Abstract
Cystathionine β-synthase (CBS) is an essential PLP-dependent enzyme of the transsulfuration pathway that condenses serine with homocysteine to form cystathionine; intriguingly, human CBS also contains a heme b cofactor of unknown function. Herein we describe the enzymatic and spectroscopic properties of a disease-associated R266K hCBS variant, which has an altered hydrogen-bonding environment. The R266K hCBS contains a low-spin, six-coordinate Fe(III) heme bearing a His/Cys ligation motif, like that of WT hCBS; however, there is a geometric distortion that exists at the R266K heme. Using rR spectroscopy, we show that the Fe(III)-Cys(thiolate) bond is longer and weaker in R266K, as evidenced by an 8 cm−1 downshift in the ν(Fe-S) resonance. Presence of this longer and weaker Fe(III)-Cys(thiolate) is correlated with alteration of the fluorescence spectrum of the active PLP ketoenamine tautomer. Activity data demonstrate that, relative to WT, the R266K variant is more impaired in the alternative cysteine-synthesis reaction than in the canonical cystathionine-synthesis reaction. This diminished cysteine synthesis activity and a greater sensitivity to exogenous PLP correlates with the change in PLP environment. Fe-S(Cys) bond weakening causes a nearly 300-fold increase in the rate of ligand switching upon reduction of the R266K heme. Combined, these data demonstrate cross talk between the heme and PLP active sites, consistent with previous proposals, revealing that alteration of the Arg266-Cys52 interaction affects PLP-dependent activity and dramatically destabilizes the ferrous thiolate-ligated heme complex, underscoring the importance of this hydrogen-bonding residue pair.
Homocysteine (Hcy) is a toxic metabolite of the methionine metabolic cycle. In addition to its role in proteins, the amino acid methionine is a fundamental building block for the biological methylating agent, S-adenosylmethionine (AdoMet). Homocysteine is generated from the de-adenosylation of S-adenosylhomocysteine (AdoHcy), the by-product of methylation reactions that use AdoMet (1). It has been found that Hcy in its cyclic form (Hcy thiolactone) reacts readily with proteins causing deleterious post-translational modifications, providing a possible molecular explanation for Hcy toxicity (2, 3). At the cellular level, increased levels of Hcy are correlated with an elevated risk of atherosclerosis (a primary cause of both cardiovascular disease and stroke) as well as thrombosis (4). Elevated levels of Hcy may be caused by nutritional abnormalities, such as dietary deficiencies in vitamins B2, B6 and B12, or by genetic abnormalities, most commonly caused by mutations in the gene that encodes for the Hcy-metabolizing enzyme cystathionine β-synthase (CBS) (4, 5).
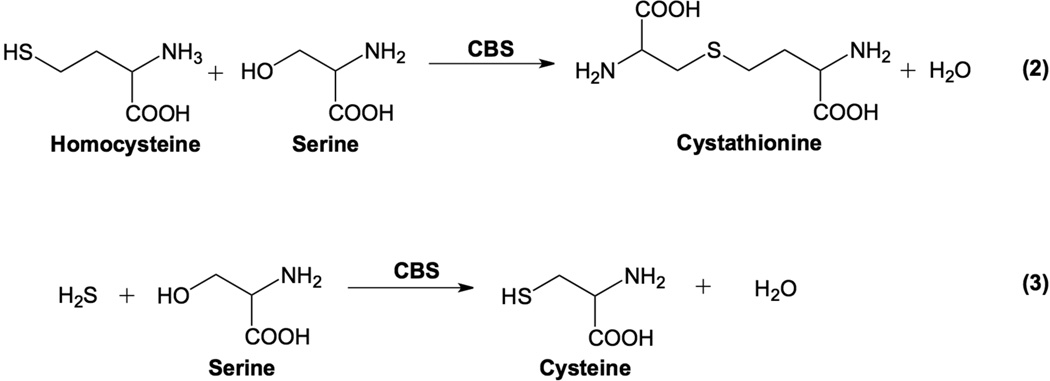
CBS is an essential pyridoxal-5’-dependent (PLP) enzyme that catalyzes the β-replacement reaction of serine with homocysteine to form cystathionine and water (Scheme 1). Elevated levels of Hcy due to CBS deficiency, chiefly caused by abnormalities in the CBS gene, result in a medical condition called classical homocystinuria or CBS-deficient homocystinuria (CBSDH). Increased plasma Hcy results in a variety of physiological symptoms including cardiovascular, skeletal and cognitive defects. Additionally, patients with CBSDH show increased risk of developing Alzheimer’s and Parkinson’s diseases (5–9).
