the Trx–(S)2 intramolecular disulfide. Trx disulfide is then reduced by Trx reductase (TR), coupled to oxidation of NADPH. P, protein.
A second mammalian glutaredoxin isoform (Grx2) was discovered more recently by homology search analysis of human and rodent EST libraries (21, 22). Although it exhibits <35% sequence homology with Grx1, Grx2 shares distinguishing structural features, including a thioredoxin fold, a CXXC active site motif (CSYC in Grx2 vs CPYC in Grx1), and conserved amino acids implicated in the stabilization of the adducted glutathionyl moiety (18, 21–23). To date, three subforms of Grx2 (Grx2a,b,c) have been described that appear to result from alternative splicing of the gene’s first exon (21, 22, 24), resulting in distinct N-terminal sequences. The N-terminus of Grx2a contains a mitochondrial localization sequence sufficient to target to the mitochondria either overexpressed Grx2 or a GFP fusion protein which contains the localization sequence (21). Moreover, studies of isolated rat mitochondria have documented that Grx2 is localized exclusively in the matrix, whereas Grx1 is found only in the intermembrane space of mitochondria (15). Grx2b and Grx2c share overlapping N-terminal regions transcribed from an alternatively spliced region of exon 1 (24). The expression pattern of Grx2b and Grx2c appears to be more restricted than that of Grx2a, being detected primarily in testes, immortalized cell lines, and tumors (24). In contrast to that of Grx2a, the subcellular localization of Grx2b and Grx2c remains unresolved, with reports of nuclear, perinuclear, and diffuse cytosolic and nuclear localizations (22, 24). Within mitochondria, it has been reported that Grx2a exists in an inactive dimer bridged by an Fe2S2 cluster (25), and Grx2c is also reported to form the Fe–S cluster in a reconstitution assay in vitro (24). The dimeric Grx2 complex is reported to dissociate in the presence of certain oxidants and reductants in vitro (25), and under conditions that promote S-nitrosylation (26); however, mechanisms of regulation of reversible cluster formation in vivo are unknown.
To date, several studies of Grx2 have emphasized apparent differences in catalytic activity in comparison to Grx1 (22, 27, 28). In contrast, this report describes identical behavior for Grx2 in many of the characteristic features of the catalytic mechanism delineated previously for Grx1-mediated deglutathionylation. In particular, Grx2 displays specificity for glutathione-containing mixed disulfides, and a double displacement kinetic mechanism with strong commitment to catalysis that precludes reversible binding of substrates. In addition, we observed that Grx2, like Grx1, exhibits glutathionyl-thiyl radical (GS•)-scavenging and GS•-mediated protein S-glutathionylation activities, suggesting specific roles in redox and sulfhydryl homeostasis within the mitochondrial matrix under conditions of oxidative stress.
EXPERIMENTAL PROCEDURES
Materials
Cysteinyl–glutathione mixed disulfide was purchased from Toronto Research Chemicals, and NADPH was from Roche. Plasmid DNA (pET-24d, Novagen) encoding human Grx1 was prepared as described by Chrestensen et al. (29). Plasmids encoding mature (i.e., exons 2–4, amino acids 41–164) human or mouse Grx2a were kindly provided by V. Gladyshev (University of Nebraska, Lincoln, NE). All other routine chemicals were reagent grade or better and obtained from Sigma.
(i) Enzymes
Recombinant human Grx1 (hGrx1) and Grx2 (hGrx2) and mouse Grx2 (mGrx2) were purified as described previously (30) from their respective plasmid DNA constructs (see above). Purified hGrx2 C40S protein was kindly provided by C. Johansson and A. Holmgren (Karolinska Institute, Stockholm, Sweden). Glutathione disulfide reductase from baker’s yeast (~170 units/mg), thioredoxin reductase from rat liver (~170 units/mg), and glutathione peroxidase from bovine erythrocytes (680 units/mg) were purchased from Sigma. Horseradish peroxidase (~225 units/mg) was obtained from Roche.
(ii) Synthesis of the [3H]BSA–SS–Glutathione Substrate
Synthesis was performed as described in ref 12 with minor modifications. In brief, a 1.4-fold molar excess of N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP) (dissolved in dry dimethylformamide) was added to 3 mM S-carboxymethyl-BSA slowly with constant stirring at room temperature in phosphate-buffered saline (PBS) [137 mM NaCl, 2.7 mM KCl, 4.3 mM sodium phosphate, and 1.4 mM potassium phosphate (pH 7.4)] over 1 h. The BSA derivative was then treated with 5 mM [3H]GSH (~1 nCi/nmol) for 1 h at room temperature. Excess [3H]GSH and SPDP were separated from the [3H]BSA-SSG by Sephadex G-25 column chromatography (pre-equilibrated with PBS). The protein fractions (with radiolabel) were pooled and concentrated to ~1 mM with respect to [3H]GS equivalents. [35S]BSA–SSG was prepared by a completely analogous procedure, except that [35S]GSH was substituted for [3H]GSH. Both radiolabeled substrates reacted indistinguishably in deglutathionylation assays.
(iii) Synthesis of the BSA–SS–[14C]Cysteine Substrate
BSA–SS–[14C]cysteine (BSA-SSC) was prepared as described by Yang et al. (18).
General Methods
Deionized, distilled water used in all enzyme assays was pretreated with 5 g/L Chelex resin to chelate metal cations.
Specific Methods
(i) Spectrophotometric Assay of Grx Activity
The coupled spectrophotometric assay for Grx activity was performed as described previously (17, 18, 30). Reaction mixtures containing Na/K phosphate buffer (0.1 mM, pH 7.5), NADPH (0.2 mM), GSSG reductase (2 units/ mL), GSH (0.02–3 mM), and Grx (0–135 nM) were prepared in microwells of a 96-well plate (final volume of 200 µL) and incubated for 5 min at 30 °C. For CSSG and BSA–SSG, reaction rates were determined within a range of concentrations from 0.25 to 4 times the apparent KM, while for GSH, the high nonenzymatic rate observed at high substrate concentrations prevented determinations at GSH concentrations greater than ~2KM. For the cysteine–SS–glutathione substrate (CSSG), reactions were initiated by addition of CSSG (0.002–0.5 mM); for the BSA–SS–glutathione substrate (BSA–SSG), initial mixtures contained BSA–SSG (5–40 µM), and deglutathionylation reactions were initiated by addition of GSH (0.1–1 mM). NADPH oxidation (equivalent to GSSG formation) was monitored according to the decreasing absorbance at 340 nm over 5 min (during which reaction rates remained linear with time) using a ThermoMax microplate reader (Molecular Devices). Non-enzymatic rates were subtracted from total rates to give enzymatic rates, and the number of nanomoles of NADPH oxidized per minute per nanomole of enzyme (i.e., turnover, min−1) was calculated using the standard extinction coefficient of NADPH (ε = 6.2 mM−1 cm−1), along with factors correcting for path length and actual NADPH absorptivity in the plate reader. Specific activity determinations were performed in the presence of 0.1 mM CSSG and 0.5 mM GSH; units of activity per milliliter were divided by the Grx protein concentration to yield units per milligram. For the BSA–SS–glutathione substrate (BSA–SSG), experiments were conducted as described except that BSA–SSG was substituted for CSSG. All velocity versus substrate plots were fit to the Michaelis–Menten equation, and apparent kinetic constants for CSSG, BSA–SSG, and GSH were calculated accordingly. Standard error values for kinetic constants were calculated using Prism (GraphPad Software). Aliquots of standard hGrx1 protein were assayed under standard conditions periodically for monitoring the consistency of assay solutions, temperature, and plate reader conditions during the accumulation of other kinetic data. When small deviations from the established values for standard Grx1 activity were noted, the companion data were normalized accordingly.
(ii) pH Dependence of Grx2 Inactivation by Iodoacetamide (IAM)
The pH dependence of inactivation of Grx2 by IAM was determined out as described previously (19, 30) with minor modifications. Grx2 (3 µM) was preincubated in the following buffers with or without IAM (0.3 mM) for 3 min: sodium citrate at pH 3.5; sodium acetate at pH 3.5, 4.0, 4.5, and 5.0; and MES at pH 5.0, 5.5, and 6.0. All buffers were at a concentration of 10 mM, and the ionic strength was adjusted to 0.5 M in all cases by addition of the appropriate amount of NaCl or KCl. Following preincubation, Grx activity was determined by adding an aliquot (4 µL) of the preincubation mixture to the spectrophotometric assay system described above. Percent activity remaining was calculated by dividing rates of deglutathionylation after preincubation with IAM by rates in the absence of IAM and multiplying the result by 100%. Percent activity was plotted versus pH, and an adaptation of the Henderson–Hasselbalch equation was used to solve for pKa:
(iii) Relative Rates of Dethiolation of Radiolabeled BSA Mixed Disulfide Substrates by the Grx Isoform
The standard radiolabel assay for Grx activity was performed as described previously (17, 18, 31). Reaction mixtures containing Na/K phosphate buffer (0.1 M, pH 7.5), GSH (0.5 mM), and Grx2 (0–110 nM) were prepared in Eppendorf tubes and placed in a 37 °C water bath for 5 min. Following preincubation, reactions were initiated by addition of [3H]BSA–SSG or [14C]BSA–SS–Cys (0.1 mM final concentration). Aliquots of the reaction mixture were removed at 0.5, 1, 2, and 3 min, and reactions were quenched by addition to an equal volume of 20% ice-cold TCA. Precipitated protein was sedimented by centrifugation at 13000g for 5 min at 4 °C, and supernatants were analyzed for non-protein-associated radioactivity (i.e., radiolabeled GSSG or GSS–Cys). Background radioactivity contributed by the nonprecipitated BSA–SSR substrate (<0.2% total) was measured independently and subtracted from each time point. Enzyme-mediated dethiolation rates were determined by calculating the difference in time-dependent radiolabel release in the presence and absence of Grx. To measure the pH dependence of hGRx2-catalyzed deglutathionylation (pH–rate profile), assays were performed as described above, except that the concentration of GSH was reduced to 0.25 mM to minimize nonenzymatic rates of deglutathionylation. Also, Na/K phosphate buffer was replaced with MES (pH 5.5–6.5), HEPES (pH 7–8), HEPPSO (pH 8.5), or glycine (pH 9–10.5). All buffers were at 0.1 M, and the ionic strength was adjusted to 0.3 M using NaCl. Rates of hGrx2-dependent deglutathionylation (expressed as turnover, min−1) were expressed as % maximal activity, and plots of activity versus pH were fit to a derivation of the Henderson–Hasselbalch equation:
For comparison of deglutathionylation rate constants in the presence of alternative RSH substrates, i.e., GSH or cysteinylglycine, the concentration of RSH was 0.25 mM, and reactions were carried out in AMPSO buffer (0.1 M, pH 9.5). Second-order rate constants for formation of the respective R-SSG products (GSSG or Gly–Cys–SSG) were calculated by dividing the concentration of R–SSG produced per minute by the concentration of BSA–SSG substrate for the uncatalyzed rate, or by dividing by the concentration of Grx for the catalyzed reaction, as described previously (17). In both cases, the quotients were then divided by the concentration of RSH, and the second-order rate constants (knonenz and kGrx) are expressed in units of M−1 min−1.
(iv) Comparison of Grx-Mediated Rates of Deglutathionylation of CSSG in the Presence of TR or GSH and GR
hGrx activity was determined by the spectrophotometric assay described above with minor changes in component concentrations. All assays contained Na/K phosphate buffer (0.1 M, pH 7.5) and NADPH (0.2 mM),and were initiated by addition of CSSG (0.5 mM final concentration). To measure catalytic rates in the presence of TR, the hGrx1 or -2 concentration was 3.7 µM and the TR concentration 230 nM. TR activity was confirmed independently by assessing NADPH-dependent reduction of DTNB in Na/K phosphate buffer (0.1 M, pH 7), and this activity matched the value reported by the supplier (reported, 29.4 units/mL; observed, 29.3 ± 1.9 units/mL; 1 unit = Δ1 AU412/min). For measurement of turnover under low-GSH conditions, hGrx1 concentration was adjusted to 0.37 µM (to achieve a linear dependence on enzyme concentration) the Grx2 concentration was 3.7 µM, the GSH concentration was 0.1 mM, and the GR concentration was 0.01 µM. Concentrations of GSH and GR were deliberately chosen to represent one-tenth of estimated intracellular concentrations (as described in ref 32).