Scheme 1.
CBS-catalyzed reactions: (2) condensation of serine with homocysteine to form cystathionine and (3) condensation of serine with hydrogen sulfide to form cysteine.
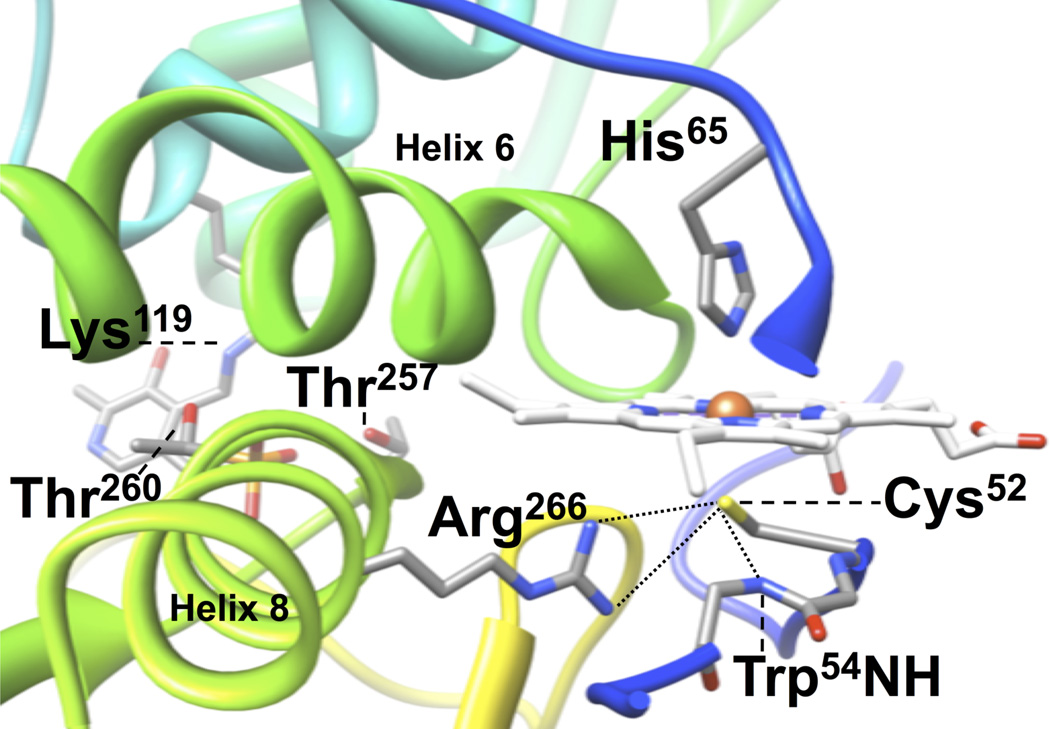
CBS from higher organisms is the only known PLP-dependent enzyme to contain a heme b cofactor (10). In mammals, CBS is an α4 homotetramer of 63-kDa subunits, each of which contains an N-terminal heme-binding domain, a central catalytic PLP-binding domain, and a C-terminal AdoMet-binding domain (10, 11). Binding of AdoMet allosterically regulates the enzyme by increasing activity 3- to 5-fold (12). Activation of CBS may also be achieved by removal of the C-terminal AdoMet-binding domain, which causes a concomitant change in protein oligomeric status from homotetrameric to homodimeric; the 45-kDa truncated form of the enzyme is known as CBS-45 (13). This observation suggests that the regulatory domain is autoinhibitory, and the inhibition may be relieved by AdoMet binding, limited proteolysis, or partial thermal denaturation (12). Crystal structures of human CBS-45 (14) and the full-length Drosophila melanogaster CBS (DmCBS) (15) show that the heme is unusually solvent exposed with its nearest edge located 14 Å from the anchoring phosphate moiety of the active site PLP. These structures confirmed spectroscopic results that showed that the ferric heme in CBS is ligated by an uncommon Cys/His motif that involves the thiolate of Cys52 trans to the Nε2 atom of His65 (human enzyme numbering, Figure 1); both ligands are provided by amino acids on the N-terminal portion of the polypeptide (16, 17). Additionally, the crystal structures reveal a complex hydrogen-bonding network that connects the heme macrocycle to the PLP active site. Of particular interest is the interaction of the positively charged guanidinium group of Arg266 with that of the negatively charged thiolate moiety of Cys52; this interaction connects the heme to the active site via helix 8, which contains Arg266, Thr257 and Thr260. The hydroxyl side chains of Thr257 and Thr260 hydrogen bond directly to the PLP phosphate; thus, this H-bond network may provide a direct means of communication between the heme cofactor and the enzyme active site (14, 15). While there is no definitive consensus on the role of heme in CBS, it is clear that heme is necessary for maximal activity of CBS from higher organisms (10, 18–21).