(v) Glutathionyl–Thiyl Radical (GS•) Scavenging Activity of hGrx2
GS• scavenging activity was measured as described by Starke et al. (33). Reaction mixtures were prepared as described for the standard spectrophotometric assay [0.1 M Na/K phosphate buffer (pH 7.5), 0.2 mM NADPH, 0.5 mM GSH, 2 units/mL GSSG reductase, and 0.01–1 µM hGrx1 or 0.03–0.1 µM hGrx2] and preincubated for 5 min at 30 °C. Then a premade complex containing FeCl2 (0.5 mM) and ADP (2.5 mM) was added to each, and the reactions were initiated by addition of H2O2 (50 µM). Reactions were monitored according to the decrease in A340 over 5 min (within the linear range of time dependence). Rates of GSSG formation were determined by measuring the time-dependent decrease in A340 (i.e., NADPH oxidation by GSSG reductase), and enzyme-catalyzed scavenging was calculated by determining the difference in reaction rates in the presence and absence of Grx and expressed as units per milliliter in the reaction mixture (1 unit = 1 µmol of NADPH oxidized/min).
(vi) GSH Peroxidase Activity of hGrx Isoforms
GSH-dependent peroxidase activity was measured via a coupled, spectrophotometric assay identical to that of CSSG deglutathionylation, except that reactions were initiated by addition of H2O2 (500 µM final concentration). Enzyme concentrations were as follows: 1 µM hGrx1, 1 µM hGrx2, and 5 nM bovine glutathione peroxidase (GPx).
(vii) Determination of Grx-Mediated Protein Glutathionylation in the Presence of GS•, GSNO, and GSSG
To ensure that the GAPDH used as a substrate for glutathionylation was fully reduced, DTT (5 mM final concentration) was added to stock solutions [~5 mg/mL GAPDH in 0.1 M Na/K phosphate buffer (pH 7.5)] followed by incubation at 4 °C for 1 h. To remove DTT, the mixture was passed over a DG-10 size exclusion column pre-equilibrated with Na/K phosphate buffer. Protein-containing fractions (identified by the absorbance at 280 nm) were pooled and concentrated to 150–450 µM for use in glutathionylation assays. Glutathionyl radicals were generated by horseradish peroxidase utilizing H2O2 and GSH according to an adaptation of the method of Harman et al. (34), who confirmed formation of GS• by EPR spectroscopy. Briefly, [35S]GSH (0.5 mM) and horseradish peroxidase (HRP, 0.2 mg/mL) were included in reaction mixtures (see below), and GS• formation was initiated by addition of H2O2 (50 µM) as described by Starke et al. (33). The GS• concentration achieved by this radical-generating system (i.e., HRP, GSH, and H2O2) was estimated previously (33) to be ≤10 µM based on the amount of GSSG that accumulated (GSSG reductase assay); formation of GS-radicals was confirmed by EPR spectroscopy (data not shown). For GSNO and GSSG, the radiolabeled GSH derivatives were prepared in advance and used to initiate each reaction. S-Nitrosoglutathione ([35S]GSNO) was prepared as described by Starke et al. (33) by combining equimolar amounts of HCl, NaNO2, and [35S]GSH and incubating the mixture at room temperature for 15 min. Essentially stoichiometric conversion of GSH to GSNO was verified by diluting the stock solution into 15 mM HEPES (pH 7.6), conditions under which ε338 was previously reported for GSNO [980 M−1 cm−1 (35)]. GSH disulfide ([35S]GSSG) was prepared by incubating GSSG with trace amounts of [35S]GSH for 30 min at room temperature. For kinetic assays, reaction mixtures contained Na/K phosphate buffer (0.1 M, pH 7.5), rabbit muscle GAPDH (25 µM), Grx (0.02–0.25 µM), and S-carboxymethyl BSA (7 mg/mL) as a coprecipitant to facilitate precipitation of the radiolabeled products. For GS•-mediated glutathionylation, HRP (0.2 mg/mL) and GSH (0.5 mM) were also included. Reactions were initiated with H2O2 (0.05 mM) (for GS•-mediated glutathionylation), [35S]GSNO (0.05–0.5 mM), or [35S]GSSG (0.1–0.3 mM). All reactions took place at room temperature and were quenched by addition of an equal volume of 20% ice-cold TCA. Precipitated proteins were sedimented by centrifugation at 10000g for 5 min at 4 °C, washed, and analyzed for bound radioactivity. Grx-mediated GAPDH-SSG formation was calculated by comparing the bound radioactivity in the presence and absence of enzyme. For determination of the rate of turnover, rates of protein glutathionylation with and without Grx were determined within the initial, linear phase of the reaction (i.e., first 30 s), by dividing the number of nanomoles of protein–SSG formed per second by the number of nanomoles of Grx in each reaction (i.e., turnover number, s−1). All reported turnover values were determined under conditions in which reaction rates exhibited a linear dependence on Grx concentration.
(viii) Estimation of Concentrations of Grx Isoforms in Mitochondrial Subcompartments
The Grx content (nanograms per milligram of total mitochondrial protein) determined previously (15) was divided by the estimated volume of the mitochondrial space per milligram of mitochondrial protein [i.e., 1 µL/mg for the matrix (36) or 1.5 µL/mg for the intermembrane space (37)] and then by the molecular mass of hGrx1 (11.7 kDa) (38) in the case of the intermembrane space or mature hGrx2a [15 kDa (24)] for the matrix. The Grx concentration in milligrams per milliliter was used to calculate the total potential deglutathionylation activity by dividing by specific activity [units per mg (Table 1)].
Table 1.
Specific Activities of Grx Isoforms for Model Substrate Cysteine–SSG (CSSG)a
| Grx isoform | specific activity (units/mg) | ratio (Grx1:Grx2) |
|---|---|---|
| hGrx1 | 106.8 ± 2.8 | – |
| hGrx2 | 11.2 ± 0.05 | 9.5 |
| mGrx2 | 12.6 ± 0.9 | 8.5 |
| hGrx2 (C40S) | 20.9 ± 1.4 | – |
Rates of CSSG deglutathionylation by recombinant human Grx1 (hGrx1) and truncated human and mouse Grx2 (hGrx2 and mGrx2) were determined using the spectrophotometric coupled assay described previously (12,17,30). Reaction mixtures contained sodium/potassium phosphate buffer (0.1 M, pH 7.4), NADPH (0.2 mM), GSH (0.5 mM), GSSG reductase (2 units/mL), and Grx (15 nM Grx1 or 64–75 nM Grx2), and reactions were initiated by addition of CSSG (0.1 mM final concentration) and monitored over 5 min. Rates of deglutathionylation were determined by measuring the rate of NADPH-dependent reduction of the reaction product GSSG and subtracting the nonenzymatic rate. Data are expressed as means ± the standard error (n = 4–12).
(ix) Calculation of R-Helix 2 Dipole Moments of hGrx1 and hGrx2
Partial PDB files of each hGrx isoform containing both the catalytic cysteine and helix 2 [hGrx1, PDB entry 1JHB (23), C22–S33; hGrx2, PDB entry 2FLS (39), C37–M50] were submitted to the Protein Dipole Moments Server (40) for calculation of the helix 2 dipole moment and helix–thiolate angle (i.e., the angle θ formed by the intersection of the helix 2 axis with a line projecting from the base of the helix to the catalytic cysteine). By representing the component of the helix dipole directed toward the cysteine thiolate as the base of a right triangle, with the helix axis (full dipole vector) as the hypotenuse, the magnitude of this component is determined by multiplying the value of the full dipole by cos θ (adjacent = hypotenuse × cos θ). For hGrx1, we submitted four of 20 total NMR structural solutions [structures 1, 6, 16, and 20 (23)] shown in a separate study (30) to predict a C22 pKa close to the empirical value of 3.5 (19, 20). The structure of hGrx2, obtained by X-ray crystallography, contained an associated GSH molecule, which was not included in our calculations.
RESULTS
hGrx2 Is Selective for Glutathione-Containing Mixed Disulfides
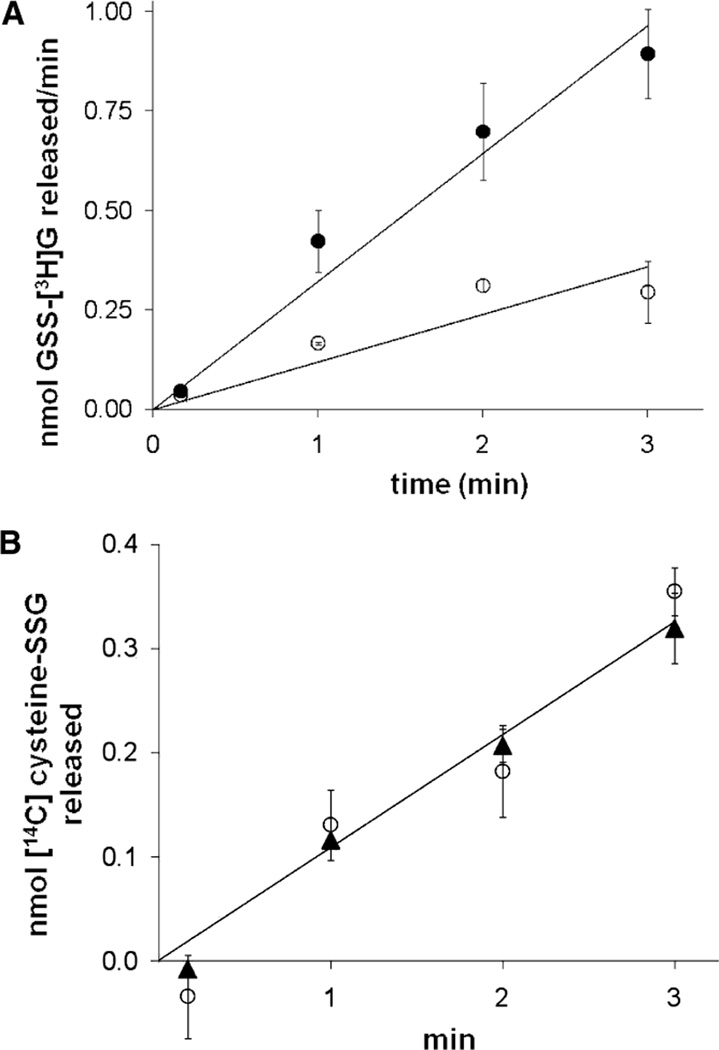
To test whether Grx2, like Grx1, is selective for glutathione-containing protein mixed disulfides (i.e., protein–SSG), we compared the rates of GSH-dependent reduction of two mixed disulfides of bovine serum albumin (BSA) in the absence and presence of Grx2 or Grx1. The first, BSA–SS–glutathione (BSA–SSG), was specifically prepared to contain a single disulfide-adducted glutathionyl moiety (see Experimental Procedures), and this has been our prototype protein–SSG substrate for kinetic analysis of glutaredoxin (16–18). The second, BSA–SS–cysteine (BSA–SSC), has the same disulfide bond to the BSA protein (but not the γ-glutamyl and glycyl moieties of GSH), yet it does not serve as a substrate for Grx1 (16, 18). Like hGrx1, hGrx2 (Figure 1) does catalyze GSH-dependent dethiolation of BSA–SSG but does not catalyze dethiolation of BSA–SSC even when a 10-fold excess of enzyme is added, documenting high selectivity for glutathione-containing mixed disulfide substrates. Notably, the rate of turnover of BSA–SSG by Grx2 (39 ± 5 min−1) was ~10-fold lower than the rate for hGrx1 (457 ± 91 min−1), analogous to the 10-fold lower activity of hGrx2 reported previously (22) for the prosubstrate hydroxyethyl disulfide (which converts to hydroxyethyl-SSG).
FIGURE 1.
hGrx1 and hGrx2 are selective for protein–GSH mixed disulfide substrates. Rates of Grx-dependent reduction of BSA–SS–[14C]cysteine ([14C]BSA–SSC) and BSA–SS–[3H]glutathione ([3H]BSA–SSG) were determined in 0.1 M Na/K phosphate buffer (pH 7.5) containing GSH (0.5 mM) and 0.1 mM RSSG substrate (see Experimental Procedures for details). (A) Time-dependent release of [3H]GSSG from [3H]BSA–SSG: (○) no hGrx2 and (●) 9 nM hGrx2 (turnover rate of 39 ± 5). (B) Time-dependent release of [14C]cysteine–SSG from [14C]BSA–SSC with or without hGrx2: (○) no hGrx2 and (▲) 110 nM Grx2 (turnover rate of 0). Each symbol represents the average of three or four experiments ± the standard error. When error bars are not visible, they are within the symbol.
Grx2 Utilizes a Nucleophilic, Double-Displacement (Ping-Pong) Catalytic Mechanism
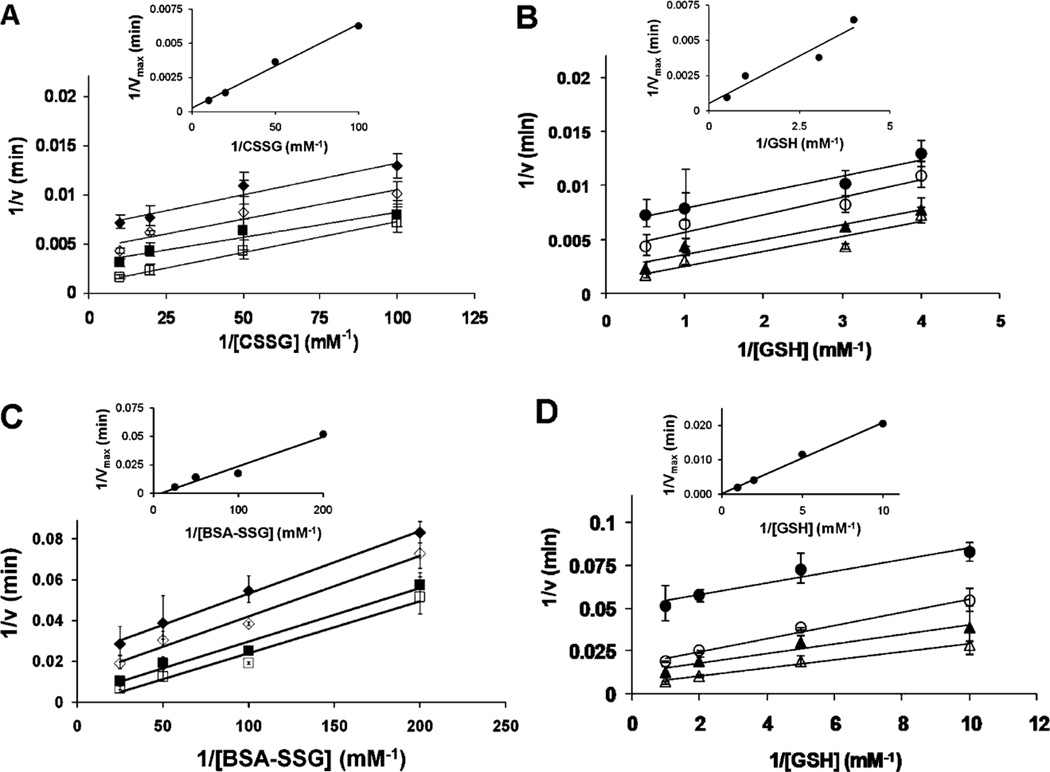
To determine the catalytic mechanism of Grx2, we performed two-substrate kinetic analysis in which the concentrations of CSSG or BSA–SSG were varied at several fixed concentrations of the second substrate, GSH, and vice versa, and double reciprocal plots (v vs 1/[RSSG] or 1/[GSH]) were generated for each fixed concentration of the other substrate. As observed for hGrx1 (16, 17), the kinetic analyses of hGrx2 (Figure 2) and mGrx2 (data not shown) produced parallel line patterns, characteristic of a nucleophilic, double-displacement (i.e., ping-pong) mechanism. Secondary plots of 1/Vmax versus 1/[S] revealed x-intercepts approaching zero (Figure 2, insets), indicative of “true KM” values approaching infinity for each of the substrates. These observations reflect a high commitment to catalysis, precluding reversible enzyme–substrate complexes in the catalytic mechanism, corresponding to the kinetic behavior characteristic of hGrx1 (16). Additional support for this essentially irreversible encounter-type mechanism was the observation that neither hGrx2 or mGrx2 was inhibited by the GSH analogue S-methylglutathione at concentrations of up to 3 mM [in 6-fold excess of GSH (data not shown)], a behavior also reported for hGrx1 (17).
FIGURE 2.
Two-substrate kinetic analysis of hGrx2-catalyzed deglutathionylation demonstrates a ping-pong kinetic pattern. The substrate dependence of Grx2-mediated reduction of R–SSG was determined using the standard spectrophotometric coupled assay (see Experimental Procedures). Reactions were initiated by addition of CSSG (for panels A and B) or GSH (for panels C and D). (A and B) Deglutathionylation of CSSG with or without hGrx2 in the presence of a fixed concentration of GSH with a varied CSSG concentration (A) or a fixed CSSG concentration with a varied GSH concentration (B): (A) (◆) 0.25 mM GSH, (◇) 0.3 mM GSH, (■) 1 mM GSH, and (□) 2 mM GSH and (B) (●) 0.01 mM CSSG, (○) 0.02 mM CSSG, (▲) 0.05 mM CSSG, and (△) 0.1 mM CSSG. (C and D) Deglutathionylation of BSA–SSG with or without hGrx2 in the presence of a fixed GSH concentration and a varied BSA–SSG concentration (C) or a fixed BSA–SSG concentration and a varied GSH concentration (D): (C) (◆) 0.1 mM GSH, (◇) 0.2 mM GSH, (■) 0.5 mM GSH, and (□) 1 mM GSH, and (D) (●) 5 µM BSA–SSG, (○) 10 µM BSA–SSG, (▲) 20 µM BSA–SSG, and (△) 40 µM BSA-SSG. Insets show secondary plots of the reciprocal of Vmax vs the reciprocal of RSSG concentration. Vmax values were calculated from fitting individual V vs [S] curves to the Michaelis–Menten equation. Data points represent the average of at least three experiments ± the standard error, and when error bars are not visible, they are within the symbol.
Grx2 Exhibits Decreased Catalytic Efficiency toward Cysteine–SSG
Previously, hGrx2 was reported to exhibit ~5-fold lower apparent KM values for multiglutathionylated BSA and RNase substrates relative to hGrx1, leading to the proposal that Grx2 exhibits a high affinity for glutathionylated proteins (27). On the other hand, 1/v versus 1/[S] curves from double-reciprocal plots (above, Figure 2) indicated that apparent KM values for the monoglutathionylated BSA–SSG substrate remained within a factor of 2 for both Grx isoforms (Table 2A). On the other hand, a marked decrease in the apparent kcat of ~10-fold (Grx2 < Grx1) was observed when either BSA-SSG or GSH was used as the substrate whose concentration was varied. This diminution in kcat corresponds to a diminished catalytic efficiency for Grx2 (human and mouse) relative to that for Grx1 (Table 2).
Table 2.
Catalytic Efficiencies of Grx Isoformsa
| Grx isoform | kcat (min−1) | KM (mM) | kcat/KM (min−1 mM−1) |
|---|---|---|---|
| (A) BSA–SSG | |||
| hGrx1 | 513 ± 19 | 0.034 ± 0.004 | (1.5 ± 2.6) × 104 |
| hGrx2 | 46 ± 2 | 0.016 ± 0.003 | (2.8 ± 0.6) × 103 |
| mGrx2 | not determined | not determined | not determined |
| (B) GSH | |||
| hGrx1 | (4.8 ± 0.23) × 103 | 0.81 ± 0.1 | (6.0 ± 1) × 103 |
| hGrx2 | 675 ± 71 | 1.6 ± 0.4 | 433 ± 145 |
| mGrx2 | 804 ± 81 | 1.3 ± 0.3 | 614 ± 203 |
| (C) CSSG | |||
| hGrx1 | (2.3 ± 0.15) × 103 | 0.055 ± 0.01 | (4.2 ± 1) × 104 |
| hGrx2 | 217 ± 16 | 0.036 ± 0.009 | (6.1 ± 2) × 103 |
| mGrx2 | 286 ± 17 | 0.055 ± 0.01 | (5.2 ± 1.3) × 103 |
Rates of deglutathionylation by recombinant hGrx1, hGrx2, and mGrx2 were determined using the spectrophotometric coupled assay (see Experimental Procedures). For part A, the BSA–SSG concentration was varied (from 0.005 to 0.25 mM) and the GSH concentration was maintained at 0.5 mM. For part B, the GSH concentration was varied (from 0.1 to 1 mM) and the CSSG concentration was maintained at 0.1 mM. For part C, the CSSG concentration was varied (from 0.002 to 0.5 mM) and the GSH concentration was maintained at 0.5 mM. Three separate experiments were performed to generate plots of v (min−1) vs [S]. These data were fit to the Michaelis–Menten equation, and apparent kcat and KM values were calculated accordingly. Apparent KM and kcat values for each substrate were consistent across additional concentrations of the second substrate (data not shown).
The reason for the discrepancy between KM,app values in this study compared to a previous report (27) is not clear; however, it is conceivable that the observed differences in KM,app reflect differences in the glutathionylated protein substrates used by each group. To eliminate steric considerations contributed by glutathionylated protein substrates, we determined apparent kinetic constants for each Grx isoform using the prototype substrate cysteine–SSG (CSSG). An additional advantage to the CSSG substrate is that it represents the conserved moiety among all protein–cysteinyl–SS–glutathione (protein–SSG) substrates, making it the logical focus of the “high affinity” of protein–SSG for Grx2, if this is an intrinsic property of the enzyme. Under standard assay conditions (see Experimental Procedures), human and mouse Grx2 both exhibited ~9-fold lower kcat values for CSSG compared to hGrx1, while apparent KM values differed by only 1.2-fold, resulting in a decreased catalytic efficiency of Grx2 similar to that observed for BSA–SSG (Table 2C). Thus, for all substrates tested, the ratio of kcat,app to KM,app (i.e., catalytic efficiency) was decreased by nearly 10-fold for human and mouse Grx2 compared to Grx1, with the decrease driven primarily by a diminished apparent kcat.
Grx2-Mediated Deglutathionylation Operates via a Monothiol Mechanism
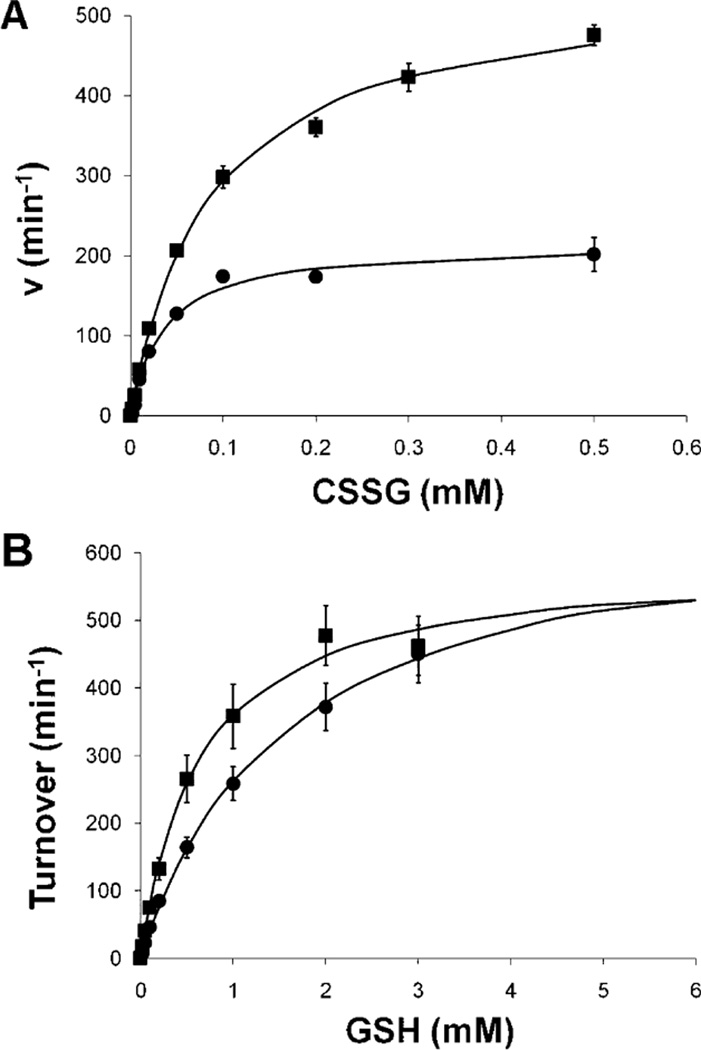
Mammalian thiol–disulfide oxidoreductase enzymes utilize distinct catalytic mechanisms, as exemplified by Grx1 (i.e., monothiol mechanism) and Trx (i.e., dithiol mechanism), as illustrated in Scheme 1 [reviewed by Mieyal et al. (5)]. To determine which mechanism is utilized by Grx2, we compared the activity of the wild-type enzyme to the activity of one in which the second cysteine at the active site was mutated to serine (i.e., C40S). Like the analogous hGrx1 mutant (i.e., C25S), hGrx2 C40S exhibited increased specific activity compared to wild-type hGrx2 (Table 1), suggesting that C40 is not required for catalysis, and it actually detracts from the catalytic rate. Under standard assay conditions, hGrx2 C40S exhibits a lower KM,app for GSH (Figure 3A) than the wild-type enzyme and approximately 2-fold increases in kcat,app and KM,app for CSSG (Figure 3B). These results suggest that the C-terminal cysteine at the active site of Grx2, like that of hGrx1 (18), competes with GSH for nucleophilic attack on the hGrx2–SSG intermediate and forms the hGrx2–intramolecular disulfide as shown in Scheme 1A. Notably, the magnitude of the increases in kcat and KM for CSSG exhibited by hGrx2 C40S is similar to that observed for the analogous hGrx1 mutant [Grx1 C25S (18)], suggesting the relative steady-state concentrations of the Grx–SSG and intramolecular disulfide forms are approximately the same for the two Grx isoenzymes.