Figure 1.
Location of key residues that interact with the heme and PLP in hCBS. Labeled are: the Fe(III) heme ligands Cys52 and His65; the cysteine(thiolate) hydrogen bonding partners Arg266 and the amide backbone of Trp54; the PLP phosphate hydrogen bonding partners Thr257 and Thr260; and the PLP internal aldimine forming Lys119. Data taken from PDB file 1JBQ (14). The polypeptide backbone is colored using a rainbow scheme from N-terminus (blue) to C-terminus (red).
In this study, we examine the effect of the disease-associated R266K mutation on the spectroscopic and catalytic properties of full-length hCBS. We show that the R266K variant is enzymatically competent but displays differences in reaction specificity, as well as PLP- and AdoMet-responsiveness compared to WT hCBS. We show that there are subtle differences in the electronic absorption, EPR and rR spectroscopic signatures of Fe(III) R266K hCBS, which may be attributed to geometric distortions at the heme iron atom, including a lengthening of the Fe(III)-S(Cys) bond due to a change in hydrogen bonding at Cys52. We use fluorescence spectroscopy to demonstrate that these spectroscopic changes at the heme cofactor correlate with spectroscopic and enzymatic changes at the PLP active site. Additionally, we show that Fe(II) R266K undergoes a thermally-induced ligand switch that is more facile than that of WT hCBS. Taken together, these data suggest that the R266K variation destabilizes the Fe(III)-Cys52(thiolate) interaction, and that this change at the heme is communicated to the enzyme active site.
Materials and Methods
Materials
Buffers and glycerol were purchased from Sigma-Aldrich and used as received. High-purity sodium dithionite was purchased from Fluka and stored under Ar(g) at −20°C until used.
Isolation and Purification of WT hCBS and R266K hCBS
Both the WT and variant proteins were expressed and purified to homogeneity as described previously (22). Briefly, E. coli Rosetta2(DE3) cells were transformed with pET-28-C-hCBS plasmid carrying either WT or the R266K human CBS sequence with a non-removable 6xHis tag at its C-terminal end. The bacterial cells were grown at 30°C, 275 rpm in 2.8 L Fernbach flasks containing 1 L of LB medium supplemented with 0.001% thiamine-HCl, 0.0025% pyridoxine-HCl, 0.1 mM FeCl3, 0.3 mM δ-aminolevulinic acid, and 30 µg/mL kanamycin (all final concentrations). The expression of hCBS was induced by adding in IPTG to a final concentration of 1 mM once the cell density reached A600 ~ 0.8; cell growth was then continued overnight. Cells were harvested and then resuspended in lysis buffer (50 mM sodium phosphate at pH 7.4, 300 mM NaCl, 0.1 mM PLP and protease inhibitor cocktail VII [A.G. Scientific]) and treated with 2 mg/mL lysozyme for 1 hr at 4°C prior to sonication. After removal of any non-soluble and particulate material, the soluble fraction was loaded on a TALON column (Contech) equilibrated in 50 mM sodium phosphate, pH 7.4, 300 mM NaCl. The resin with bound CBS was extensively washed (50 mM sodium phosphate, pH 7.4, 300 mM NaCl, 10 mM imidazole); bound protein was eluted by using 200 mM imidazole (final concentration) in the wash buffer. Eluted protein was immediately desalted on a Sephadex G-25 resin (GE Healthcare), and the buffer was exchanged with DEAE loading buffer (15 mM potassium phosphate, pH 7.2, 1 mM EDTA, 1 mM DTT, 10% ethylene glycol). The desalted sample was bound to a DEAE Sepharose resin (GE Healthcare), washed, and hCBS protein was eluted with 300 mM potassium phosphate in the DEAE loading/wash buffer. The hCBS protein was concentrated, buffer exchanged into 20 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM TCEP, and 0.01% Tween20, and stored in aliquots at −80°C. Protein was buffer exchanged into the appropriate buffer as described in the activity and spectroscopy sections (vide infra).