FIGURE 3.
hGrx2 (C40S) exhibits higher KM and Vmax values for CSSG, but a lower KM for GSH, than the wild-type enzyme does. Turnover of CSSG by wild-type [WT, 135 nM (●)] and C40S [135 nM (■)] hGrx2 was assessed via the spectrophotometric coupled assay (see Experimental Procedures) with a varied CSSG (0.002–0.5 mM) or GSH (0.02–3 mM) concentration, and velocity vs substrate curves were fit to the Michaelis–Menten equation. For wild-type hGrx2, apparent kcat and KM values for RSSG and GSH are listed in Table 2. For hGrx2 (C40S), kcat,app,CSSG = 543 ± 14 min−1, KM,app,CSSG = 0.083 ± 0.009 mM, kcat,app,GSH = 589 ± 46 min−1, and KM,app,GSH = 0.63 ± 0.1 mM. Each point represents the average of at least three determinations ± the standard error.
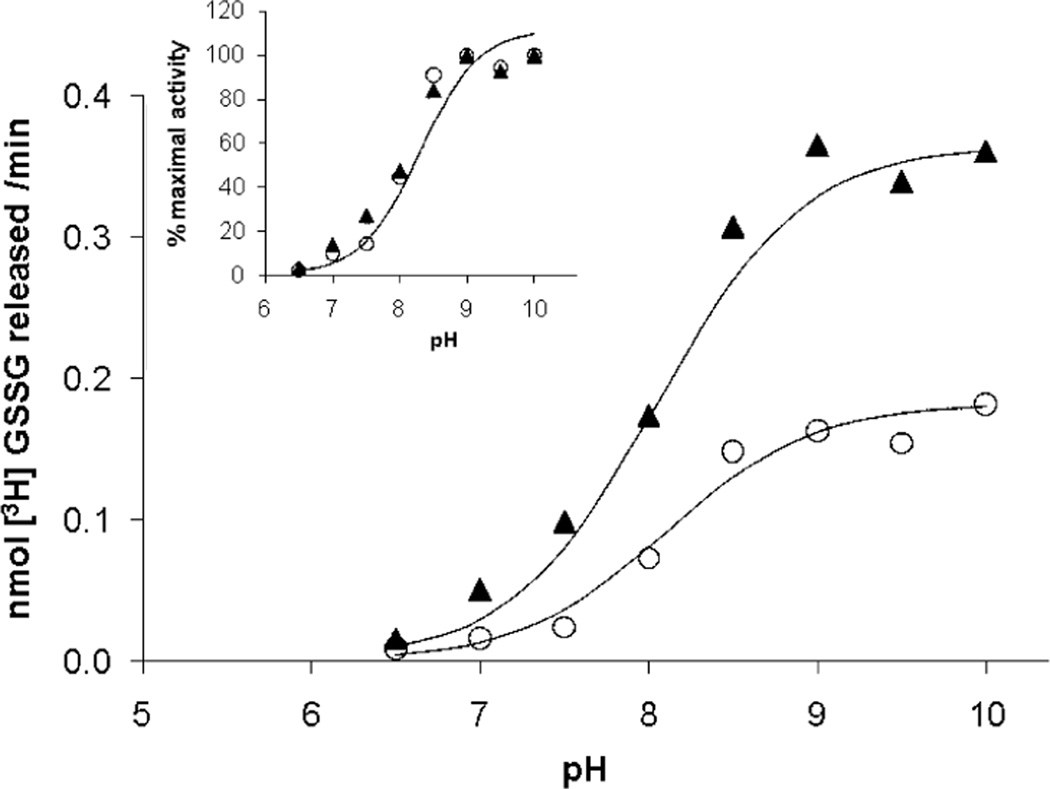
pH–Rate Profile of hGrx2-Dependent Deglutathionylation
The pH–rate profile of hGrx1-catalyzed deglutathionylation of BSA–SSG displays a typical sigmoid titration curve with an inflection point near pH 8.5, corresponding to the pKa of GSH and indicating the rate-determining step of catalysis as nucleophilic attack of the GS–thiolate on the hGRx1–SSG intermediate [i.e., Scheme 1A, step 2 (17)]. Likewise, the pH dependence of hGrx2-mediated deglutathionylation of BSA–SSG also exhibits an inflection point near 8.5 (Figure 4), which can be superimposed on the pH–rate profile of the nonenzymatic reaction of GSH with BSA–SSG (Figure 4, inset), indicating the limiting factor in both reactions is the titration of the GSH–thiol, as observed previously for Grx1 (17). Thus, the Grx2-mediated reaction displays the same rate-determining step (Scheme 1A, step 2). This observation differs from previous reports of maximal activity at pH 8.5 (i.e., a bell-shaped curve) for rat liver Grx1 (41), and for human Grx1 and Grx2 (27). This discrepancy likely relates to an artifact of the assay for Grx activity in these previous reports. In the GR-coupled assay, an apparent maximum near pH 8.5 is observed because of the pH sensitivity of GR activity. If enough GR is added at higher pH, a typical single-component titration curve is achieved (as described for Grx1 in ref 16). To avoid this artifact here, we used the radiolabel assay which monitors formation of GSSG in the absence of GR.
FIGURE 4.
pH–rate profile of BSA–SSG deglutathionylation in the absence and presence of hGrx2. Deglutathionylation of [3H]-BSA–SSG was assessed at pH 6.5–10.5, using the standard radiolabel release assay described in Experimental Procedures. Buffers (0.1 M) were MES (pH 6.5), HEPES (pH 7–8), HEPPSO (pH 7.5–8.5), and glycine (pH 9–10.5), and the final ionic strength of each buffer was adjusted to 0.3 M using NaCl. Nonenzymatic rates were subtracted from rates in the presence of hGrx2 (15–30 nM), and rates of deglutathionylation with or without hGrx2 are expressed as percentages of the maximum rate. hGrx2 concentrations were within the linear range of concentration dependence at each pH. Data from a single experiment that is representative of results from five analogous experiments are displayed.
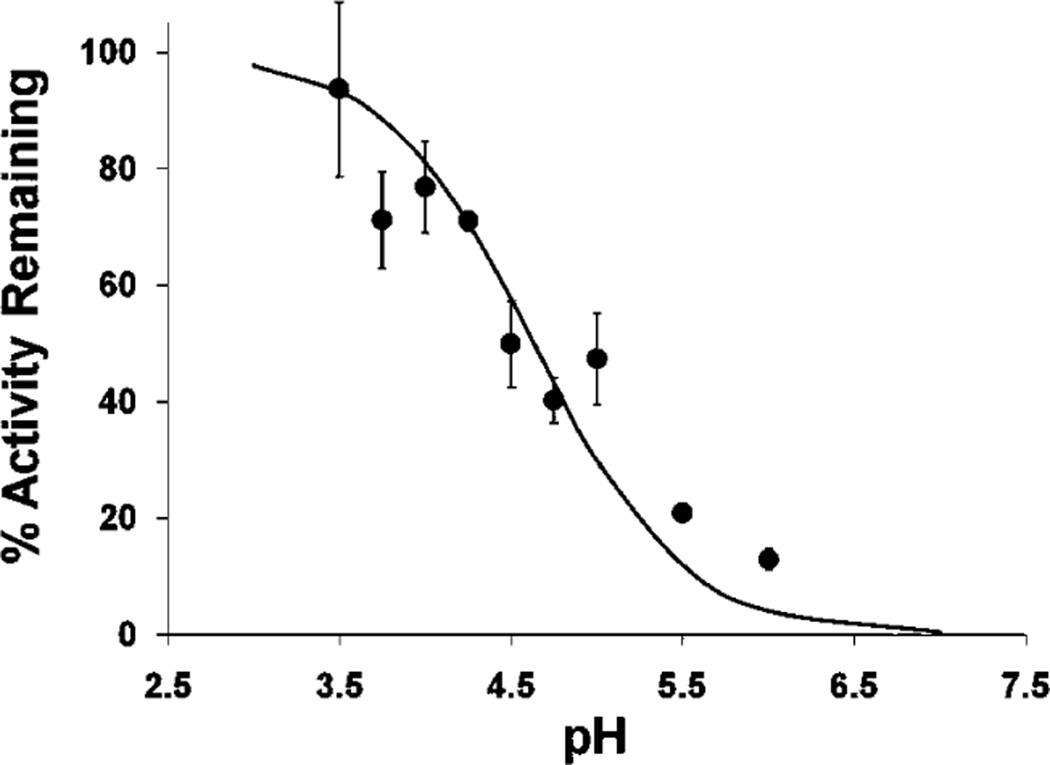
The pKa of Grx2’s Catalytic Cys Is 4.6
For typical thiol–disulfide exchange reactions, each 1 pH unit decrease in the pKa of the leaving group thiol predicts an ~4-fold increase in the second-order rate constant (42). For Grx1-catalyzed protein deglutathionylation, the rate-determining step is thiol–disulfide exchange between the Grx1–SSG intermediate and GSH (17), and the difference in pKa between the leaving group in that reaction (i.e., ~3.5 for Grx1–SH) and the leaving group of the uncatalyzed reaction (i.e., ~8.5 for a typical protein–SH) accounts for the majority of the rate enhancement of Grx1-mediated deglutathionylation [4ΔpKa = 45 = 1024-fold (17)]. Here, we have shown that Grx2 operates via an analogous ping-pong mechanism (Figure 2), in which attack of GS− on the Grx2–SSG intermediate is also rate-determining (Figure 4). To investigate whether the diminished kcat of hGrx2 toward glutathionylated substrates could be explained by a higher pKa of its catalytic cysteine, we determined its pKa according to the pH dependence of hGrx2 inactivation by iodoacetamide (IAM). hGrx2’s catalytic Cys was found to have a pKa of 4.6 (Figure 5), approximately 1 pH unit higher than that of Grx1, accounting for approximately half of the ~10-fold decrease in specific activity (see Discussion).
FIGURE 5.
Determination of the pKa of the active site Cys–SH of hGrx2. hGrx2 (3 µM) was incubated with iodoacetamide (IAM, 0.3 mM) for 3 min in the following incubation buffers: sodium citrate at pH 3.5; sodium acetate at pH 3.5, 4.0, 4.5, and 5.0; and 2-(N-morpholino)ethanesulfonic acid (MES) at pH 5.0, 5.5, and 6.0. All buffers were at a concentration of 10 mM, and the ionic strength was adjusted to 0.5 M by addition of the appropriate amount of NaCl or KCl. Following incubation, hGrx2 activity was determined using the standard spectrophotometric coupled assay (Experimental Procedures) after a 50-fold dilution into the assay mix [0.1 M Na/K phosphate (pH 7.5), 0.2 mM NADPH, 0.5 mM GSH, and 2 units/mL GSSG reductase]. The percent activity remaining at each pH was determined from the ratio of Grx2-mediated deglutathionylation rates for enzyme preincubated with IAM versus no IAM (see Experimental Procedures). Symbols represent the mean of at least three determinations of hGrx1 activity with or without IAM preincubation ± the standard error. When error bars are not visible, they are within the symbol.