Activity Measurements
The cysteine-synthesis activity of WT and R266K Fe(III) hCBS was determined as previously described (23). Briefly, the reaction mixture (600 µL) contained 0.5 mg/mL BSA, 10 mM DTT, 0.5 mM PLP, 0.35 mM AdoMet, 0.020 mg/mL enzyme, 10 mM L-serine, and 10 mM Na2S in 200 mM Tris buffer at pH 8.6 (the pH at which CBS is most active). This solution was incubated for 12 min at 37°C, and then an aliquot of the reaction mixture (100 µL) was taken and mixed with 50% (w/v) trichloroacetic acid. Precipitated protein was removed by centrifugation, and the supernatant (100 µL) was combined with acetic acid (100 µL) and ninhydrin (100 µL), and the mixture was heated in a boiling water bath for 3 min and then immediately cooled on ice. The absorbance at 560 nm was measured to determine the amount of cysteine generated, and the value was interpolated from a standard curve generated by the same method using cysteine solutions of known concentrations that contained all other reagents except enzyme (24). Cystathionine-synthesis activity was determined as reported in (22).
Electronic Absorption Spectroscopy
Electronic absorption spectra were recorded on a double-beam Varian Cary 4 Bio spectrophotometer with a temperature controller, set to a spectral bandwidth of 0.5 nm. Spectra of protein samples were recorded in 100 mM CHES buffer, 100 mM NaCl, pH 8.6. In all spectroscopic studies, non-coordinating CHES buffer was used in lieu of Tris buffer to prevent adventitious coordination of the buffer to the heme iron center. Samples were purged of oxygen by flowing Ar(g) through the headspace of a septum-sealed cuvette for 20 min. Reduction of Fe(III) protein samples was accomplished by adding an anaerobically prepared stock solution of sodium dithionite to achieve a final sample concentration of 1–5 mM. For spectral measurements, solutions of dithionite and Fe(III) protein were allowed 20 min to equilibrate at 4°C before anaerobic addition of stock dithionite (40 mM, 10 µL) to the Fe(III) protein. For kinetic measurements, solutions of dithionite and Fe(III) protein were allowed 20 min to equilibrate at 10, 20 or 30°C before anaerobic addition of the dithionite solution (40 mM, 10 µL) to the Fe(III) protein. The rate at which Fe(II) R266K hCBS (Soret 447 nm) was converted to Fe(II) R266K hCBS424 (Soret 424 nm) was fitted by a single-wavelength method utilizing the Solver function in Microsoft Excel 2004; the values for the rate constants with minimal residuals between experimental and predicted absorbance measurements are reported. Loss of the Soret at 447 nm was best fitted using a biexponential decay, as described in eq 1. The absorbance at time infinity (Abs∞) was measured experimentally by forcing full conversion of Fe(II) R266K hCBS to Fe(II) R266K hCBS424 using thermal treatment (25). Briefly, Fe(III) protein was equilibrated at 4°C; a solution of sodium dithionite (40 mM, 10 µL) was added to the Fe(III) protein, and reduction was monitored spectrophotometrically until no further changes were observed at 447 nm. The temperature was then ramped to 37°C, and conversion from the 447 nm Soret to the 424 nm Soret was monitored spectrophotometrically; thermal conversion was considered complete when no further changes in the intensity of the 424 nm Soret were observed. The constants α and β represent the collection of values that include the absorbance at time zero and the extinction coefficients of each species, respectively.
| (1) |
Fluorescence Spectroscopy
Fluorescence measurements were taken on an ISS PC1 photon counting fluorometer (ISS Instruments, Inc., Champaign, IL) at room temperature. Spectra of protein samples were recorded in 100 mM CHES buffer, 100 mM NaCl, pH 8.6. Protein samples (7–10 µM, 400 µL) were placed in a quartz cuvette with a 2 mm excitation path length and a 1 cm emission path length. Emission spectra were recorded from 425 nm to 635 nm with an excitation wavelength of 410 nm and excitation and emission slit widths of 4 mm and 2 mm, respectively. Corrections for buffer fluorescence were made by subtracting the emission spectrum of buffer from spectra of samples containing buffer and protein.