Contribution of the Grx Helix 2 Dipole Moment to Stabilization of the Catalytic Cysteine Thiolate
The basis for the low pKa of hGrx1’s catalytic cysteine is uncertain, but interaction with the dipole of helix 2 has been suggested to contribute (30, 43). To determine whether a weakened ion–dipole interaction between helix 2 and the catalytic cysteine thiolate explained the higher pKa of hGrx2, we used published structures of hGrx1 and hGrx2, in combination with a dipole calculation program [http://bioportal.weizmann.ac.il/dipol/ (40)], to determine the component of the helix 2 dipole moment directed toward each enzyme’s catalytic cysteine (see Experimental Procedures). Surprisingly, the component of the helix 2 dipole was greater for hGrx2 than for each of the hGrx1 structures analyzed (Table 3), predicting greater stabilization of its catalytic cysteine thiolate by the helix. Therefore, the increased pKa of hGrx2 cannot be explained by a weakened ion–dipole interaction with helix 2 and is likely explained by other electrostatic interactions (see Discussion).
Table 3.
Helix 2 Dipole Moments for hGrx1 and hGrx2a
| Grx structure |
dipole moment (D) |
helix–thiolate angle (deg) |
component of dipole (D) |
|---|---|---|---|
| hGrx1 (1) | 182 | 24.4 | 166 |
| hGrx1 (6) | 170 | 19.7 | 160 |
| hGrx1 (16) | 177 | 19.2 | 167 |
| hGrx1 (20) | 168 | 24.4 | 153 |
| hGrx2 | 213 | 10.6 | 209 |
Partial PDB files of hGrx1 [1JHB (23)] and hGrx2 [2FLS (39)] were submitted to the Protein Dipole Moments Server [http://bioportal.weizmann.ac.il/dipol/ (40)] to calculate the dipole moment of helix 2, as well as the angle between the helix and catalytic cysteine (see Experimental Procedures). For hGrx1, the number in parentheses indicates the specific NMR structure shown previously to predict the empirically observed catalytic cysteine pKa of approximately 3.5 (30). The component of each dipole directed toward the catalytic cysteine was calculated by multiplying the dipole moment by the cosine of the angle formed by the intersection of the helix axis with a line projecting from the base of the helix to the catalytic cysteine.
Grx2 Exhibits Decreased Enhancement of the Nucleophilicity of GSH Compared to Grx1
Rate enhancement of protein deglutathionylation by Grx1 is attributed both to the low pKa of its catalytic cysteine (see above) and to its ability to enhance the nucleophilicity of the second substrate, GSH, beyond that predicted by its pKa, as shown by Bronsted analysis (17). The preference for GSH as the second substrate appears to be due mainly to the γ-glutamylcysteine moiety as demonstrated in previous studies of hGrx1 (17) and Escherichia coli Grx1 (44). To determine whether hGrx2, like hGrx1, enhances the nucleophilicity of GSH, we determined the rate enhancement of hGrx1- and hGrx2-mediated deglutathionylation of BSA–SSG using GSH, and the closely related cysteinylglycine, the nucleophilicity of which is not enhanced by human Grx1 (17). Indeed, second-order rate constants for deglutathionylation by cysteinylglycine reflected the rate enhancement predicted by the pKa of each GRx enzyme’s catalytic cysteine (Table 4 and Discussion). When cysteinylglycine was replaced with GSH, rate enhancement was further augmented by ~20-fold for hGRx1, but by less than 10-fold for hGRx2 (Table 4). Thus, hGRx1 appears to enhance GSH’s ability to serve as a second substrate to a greater degree than does hGRx2.
Table 4.
Enhancement of GSH Nucleophilicity by Grx isoformsa
| substrate | Grx isoform |
k(−Grx) (µM−1min−1) |
k(+Grx) (µ M−1 min−1) |
rate enhancement |
enhancement due to ΔpKa (4ΔpKa) |
additional enhancement of nucleophilicity |
|---|---|---|---|---|---|---|
| GSH | hGrx1 | 143 ± 8 | (2.58 ± 0.8) × 106 | (1.85 ± 0.9) × 104 | 1024 | 18.1 |
| hGrx2 | 140 ± 8 | (2.86 ± 0.5) × 105 | (2.0 ± 0.3) × 103 | 256 | 7.8 | |
| Cys–Gly | hGrx1 | 98.1 ± 9 | (1.51 ± 0.03) × 105 | (1.54 ± 0.03) × 103 | 1024 | 1.5 |
| hGrx2 | 98.1 ± 9 | (3.11 ± 0.6) × 104 | 318 ± 59 | 256 | 1.2 |
Time-dependent release of [3H]GSSG from [3H]BSA–SSG was assessed as described in Experimental Procedures. Reaction mixtures contained 0.1 M AMPSO buffer (pH 9.5), GSH or cysteinyl glycine (Cys–Gly) (0.25 mM final concentration), and Grx (3–9 nM). Enzyme and substrate concentrations were within the range of linear dependence (determined in separate experiments). Second-order rate constants (M−1 min−1) for deglutathionylation were calculated as described by Srinivasan et al. (17), i.e., rate of GSSG release = k[BSA–SSG][RSH] (for the uncatalyzed reaction) or k[Grx–SSG][RSH] (for the catalyzed reaction). Final GSH and Cys–Gly concentrations at pH 7.5 were calculated according to their pKa values [GSH = 8.7, Cys–Gly = 8.8 (17)]. Rate constants represent averages of three to seven determinations ± the standard error.
The GSH/GR System Is the Preferred Coupling System for Grx1 and Grx2 Even under “Oxidative Stress” Conditions
In the absence of GSH, hGrx2-mediated reduction of CSSG was shown to be augmented somewhat by NADPH and thioredoxin reductase (TR) (27). Under GSH-free conditions, we observed TR-facilitated CSSG reduction by both Grx1 and Grx2, displaying similar rates for the two isoforms, comparable to the rate reported previously for hGrx2 (27). Remarkably, replacement of TR with GSH and GSSG reductase (GR), at concentrations 10-fold lower than those found in cells (45), increased CSSG deglutathionylation rates by 20-fold for Grx2 and >200-fold for Grx1 (Table 5). Addition of TR (with or without Trx) to the reaction mixtures containing GSH and GR did not further augment CSSG turnover in either case. Therefore, while TR can assist deglutathionylation by both Grx isoforms to some degree, it is substantially less efficient than the GSH/GR system, even under conditions of depleted GSH, which might occur under oxidative stress conditions in vivo (see Discussion).
Table 5.
Contribution of TR to Grx-Mediated Deglutathionylation of CSSGa
| reducing system |
CSSG turnover by hGrx1 (min−1) |
CSSG turnover by hGrx2 (min−1) |
|---|---|---|
| GSH, GR | 196.9 ± 14.5 | 12.98 ± 0.69 |
| TR | 0.94 ± 0.13 | 0.62 ± 0.08 |
Rates of deglutathionylation of CSSG were measured using the spectrophotometric assay (see Experimental Procedures). Reaction mixtures contained Na/K phosphate buffer (0.1 M, pH 7.5), NADPH (0.2 mM), and hGrx1 (0.37 µM in experiments with GSH and 3.7 µM in experiments without GSH) or hGrx2 (3.7 µM). When present, other components included GSH (0.1 mM), GSSG reductase (0.01 µM), Trx (10 µM), and TR (230 nM). Reactions were initiated by addition of CSSG (0.001–0.5 mM final concentration) and monitored over 5 min. Turnover rates represent the average of at least three experiments and are expressed as the mean ± the standard error.
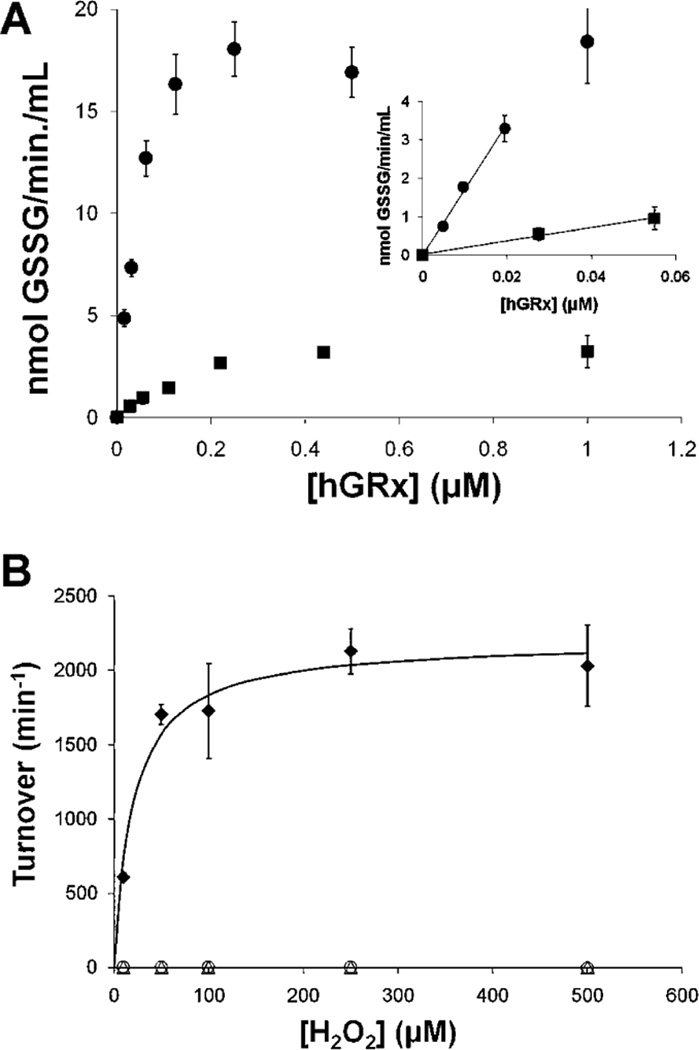
Like Grx1, Grx2 Exhibits Glutathione–Thiyl Radical Scavenging Activity
Under conditions of glutathione-thiyl radical (GS•) generation, Grx1 catalyzes GSSG formation (33). Likewise, in this study, we observed that hGrx2 catalyzed GSSG formation when GS• was produced; however, turnover was ~10-fold slower than that of hGrx1 (Figure 6A). As shown previously for hGrx1 (33), hGrx2-mediated formation of GSSG under GS radical generating conditions could not be explained by the enzyme acting as a GSH peroxidase (Figure 6B).
FIGURE 6.
Thiyl radical (GS•) scavenging activity of Grx2. (A) hGrx (0–1 µM) was preincubated with 0.5 mM GSH, 0.2 mM NADPH, and 2 units/mL GSSG reductase in 0.1 M Na/K phosphate buffer (pH 7.5) at 30 °C for 5 min (see Experimental Procedures and ref 33). Then a premade complex containing FeCl2 (0.5 mM) and ADP (2.5 mM) was added to each, and the reactions were initiated by addition of H2O2 (50 µM). Reactions were monitored according to the decrease in A340 over 5 min (within the linear range of time dependence). The amount of GSSG formed per minute was calculated according to the extinction coefficient of NADPH, corrected for the plate reader assay (see Experimental Procedures). The inset shows a region of linear dependence on hGrx concentration. GS radical scavenging activities in this time period were used to calculate turnover [hGrx1 (●), 170.0 ± 7.7; hGrx2 (■), 18.7 ± 1.2; 9.1 hGrx1:hGrx2 ratio]. Symbols represent the means of four to six determinations ± the standard error. (B) GSH-dependent peroxidase activity was measured in 0.1 M Na/K phosphate buffer (pH 7.5) containing GSH (0.5 mM) and GSSG reductase (2 units/ mL). Reactions were initiated by addition of H2O2, and A340 was monitored over 5 min. Rates of GSSG formation (i.e., NADPH oxidation) were calculated using the extinction coefficient of NADPH. Glutathione peroxidase (GPx) was used as a positive control: (◆) GPx (5 nM), (○) hGrx1 (1 µM), and (▵) hGrx2 (1 µM). Each symbol represents the average of at least three determinations ± the standard error. When error bars are not visible, they are within the symbol.
Like Grx1, Grx2 Can Promote Protein S-Glutathionylation in the Presence of Various Activated Glutathionyl Donors
Previously, we demonstrated that hGrx1 promotes protein S-glutathionylation of model protein substrates in the presence of GS•, GSNO, or GSSG, with GS• being the most efficient GS donor (33). Here we compared rates of glutathionylation of the model protein GAPDH by hGrx1 and hGrx2. For all substrates tested, the majority of GAPDH glutathionylation occurred within the initial, linear phase of the reaction time course [≤30 s (data not shown)]. Like hGrx1, hGrx2 promoted GAPDH–SSG formation in the presence of all three GS donors, with GS• being far superior as a substrate compared to GSNO and GSSG (Table 6). With GSSG, hGrx1 was a better catalyst of GAPDH glutathionylation than Grx2 (i.e., ~4-fold higher turnover); however, equivalent rates of GAPDH glutathionylation for the two Grx isoforms were observed with GS• and GSNO.
Table 6.
hGrx1 and hGrx2 Exhibit Equal Rates of Protein Glutathionylation in the Presence of GS•, GSNO, and GSSGa
| protein–SSG formation |
||
|---|---|---|
| substrate | Grx1-mediated (s−1) | Grx2-mediated (s−1) |
| GS• [≥10 µM (33)] | 0.43 ± 0.03 | 0.38 ± 0.04 |
| GSNO (500 µ M) | 0.21 ± 0.07 | 0.24 ± 0.08 |
| GSNO (250 µM) | 0.09 ± 0.04 | 0.11 ± 0.07 |
| GSSG (250 µM) | 0.38 ± 0.01 | 0.12 ± 0.007 |
| GSSG (100 µM) | 0.24 ± 0.03 | 0.06 ± 0.003 |
Reaction mixtures contained Na/K phosphate buffer (0.1 M, pH 7.5), rabbit muscle GAPDH (25 µM), Grx (0.02–0.25 µM), S-carboxymethyl BSA (7 mg/mL), and [35S]GSNO (500 µM), [35S]GSSG (500 µM), or GSH (0.5 mM) and GS• (approximately 10 µM; see ref 33). Reactions took place at room temperature and were quenched by addition of 20% ice-cold TCA. Precipitated proteins were pelleted by centrifugation, washed, and analyzed for bound radioactivity. Grx-mediated GAPDH–SSG formation was assessed by comparing the bound radioactivity in the presence and absence of the enzyme. Rates are expressed as nanomoles of protein–SSG per minute per nanomole of Grx (i.e., turnover, min−1). Turnovers were determined within the linear range of Grx dependence for each substrate and are expressed as the mean ± the standard error (n = 3–12).
DISCUSSION
Kinetic Comparison of Mammalian Grx Isoforms
Earlier reports have emphasized differences between the human Grx isoforms (26–28). In contrast, we have documented remarkable catalytic similarities between Grx2 and Grx1, including a ping-pong deglutathionylation mechanism (Scheme 1A) with the same rate-determining step, which cycles through an intermediate involving only the N-terminal active site cysteine residue (monothiol mechanism) and exhibits strong commitment to catalysis. An important consequence of this mechanism in vivo is that kcat and KM values for protein–SSG substrates increase or decrease in parallel with the concentration of GSH; therefore, maximal rates of deglutathionylation by both enzymes will depend up on the redox balance of the local environment. Another similarity between the Grx enzymes is that the GSH/GR system is superior to the Trx system in supporting deglutathionylation, even when GSH and GR were diminished to one-tenth of their predicted intracellular concentrations. Finally, hGrx2 (like hGrx1) exhibited GS• scavenging activity, as well as catalysis of protein–SSG formation, with GS• acting as the best activated glutathionyl cosubstrate.
A striking distinction between mammalian Grx1 and Grx2 is their difference in catalytic efficiency, reflecting ~10-fold lower kcat values for hGrx2 toward CSSG, BSA–SSG, and GSH, accompanied by only minor changes in KM,app. We used our understanding of the basis for Grx1-mediated rate enhancement to investigate why Grx2 exhibits a lower kcat and found that the ~10-fold difference in kcat between hGrx1 and h/mGrx2 can be explained by two factors: (1) a 1 pH unit increase in the pKa of Grx2’s catalytic cysteine, which predicts an ~4-fold smaller rate enhancement versus hGrx1 (i.e., 4ΔpKa = 44, or 256-fold vs 1024-fold), and (2) a diminished enhancement of the nucleophilicity of GSH.
For Grx1, the basis for enhancement of GSH nucleophilicty, as well as for the low pKa of its catalytic cysteine, is not fully understood; however, structural and site-directed mutagenesis studies provide some insight. For example, a recent analysis of the role of K19 in rate enhancement by hGrx1 (30) suggested that the primary role of this moiety is not to stabilize the catalytic cysteine thiolate [as was inferred from its proximity to C22 in the average NMR structure (23)] but rather to enhance deglutathionylation by another mechanism, such as enhancing the nucleophilicity of GSH. Interestingly, structural comparison of reduced hGrx1 (23) and hGrx2 (39) indicates a substantial shift in the orientation of hGrx2’s K34 (homologous to K19 of hGrx1) to a position farther from the catalytic cysteine (~3 Å for hGrx1 vs 8.2 Å for hGrx2). The effect of this shift in orientation on enzyme–substrate interactions is unknown but could be explored by determining the effect of the K34 mutation on the rate enhancement (and pKa) of hGrx2.
Several hypotheses have been proposed to explain the low pKa of Grx1’s catalytic cysteine (C22) besides the proximity of positively charged residues, including ion–dipole interactions between C22 and helix 2 (30, 43), H bonding between the C22-thiolate and C25 and/or T21 side chains (23, 30), and H bonds between the C22-thiolate and backbone residues within the active site (46). Calculation of the helix 2 dipole moment for each hGrx isoform revealed that the dipole vector component was greater for Grx2 than for hGrx1, suggesting that a diminished helix 2 dipole does not explain the higher pKa of the Grx2 catalytic thiolate.
Increased H bonding distances within the hGrx2 active site also do not explain its higher pKa relative to that of hGrx1. Foloppe et al. (46) proposed that for pig Grx1 (pGrx), H bonds between the catalytic cysteine thiolate and amide hydrogens of active site residues P23 and C25 account for the low catalytic cysteine pKa (~3.5). However, calculated distances between Grx2’s C37 and homologous Y39 and C40 amide nitrogens are all shorter than the distances between hGrx1’s C22, Y24, and C25 backbone nitrogens. In fact, the only interaction proposed to stabilize the hGrx1 C22-thiolate that appears to be diminished for hGrx2 is the H bond between the catalytic cysteine thiolate and the hydroxyl group of T21, which is supported by only one of 20 hGrx1 NMR structures (2.2 Å for hGrx1, structure 16; 9.4 Å for hGrx2). Importantly, these comparisons are complicated by the fact that the protein structures were determined using different methodologies (i.e., NMR vs X-ray crystallography) and that the structure of hGrx2 contains an associated GSH that may alter active site orientation. Determination of the structure of reduced hGrx2 will facilitate more direct structural comparison, providing additional insight into the basis for the unique pKa values of hGrx1 and hGrx2.
Additional Activities of Grx2 and Implications of Glutathionyl Transfer Activities
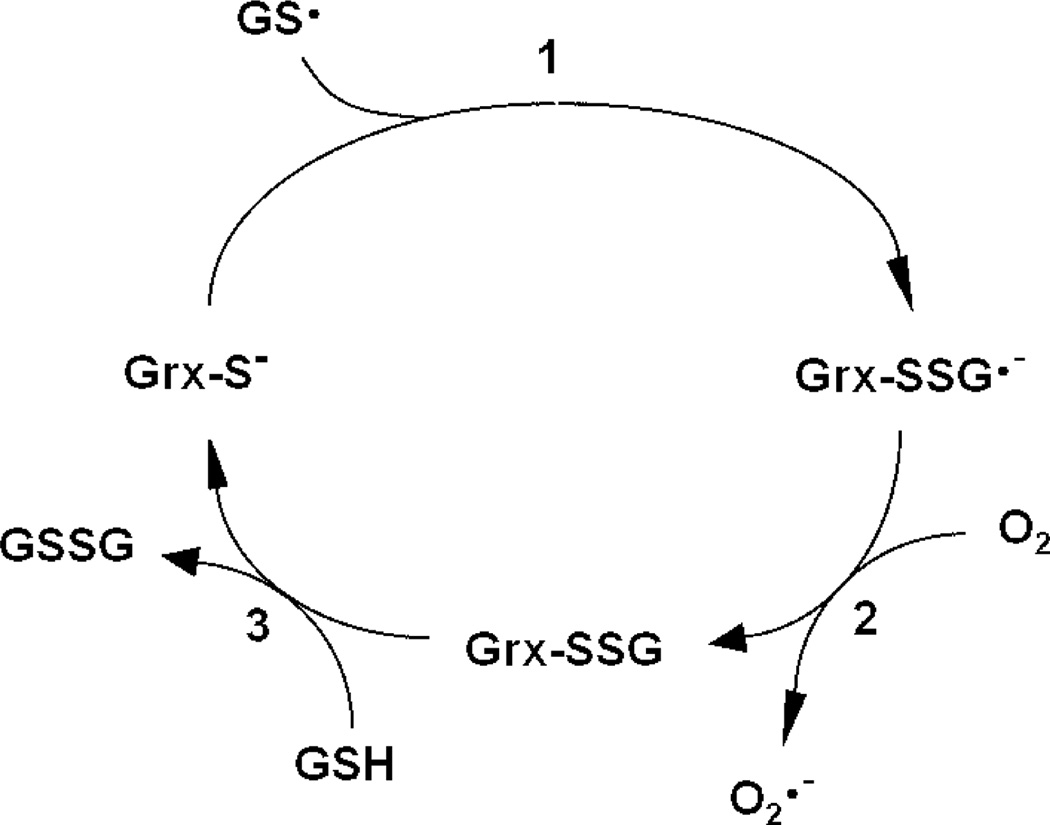
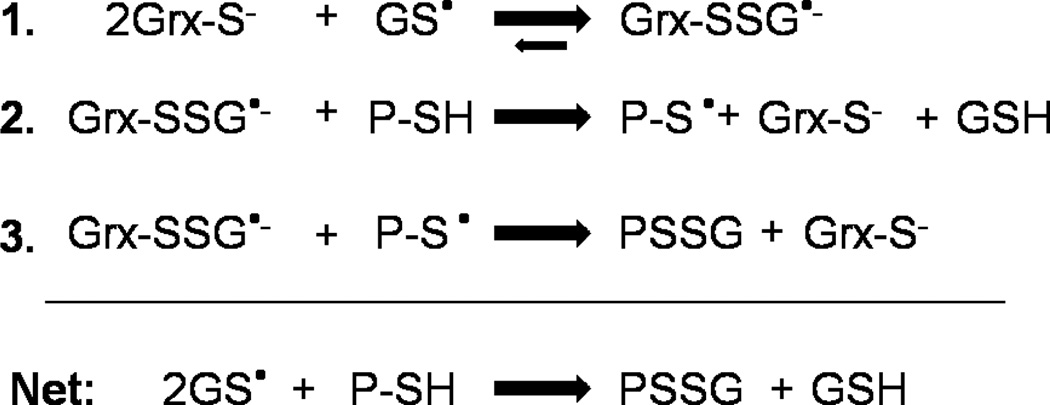
hGrx1 was previously shown to scavenge glutathione thiyl radicals (GS•), likely via initial stabilization of the radical by formation of a Grx–SSG•− disulfide anion radical intermediate [Scheme 2 (33)]. Rate enhancement by hGrx1 was explained by the low pKa of its catalytic cysteine (which serves as a nucleophile in the first step of the reaction and is >99% ionized at physiological pH), as well as its ability to stabilize a covalently bound glutathionyl moiety (18). On the basis of its low pKa (4.6, >99% ionized at physiological pH) and conserved residues implicated in stabilization of the adducted glutathionyl moiety (21, 22), we hypothesized that hGrx2 would also exhibit GS• scavenging activity. The 10-fold lower rate of GS• scavenging by hGrx2 compared to that of hGrx1 suggests that the final step of the reaction (i.e., Scheme 2, step 3), which is identical to the rate-limiting step of deglutathionylation (i.e., Scheme 1A, step 2), is also rate-determining for GS• scavenging. As shown previously for hGrx1 (33), hGrx2-mediated GS• scavenging cannot be explained by the enzyme acting as a GSH peroxidase. Although GSH peroxidase activities were reported recently for hGrx2 (28), >5 µM Grx2 was required for detection of activity, a concentration more than 100 times what was effective in GS• scavenging and more than 5 times the predicted Grx2 concentration within the mitochondrial matrix (see below).
Scheme 2.
Catalytic Mechanism of GS• Scavenging by Mammalian Grx1 (33)a
a In the first step of the reaction, the catalytic cysteine of Grx attacks the sulfur of GS•, forming a Grx–SSG•− disulfide anion radical intermediate. The free electron of this intermediate then reacts with O2, forming O2•− and Grx–SSG. In the third step of the reaction, the Grx–SSG intermediate is reduced by GSH, forming GSSG. GS•, GSH thiyl radical; O2•−, superoxide.