MCD Spectroscopy
Magnetic circular dichroism (MCD) spectra were recorded on a Jasco J-715 CD spectropolarimeter with the sample compartment modified to accommodate an SM-4000-8T magnetocryostat (Oxford instruments). Spectra of protein samples were recorded in 100 mM CHES buffer, 100 mM NaCl, pH 8.6. For each protein sample in buffer, approximately 55% (v/v) glycerol was present. Glycerol was introduced to the Fe(III) form of the protein and stirred with a syringe until the solution was homogeneous; the final protein concentration was 16 µM in a total volume of 150 µL. Glycerol had no effect on the electronic absorption spectra at room or liquid-helium temperatures. Samples were transferred via gastight syringe into cells, flash-frozen and stored in N2(l). MCD spectra were taken over a temperature range from 4 to 200K. The MCD signal at each temperature was recorded at ±7 T. Negative polarity data were subtracted from positive polarity data to remove CD contributions, and the resulting spectrum divided by 2.
EPR Spectroscopy
X-band electron paramagnetic resonance (EPR) spectra were collected on a Bruker ELEXSYS E500 equipped with an Oxford ESR 900 continuous flow cryostat connected to an Oxford ITC4 temperature controller. The microwave frequency was monitored using an EIP model 625A CW microwave frequency counter. Field calibration was achieved using a Varian ER 035 gaussmeter. Spectra of protein samples were recorded in 100 mM CHES buffer, 100 mM NaCl, pH 8.6. The final concentration of protein was 140–240 µM in a total volume of 150 µL. Each sample was transferred via a gastight syringe into a quartz EPR tube and frozen in N2(l). For all samples, scans of 0–10000 G revealed no signals other than those reported.
Resonance Raman Spectroscopy
Resonance Raman spectra were recorded using an excitation wavelength of 413.1 nm from a Coherent I-302C Kr+ laser in a backscattering 135° sample geometry with an Acton Research monochromator set to a grating of 2400 groves/mm. Incident powers ranged from 10–12 mW and were focused with a cylindrical lens onto the sample. A Princeton Instruments Spex 1877 triple spectrograph outfitted with a cooled, intensified diode array detector was operated under computer control using Spectrasense software. The solution samples were prepared as described for EPR spectroscopy (vide supra) and were placed in a quartz dewar cooled with ice water to reduce local heating. Peak positions were calibrated relative to a K2SO4 resonance at 981 cm−1. Assignments of key vibrational modes are noted and are based on the work of Spiro et al. and Green et al. (26–28).
Results
R266K hCBS Binds Heme Similarly to WT hCBS
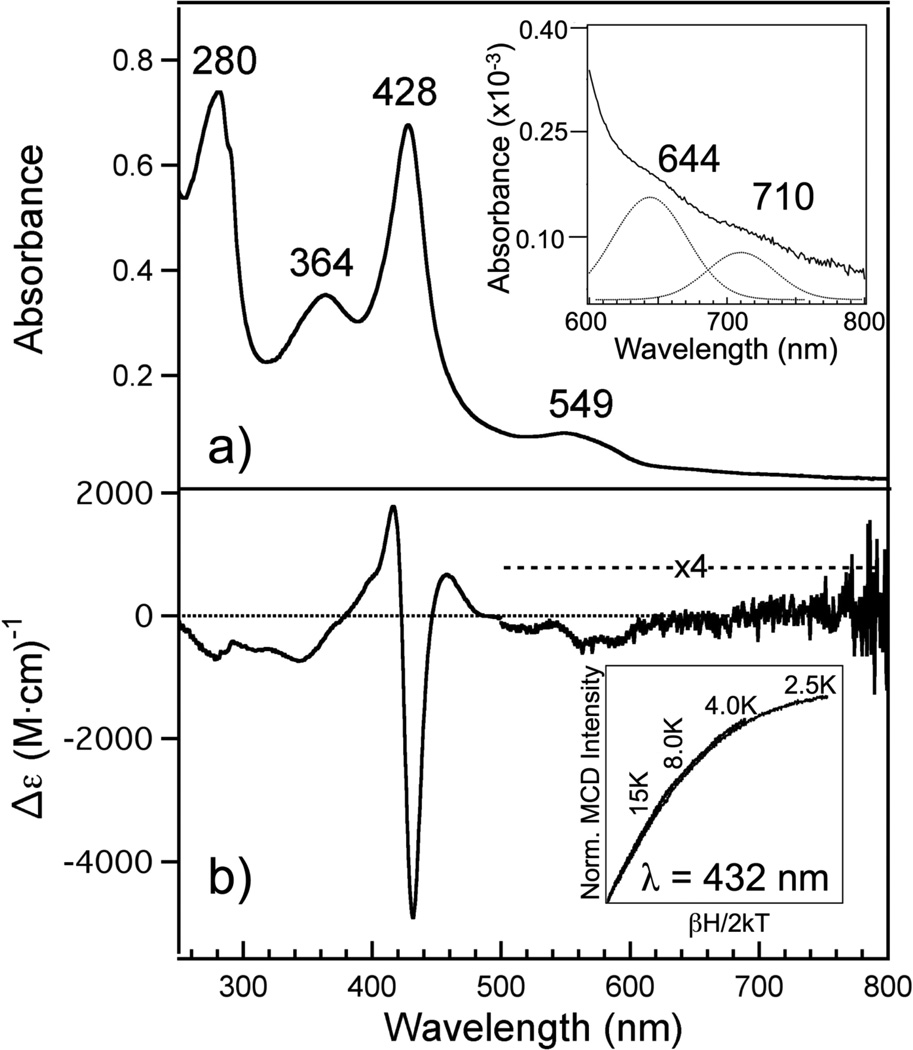
The electronic absorption spectrum of purified R266K hCBS is only subtly different from that of WT hCBS, indicating that the R266K variant binds heme in a similar manner. The spectrum of as-isolated R266K (Figure 2A) exhibits a distinct δ band at 364 nm, a sharp Soret band at 428 nm and a broad absorption envelope for the α/β region with maximal intensity at 549 nm. This electronic absorption spectrum suggests that the heme in the R266K variant is in its Fe(III) oxidation state and is bound by Cys52 and His65, like that of the WT protein. Additionally, there are two ligand-to-metal charge-transfer transitions (Cys(thiolate)→Fe(III)) evident in the 600–750 nm region of R266K (Figure 2A, inset) that are red-shifted 4 nm compared to WT, suggesting the R266K Cys(thiolate)-Fe(III) heme interaction is slightly different than that of WT hCBS.
Figure 2.
(A) Electronic absorption spectrum of Fe(III) R266K hCBS. Fe(III) R266K (7.8 µM) was in 100 mM CHES buffer and 100 mM NaCl, pH 8.6 at room temperature. Inset: close-up of the ligand-to-metal charge transfer (LMCT) transitions including the best-fit bands assuming a Gaussian peak shape (dotted). (B) MCD spectrum of Fe(III) R266K hCBS. Fe(III) R266K (15.7 µM) was in 100 mM CHES buffer, 100 mM NaCl and 55% glycerol (v/v) at 4.0 K and 7 T. Inset: the field dependence of the MCD intensity at 432 nm was recorded at 2.5, 4.0, 8.0 and 15 K. The curves were normalized to the most intense data point (2.5 K, 7 T).
MCD spectroscopy, with its unique ability to fingerprint the spin and coordination states of heme proteins, confirms the conclusion that the heme is low-spin Fe(III) and ligated by a Cys/His motif. The MCD spectrum of R266K (Figure 2B) is dominated by an intense, temperature-dependent C-term in the Soret region with peak-crossover-trough positions of 416 nm-423 nm-432 nm, similar to WT Fe(III) hCBS (29), as well as other Fe(III) Cys/His or Cys/neutral donor ligated heme proteins such as P450CAM + ImH, BxRcoM, hRev-Erbβ, DmE75 and RrCooA (30–33). The magnetic saturation behavior of the most intense wavelength of this dominating C-term (432 nm) displays an overlapping nature at different temperatures (Figure 2B, inset), indicative that the R266K heme is in a low-spin, Fe(III) S=. state. Taken together, these data suggest that the heme in R266K is low-spin, Fe(III) and bound by the native heme ligands, Cys52 and His65.