Mechanisms of protein S-glutathionylation in vivo have not yet been elucidated; however, the involvement of oxidized derivatives of GSH has been proposed (1–3, 47, 48), and Starke et al. (33) provided evidence that such reactions may be mediated by hGrx1. Like hGrx1, hGrx2 promoted transfer of a glutathionyl moiety to GAPDH in the presence of various oxidized derivatives of GSH, with GS• acting as a far superior substrate compared to GSNO and GSSG. For GSSG, hGrx2 exhibited approximately one-fourth of the glutathionylation rate of hGrx1. This observation suggests that the protein glutathionylation reaction with GSSG proceeds via a thiol–disulfide exchange mechanism analogous to the deglutathionylation reaction [i.e., Scheme 1A, with protein-SSG replaced with GSSG and GSH replaced with protein–SH, as documented previously for hGrx1 (19) and discussed in ref 5]. The decreased activity of hGrx2 in this case is ascribed entirely to the difference in pKa of the catalytic cysteine, because there is no enhancement of nucleophilicity for the GAPDH–cysteine–thiol moiety by either enzyme. Importantly, the GSSG concentration required to achieve turnover by this mechanism (≥0.1 mM) suggests that it is unlikely to contribute significantly to protein glutathionylation under redox signaling conditions in vivo, in which the GSH:GSSG ratio does not deviate substantially from the typical value of 100, corresponding to micromolar concentrations of GSSG for most cells.
For GS• and GSNO, hGrx2 promoted GAPDH glutathionylation at rates equivalent to those of hGrx1, representing a remarkable contrast to its lower deglutathionylation and GS• scavenging activities. These equivalent catalytic rates suggest that the rate-determining step of Grx-mediated protein–SSG formation, in contrast to catalysis of deglutahionylation, does not involve the catalytic cysteine serving as a leaving group. This concept is consistent with the mechanism proposed previously for GS• transfer by hGrx1 [Scheme 3 (33)]. For both Grx enzymes, GS• was a much better glutathionyl donor substrate than GSNO or GSSG and thus represents a more likely substrate for protein S-glutathionylation by Grx in the absence of overt oxidative or nitrosative stress. Interestingly, while GSNO may not serve as a direct substrate for Grx-mediated glutathionylation under resting conditions, it may still contribute to protein–SSG formation by reacting with GSH to form the superior substrate, GS• (49). The possibility of GSNO dismutation, combined with the observation that a 10-fold molar excess of GSNO disrupted inactive Grx2 dimers in vitro (26), suggests a potential role for GSNO as an activator of Grx2-mediated glutathionylation in vivo, first by releasing the enzyme from its inactive form and then by reacting with GSH to generate a more efficient substrate for protein glutathionylation.
Scheme 3.
Proposed Mechanism of GS• Transfer by Mammalian Grx1 (33)a
a The first step of the reaction is the same as in Scheme 2, i.e., the catalytic cysteine of Grx attacks GS•, forming the enzyme disulfide anion radical intermediate, Grx–SSG•−. In the second step of the reaction, the Grx–disulfide anion radical intermediate abstracts a H atom from the reduced protein, forming protein–S• and GSH and restoring the Grx anion. In step 3, the protein radical reacts with another Grx–disulfide anion radical, forming protein–SSG and another molecule of reduced Grx enzyme. GS•, GSH thiyl radical; Grx–SSG•−, glutaredoxin–SSG disulfide anion radical; P, protein.
In Vivo Implications of Grx2 Activities
A major distinction between mammalian Grx isoforms is their nearly 10-fold difference in specific activity toward glutathionylated substrates. However, deglutathionylation activity in vivo is likely to be determined by multiple additional considerations, such as intracellular enzyme concentrations, pH [~8 in the mitochondrial matrix (50)], and activating or inactivating factors. We estimated the intramitochondrial concentrations of Grx1 and Grx2a using the results of our previous study of Grx content in rat mitochondria (15) in combination with published volumes of intramitochondrial spaces (36, 37) (see Experimental Procedures). Accordingly, the concentration of Grx2 in the mitochondrial matrix is predicted to be ~1 µM, while that of Grx1 in the intermembrane space is approximately 10-fold lower (~0.1 µM). Assuming that both Grx isoforms are fully active, the 10-fold higher Grx2 concentration, balanced by its 10-fold lower kcat, predicts that total deglutathionylation and GS• scavenging activities should be similar across mitochondrial subcompartments. In contrast, Grx-mediated glutathionyl transfer activity, catalyzed at equivalent rates by both Grx isoforms, would be expected to be higher in the mitochondrial matrix, where the Grx2 concentration is 10-fold greater.
Prediction of Grx2’s intracellular activity is further complicated by the observation that the protein may be sequestered into inactive FeS cluster dimers, according to immunoprecipitation studies performed under resting conditions in cultured cells (25). Regulation of dimer integrity in vivo is not yet understood, but exposure to oxidants (e.g., GSSG and GSNO) (25, 26) in vitro results in dimmer dissociation. The concept that Grx2 may be released (and, indeed, activated) primarily under oxidative conditions suggests that its catalytic activities involving oxidized substrates (e.g., GS• scavenging, glutathionyl transfer) may represent its primary functions within the mitochondria. Alternatively, Grx2 may be released upon oxidative stress but primarily catalyze deglutathionylation, reestablishing sulfhydryl homeostasis, before being resequestered into inactive dimers. Clearly, more studies are necessary to unravel the roles of Grx2 in situ.
Knockout and overexpression studies of hGrx2 in HeLa cells (51, 52) and transgenic mice (53) suggest that Grx2 confers pro-survival effects during oxidative stress, but the mechanisms of these protective effects are unknown. Our studies suggest that Grx2 could promote protein glutathionylation or deglutathionylation depending upon the redox status of the local environment, and the presence of specific oxidized derivatives of GSH. The distinct effects of glutathionylation on mitochondrial respiratory chain components suggest that either activity could be cytoprotective. For example, glutathionylation of Complex II in vitro increases its electron transfer efficiency and decreases O2•− production, a trigger of apoptosis in disease states such as cardiac ischemia-reperfusion (54). Conversely, deglutathionylation of Complex I restores its electron transfer activity, preventing electron leakage and O2•− production (55, 56). Interestingly, hGrx2 (>6 µM) was recently shown to promote glutathionylation of the 75 kDa subunit of Complex I under oxidizing conditions (i.e., GSH:GSSG ratio of 3) in vitro (55); however, the physiological relevance of this observation is yet to be determined. Further studies focused on the relationship between Grx2 activity and glutathionylation status of established targets, or as yet unknown mitochondrial targets, will improve our understanding of the mechanism by which Grx2 confers its cytoprotective effects.
In summary, we found that mammalian Grx2 exhibits catalytic properties remarkably similar to those of hGrx1 (e.g., monothiol, nucleophilic double-displacement catalytic mechanism, high commitment to catalysis, selectivity for protein-SSG substrates, and GS• scavenging and glutathionyl transfer activities), as well as some unique features, such as a higher catalytic cysteine pKa and decreased enhancement of GSH nucleophilicity, resulting in decreased kcat,app values for both substrates in deglutathionylation reactions. The major catalytic function of Grx2 in vivo remains to be characterized, but this pursuit represents an exciting frontier in the fields of redox signaling, mitochondrial redox homeostasis, and cellular viability.
ACKNOWLEDGMENT
We thank Dr. Suparna Qanungo (Department of Pharmacology, Case Western Reserve University) and Dr. Periannan Kuppusamy and Dr. Mahmood Khan (Dorothy M. Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH) for assistance with EPR spectroscopy for detection of glutathione thiyl radical (GS•). We appreciate the technical assistance of Mary Consolo, who prepared the [3H]BSA-SSG substrate and assisted in purification of hGrx2. We are grateful to Dr. Vernon Anderson (Department of Biochemistry, Case Western Reserve University) for assistance in theoretical calculations of thiolate stabilization energy contributed by helix 2 of the Grx enzyme, and we thank Dr. Anthony Berdis (Department of Pharmacology, Case Western Reserve University) for critical review of the manuscript.
Footnotes
This work was supported by NIH Grants R01 AG024413 and PO1 AG 15885 and a Department of Veterans Affairs Merit Review Grant (J.J.M.) and NIH Grants F30 AG 029687A, T32 GM008803, and T32 GM07250 (M.M.G.).
Abbreviations: AMPSO, N-(1,1-dimethyl-2-hydroxyethyl)-3-amino-2-hydroxypropanesulfonic acid; BSA, bovine serum albumin; BSA–SSC, bovine serum albumin–cysteine mixed disulfide; BSA–SSG, bovine serum albumin–glutathione mixed disulfide; C40S, cysteine 40 to serine mutant; CSSG, cysteine–glutathione disulfide; DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); DTT, dithiothreitol; EST, expressed sequence tag; GAPDH, glyceraldehyde phosphate dehydrogenase; GFP, green fluorescent protein; GR, glutathione reductase; Grx, glutaredoxin; GS•, glutathione thiyl radical; GSH, glutathione; GSNO, S-nitrosoglutathione; GSSG, glutathione disulfide; H2O2, hydrogen peroxide; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; HEPPSO, N-(2-hydroxyethyl)piperazine-N′-2-hydroxypropanesulfonic acid; HRP, horseradish peroxidase; IAM, iodoacetamide; MES, 2-(N-morpholino)ethanesulfonic acid; NADPH, nicotinamide adenine dinucleotide phosphate; NTSP, N-succinimidyl pyridyl bis(3,3′-dithiopropionate); protein–SSG, protein–GSH mixed disulfide; TCA, trichloroacetic acid; TR, thioredoxin reductase; Trx, thioredoxin.