EPR and rR Spectroscopies Suggest a Geometric Distortion and Lengthening of the Fe(III)-S(Cys) Bond
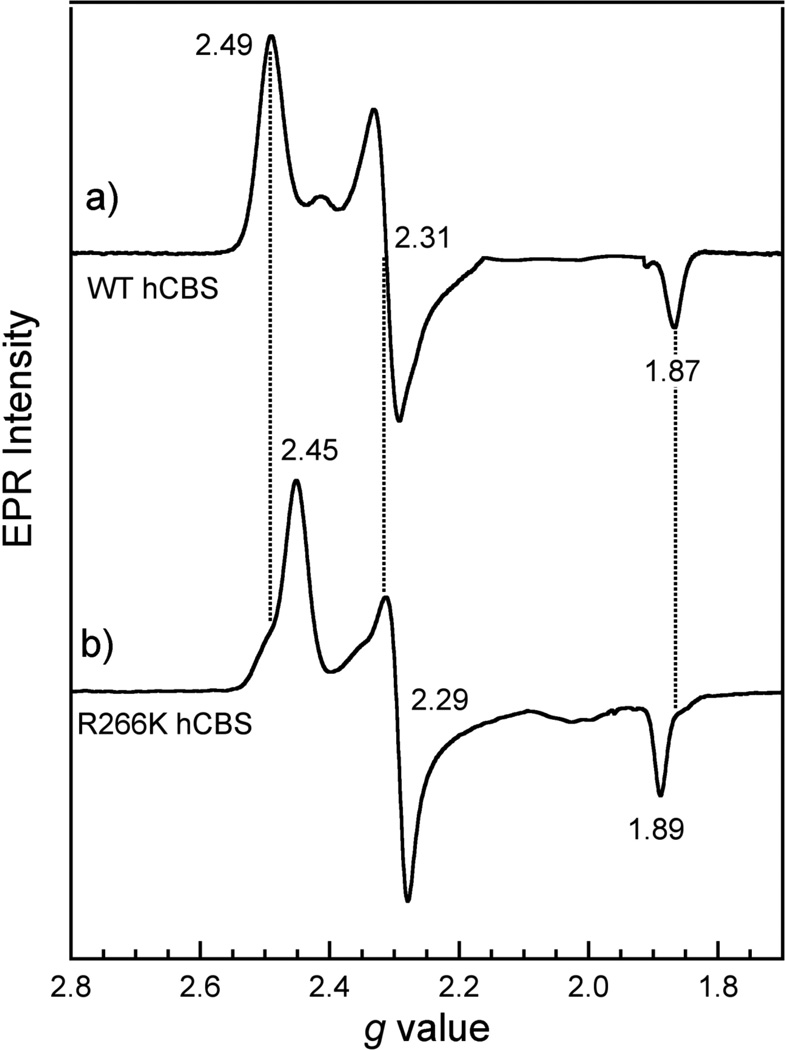
EPR spectroscopy suggests a perturbation in the relative energies of the d-orbitals of Fe(III) R266K hCBS compared to those of Fe(III) WT hCBS. The EPR spectra, which are sensitive to the environment of the paramagnetic Fe(III) center, are shown in Figure 3 for WT (A) and R266K (B) hCBS, and reveal only the presence of a rhombic, low-spin Fe(III) signal. The Fe(III) WT hCBS spectrum is essentially identical to previous reports (16, 29). However, the positions of the major g values for the Fe(III) R266K hCBS variant are different; the g values gz, gy and gx are 2.45, 2.29 and 1.89, respectively, with additional minor signals present (a common feature for rhombic EPR signals of low-spin heme thiolate-ligated proteins). Analysis of the g values for the Fe(III) R266K variant, using the method outlined by Palmer (34), yields values for rhombicity (V/Δ) and tetragonality (Δ/λ) of 0.37 and 5.50, respectively. These values place Fe(III) R266K hCBS in the “P” family of the Blumberg-Peisach diagram—the same region in which WT Fe(III) hCBS resides, as well as other ferric cysteine(thiolate)-ligated heme proteins bearing a sixth neutral donor ligand (35). While WT and R266K exhibit virtually the same rhombicity (0.38 and 0.37, respectively), their tetragonality values different significantly (5.08 and 5.50, respectively). These results suggest that both a rhombic distortion (V) and an axial distortion (Δ) take place in concert in the R266K variant; these geometric distortions are predicted to change 1) the effective overlap of the orbitals of the heme axial ligands as well as 2) the relative d-orbital energies in Fe(III) R266K with respect to Fe(III) WT hCBS.
Figure 3.
X-band EPR spectra of Fe(III) A) WT hCBS and B) R266K hCBS. Proteins (141 µM WT and 235 µM R266K) were in 100 mM CHES buffer and 100 mM NaCl, pH 8.6. Each spectrum represents an average of 10 scans taken at 10K, with 9.379 GHz microwave frequency, 8.000 G modulation amplitude, 100 kHz modulation frequency, 74 dB receiver gain, 163.84 ms time constant and a power of 1.002 mW.