REFERENCES
- 1.Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur J. Biochem. 2000;267:4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 2.Shelton MD, Chock PB, Mieyal JJ. Glutaredoxin: Role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid. Redox Signaling. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 3.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Thomas JA, Poland B, Honzatko R. Protein sulfhydryls and their role in the antioxidant function of protein S-thiolation. Arch. Biochem. Biophys. 1995;319:1–9. doi: 10.1006/abbi.1995.1261. [DOI] [PubMed] [Google Scholar]
- 5.Mieyal JJ, Srinivasan U, Starke DW, Gravina SA, Mieyal PA. In: Glutathionyl specificity of the Thioltransferases: Mechanistic and Physiological Implications. Packer L, Cadenas E, editors. Marcel Dekker, Inc; New York: 1995. pp. 305–372. [Google Scholar]
- 6.Adachi T, Pimentel DR, Heibeck T, Hou X, Lee YJ, Jiang B, Ido Y, Cohen RA. S-Glutathiolation of Ras mediates redox-sensitive signaling by angiotensin II in vascular smooth muscle cells. J. Biol. Chem. 2004;279:29857–29862. doi: 10.1074/jbc.M313320200. [DOI] [PubMed] [Google Scholar]
- 7.Pimentel DR, Adachi T, Ido Y, Heibeck T, Jiang B, Lee Y, Melendez JA, Cohen RA, Colucci WS. Strain-stimulated hypertrophy in cardiac myocytes is mediated by reactive oxygen species-dependent Ras S-glutathiolation. J. Mol. Cell. Cardiol. 2006;41:613–622. doi: 10.1016/j.yjmcc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, Cohen RA. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat. Med. 2004;10:1200–1207. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 9.Barrett WC, DeGnore JP, Keng YF, Zhang ZY, Yim MB, Chock PB. Roles of superoxide radical anion in signal transduction mediated by reversible regulation of proteintyrosine phosphatase 1B. J. Biol. Chem. 1999;274:34543–34546. doi: 10.1074/jbc.274.49.34543. [DOI] [PubMed] [Google Scholar]
- 10.Qanungo S, Starke DW, Pai HV, Mieyal JJ, Nieminen AL. Glutathione supplementation potentiates hypoxic apoptosis by S-glutathionylation of p65-NFκB. J. Biol. Chem. 2007;282:18427–18436. doi: 10.1074/jbc.M610934200. [DOI] [PubMed] [Google Scholar]
- 11.Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, Heintz NH, Heim J, Ho YS, Matthews DE, Wouters EF, Janssen-Heininger YM. Dynamic redox control of NF-κB through glutaredoxinregulated S-glutathionylation of inhibitory κB kinase β. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13086–13091. doi: 10.1073/pnas.0603290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chrestensen CA, Starke DW, Mieyal JJ. Acute cadmium exposure inactivates thioltransferase (glutaredoxin), inhibits intracellular reduction of protein-glutathionyl-mixed disulfides, and initiates apoptosis. J. Biol. Chem. 2000;275:26556–26565. doi: 10.1074/jbc.M004097200. [DOI] [PubMed] [Google Scholar]
- 13.Jung CH, Thomas JA. S-Glutathiolated hepatocyte proteins and insulin disulfides as substrates for reduction by glutaredoxin, thioredoxin, protein disulfide isomerase, and glutathione. Arch. Biochem. Biophys. 1996;335:61–72. doi: 10.1006/abbi.1996.0482. [DOI] [PubMed] [Google Scholar]
- 14.Ho YS, Xiong Y, Ho DS, Gao J, Chua BH, Pai H, Mieyal JJ. Targeted disruption of the glutaredoxin 1 gene does not sensitize adult mice to tissue injury induced by ischemia/ reperfusion and hyperoxia. Free Radical Biol. Med. 2007;43:1299–1312. doi: 10.1016/j.freeradbiomed.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pai HV, Starke DW, Lesnefsky EJ, Hoppel CL, Mieyal JJ. What is the functional significance of the unique localization of glutaredoxin 1 (GRx1) in the intermembrane space of mitochondria. Antioxid. Redox Signaling. 2007;9:2027–2033. doi: 10.1089/ars.2007.1642. [DOI] [PubMed] [Google Scholar]
- 16.Gravina SA, Mieyal JJ. Thioltransferase is a specific glutathionyl mixed disulfide oxidoreductase. Biochemistry. 1993;32:3368–3376. doi: 10.1021/bi00064a021. [DOI] [PubMed] [Google Scholar]
- 17.Srinivasan U, Mieyal PA, Mieyal JJ. pH profiles indicative of rate-limiting nucleophilic displacement in thioltransferase catalysis. Biochemistry. 1997;36:3199–3206. doi: 10.1021/bi962017t. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, Jao S, Nanduri S, Starke DW, Mieyal JJ, Qin J. Reactivity of the human thioltransferase (glutaredoxin) C7S, C25S, C78S, C82S mutant and NMR solution structure of its glutathionyl mixed disulfide intermediate reflect catalytic specificity. Biochemistry. 1998;37:17145–17156. doi: 10.1021/bi9806504. [DOI] [PubMed] [Google Scholar]
- 19.Mieyal JJ, Starke DW, Gravina SA, Hocevar BA. Thioltransferase in human red blood cells: Kinetics and equilibrium. Biochemistry. 1991;30:8883–8891. doi: 10.1021/bi00100a023. [DOI] [PubMed] [Google Scholar]
- 20.Gan ZR, Wells WW. Identification and reactivity of the catalytic site of pig liver thioltransferase. J. Biol. Chem. 1987;262:6704–6707. [PubMed] [Google Scholar]
- 21.Gladyshev VN, Liu A, Novoselov SV, Krysan K, Sun QA, Kryukov VM, Kryukov GV, Lou MF. Identification and characterization of a new mammalian glutaredoxin (thioltransferase), Grx2. J. Biol. Chem. 2001;276:30374–30380. doi: 10.1074/jbc.M100020200. [DOI] [PubMed] [Google Scholar]
- 22.Lundberg M, Johansson C, Chandra J, Enoksson M, Jacobsson G, Ljung J, Johansson M, Holmgren A. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J. Biol. Chem. 2001;276:26269–26275. doi: 10.1074/jbc.M011605200. [DOI] [PubMed] [Google Scholar]
- 23.Sun C, Berardi MJ, Bushweller JH. The NMR solution structure of human glutaredoxin in the fully reduced form. J. Mol. Biol. 1998;280:687–701. doi: 10.1006/jmbi.1998.1913. [DOI] [PubMed] [Google Scholar]
- 24.Lonn ME, Hudemann C, Berndt C, Cherkasov V, Capani F, Holmgren A, Lillig CH. Expression Pattern of Human Glutaredoxin 2 Isoforms: Identification and Characterization of Two Testis/Cancer Cell-Specific Isoforms. Antioxid. Redox Signaling. 2008;10:547–558. doi: 10.1089/ars.2007.1821. [DOI] [PubMed] [Google Scholar]
- 25.Lillig CH, Berndt C, Vergnolle O, Lonn ME, Hudemann C, Bill E, Holmgren A. Characterization of human glutaredoxin 2 as iron-sulfur protein: A possible role as redox sensor. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8168–8173. doi: 10.1073/pnas.0500735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashemy SI, Johansson C, Berndt C, Lillig CH, Holmgren A. Oxidation and S-nitrosylation of cysteines in human cytosolic and mitochondrial glutaredoxins: Effects on structure and activity. J. Biol. Chem. 2007;282:14428–14436. doi: 10.1074/jbc.M700927200. [DOI] [PubMed] [Google Scholar]
- 27.Johansson C, Lillig CH, Holmgren A. Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J. Biol. Chem. 2004;279:7537–7543. doi: 10.1074/jbc.M312719200. [DOI] [PubMed] [Google Scholar]
- 28.Fernando MR, Lechner JM, Lofgren S, Gladyshev VN, Lou MF. Mitochondrial thioltransferase (glutaredoxin 2) has GSH-dependent and thioredoxin reductase-dependent peroxidase activities in vitro and in lens epithelial cells. FASEB J. 2006;20:2645–2647. doi: 10.1096/fj.06-5919fje. [DOI] [PubMed] [Google Scholar]
- 29.Chrestensen CA, Eckman CB, Starke DW, Mieyal JJ. Cloning, expression and characterization of human thioltransferase (glutaredoxin) in E. coli. FEBS Lett. 1995;374:25–28. doi: 10.1016/0014-5793(95)01066-n. [DOI] [PubMed] [Google Scholar]
- 30.Jao SC, English Ospina SM, Berdis AJ, Starke DW, Post CB, Mieyal JJ. Computational and mutational analysis of human glutaredoxin (thioltransferase): Probing the molecular basis of the low pKa of cysteine 22 and its role in catalysis. Biochemistry. 2006;45:4785–4796. doi: 10.1021/bi0516327. [DOI] [PubMed] [Google Scholar]
- 31.Shelton MD, Kern TS, Mieyal JJ. Glutaredoxin regulates nuclear factor κ-B and intercellular adhesion molecule in Muller cells: Model of diabetic retinopathy. J. Biol. Chem. 2007;282:12467–12474. doi: 10.1074/jbc.M610863200. [DOI] [PubMed] [Google Scholar]
- 32.Starke DW, Chen Y, Bapna CP, Lesnefsky EJ, Mieyal JJ. Sensitivity of protein sulfhydryl repair enzymes to oxidative stress. Free Radical Biol. Med. 1997;23:373–384. doi: 10.1016/s0891-5849(97)00009-9. [DOI] [PubMed] [Google Scholar]
- 33.Starke DW, Chock PB, Mieyal JJ. Glutathionethiyl radical scavenging and transferase properties of human glutaredoxin (thioltransferase). Potential role in redox signal transduction. J. Biol. Chem. 2003;278:14607–14613. doi: 10.1074/jbc.M210434200. [DOI] [PubMed] [Google Scholar]
- 34.Harman LS, Carver DK, Schreiber J, Mason RP. One- and two-electron oxidation of reduced glutathione by peroxidases. J. Biol. Chem. 1986;261:1642–1648. [PubMed] [Google Scholar]
- 35.Lobysheva II, Serezhenkov VA, Vanin AF. Interaction of peroxynitrite and hydrogen peroxide with dinitrosyl iron complexes containing thiol ligands in vitro. Biochemistry (Moscow, Russ. Fed.) 1999;64:153–158. [PubMed] [Google Scholar]
- 36.Pfaff E, Klingenberg M, Ritt E, Vogell W. [Correlation of the unspecific permeable mitochondrial space with the “intermembrane space”] Eur. J. Biochem. 1968;5:222–232. doi: 10.1111/j.1432-1033.1968.tb00361.x. [DOI] [PubMed] [Google Scholar]
- 37.Cohen NS, Cheung CW, Raijman L. Measurements of mitochondrial volumes are affected by the amount of mitochondria used in the determinations. Biochem. J. 1987;245:375–379. doi: 10.1042/bj2450375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papov VV, Gravina SA, Mieyal JJ, Biemann K. The primary structure and properties of thioltransferase (glutaredoxin) from human red blood cells. Protein Sci. 1994;3:428–434. doi: 10.1002/pro.5560030307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansson C, Kavanagh KL, Gileadi O, Oppermann U. Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J. Biol. Chem. 2007;282:3077–3082. doi: 10.1074/jbc.M608179200. [DOI] [PubMed] [Google Scholar]
- 40.Felder CE, Prilusky J, Silman I, Sussman JL. A server and database for dipole moments of proteins. Nucleic Acids Res. 2007;35:W512–W521. doi: 10.1093/nar/gkm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gan ZR, Wells WW. Purification and properties of thioltransferase. J. Biol. Chem. 1986;261:996–1001. [PubMed] [Google Scholar]
- 42.Szajewski RP, Whitesides GM. Rate Constants and Equilibrium Constants for Thiol-Disulfide Interchange Reactions Involving Oxidized Glutathione. J. Am. Chem. Soc. 1980;102:2011–2026. [Google Scholar]
- 43.Kortemme T, Creighton TE. Ionisation of cysteine residues at the termini of model α-helical peptides. Relevance to unusual thiol pKa values in proteins of the thioredoxin family. J. Mol. Biol. 1995;253:799–812. doi: 10.1006/jmbi.1995.0592. [DOI] [PubMed] [Google Scholar]
- 44.Peltoniemi MJ, Karala AR, Jurvansuu JK, Kinnula VL, Ruddock LW. Insights into deglutathionylation reactions. Different intermediates in the glutaredoxin and protein disulfide isomerase catalyzed reactions are defined by the γ-linkage present in glutathione. J. Biol. Chem. 2006;281:33107–33114. doi: 10.1074/jbc.M605602200. [DOI] [PubMed] [Google Scholar]
- 45.Carlberg I, Mannervik B. Glutathione reductase. Methods Enzymol. 1985;113:484–490. doi: 10.1016/s0076-6879(85)13062-4. [DOI] [PubMed] [Google Scholar]
- 46.Foloppe N, Nilsson L. Stabilization of the catalytic thiolate in a mammalian glutaredoxin: Structure, dynamics and electrostatics of reduced pig glutaredoxin and its mutants. J. Mol. Biol. 2007;372:798–816. doi: 10.1016/j.jmb.2007.05.101. [DOI] [PubMed] [Google Scholar]
- 47.Ghezzi P. Regulation of protein function by glutathionylation. Free Radical Res. 2005;39:573–580. doi: 10.1080/10715760500072172. [DOI] [PubMed] [Google Scholar]
- 48.Hurd TR, Filipovska A, Costa NJ, Dahm CC, Murphy MP. Disulphide formation on mitochondrial protein thiols. Biochem. Soc. Trans. 2005;33:1390–1393. doi: 10.1042/BST0331390. [DOI] [PubMed] [Google Scholar]
- 49.Singh SP, Wishnok JS, Keshive M, Deen WM, Tannenbaum SR. The chemistry of the S-nitrosoglutathione/glutathione system. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14428–14433. doi: 10.1073/pnas.93.25.14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lillig CH, Lonn ME, Enoksson M, Fernandes AP, Holmgren A. Short interfering RNA-mediated silencing of glutaredoxin 2 increases the sensitivity of HeLa cells toward doxorubicin and phenylarsine oxide. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13227–13232. doi: 10.1073/pnas.0401896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enoksson M, Fernandes AP, Prast S, Lillig CH, Holmgren A, Orrenius S. Overexpression of glutaredoxin 2 attenuates apoptosis by preventing cytochrome c release. Biochem. Biophys. Res. Commun. 2005;327:774–779. doi: 10.1016/j.bbrc.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 53.Nagy N, Malik G, Tosaki A, Ho YS, Maulik N, Das DK. Overexpression of glutaredoxin-2 reduces myocardial cell death by preventing both apoptosis and necrosis. J. Mol. Cell. Cardiol. 2008;44:252–260. doi: 10.1016/j.yjmcc.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 54.Chen YR, Chen CL, Pfeiffer DR, Zweier JL. Mitochondrial complex II in the post-ischemic heart: Oxidative injury and the role of protein S-glutathionylation. J. Biol. Chem. 2007;282:32640–32654. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 55.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: Implications for mitochondrial redox regulation and antioxidant defense. J. Biol. Chem. 2004;279:47939–47951. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 56.Taylor ER, Hurrell F, Shannon RJ, Lin TK, Hirst J, Murphy MP. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J. Biol. Chem. 2003;278:19603–19610. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- 57.Holmgren A. Thioredoxin. Annu. Rev. Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]