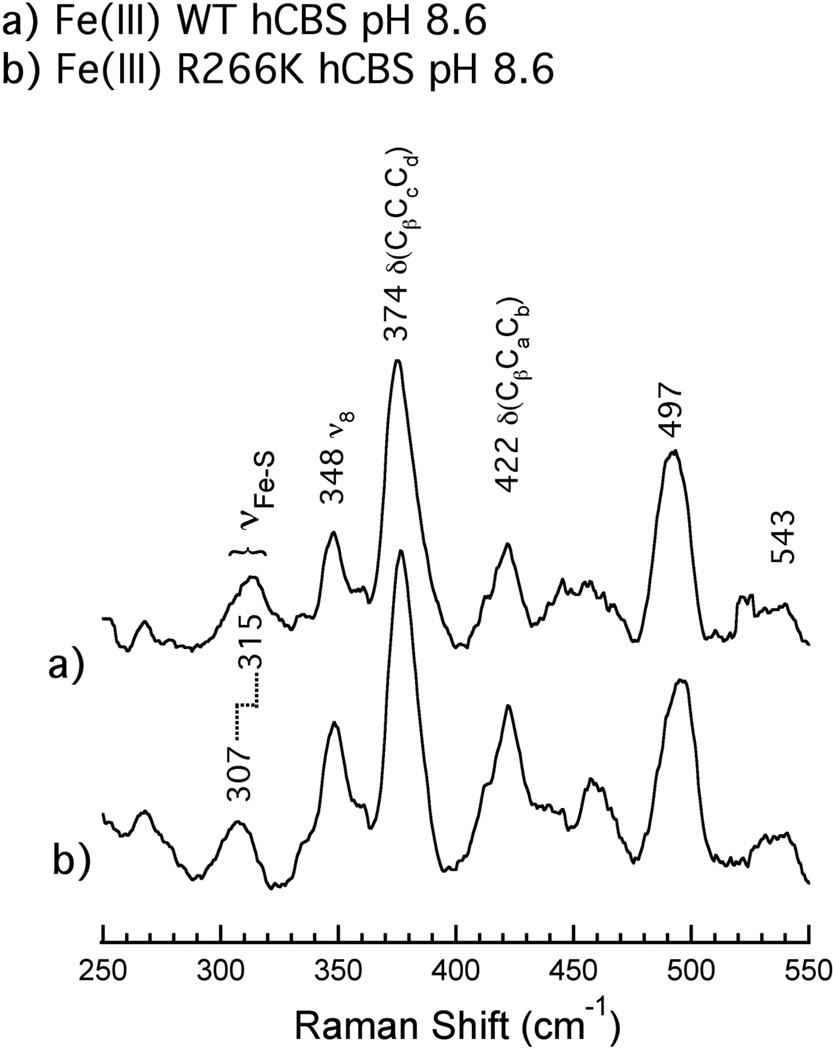
Resonance Raman spectroscopy reveals a lowering of the vibrational energy of the Fe(III)-S(Cys) resonance of R266K hCBS, suggesting a lengthening of the Fe(III)-Cys(thiolate) bond in the R266K hCBS variant. Figure 4 depicts the low-energy region of the rR spectra of Fe(III) WT (A) and R266K (B) hCBS; this region typically contains low-energy metal-ligand vibrations that are enhanced due to coupling to porphyrin vibrations. The ν(Fe-S(Cys)) resonance was previously identified at 312 cm−1 based on global 34S substitution of the C-terminal 143 amino acid-truncated version of hCBS, CBS-45 (28). In our hands, using the full-length 63 kDa WT hCBS bearing a C-term His6 tag, we observe a similar broad band centered at 315 cm−1 for ν(Fe-S(Cys)). In the Fe(III) R266K variant, this same resonance is downshifted in energy to 307 cm−1, indicative of a longer (i.e. weaker) Fe(III)-S(Cys) bond. While ν(Fe-S(Cys)) is shifted to lower energy in R266K, the relative energies and intensities of the oxidation, spin and coordination-state marker bands ν3, ν4, ν2 and ν10 remained unchanged (Figure S1), consistent with electronic absorption, MCD, and EPR spectra that indicated the same oxidation, spin and coordination states for the Fe(III) R266K variant compared to Fe(III) WT hCBS. Additionally, the rR results confirm the observations made from electronic absorption and EPR spectroscopies that suggested that the nature of the Fe(III)-Cys(thiolate) interaction had changed subtly between WT and the R266K variant (vide supra).
Figure 4.
Low frequency resonance Raman spectra of Fe(III) A) WT hCBS and B) R266K hCBS. Proteins (141 µM WT and 235 µM R266K) were in 100 mM CHES buffer and 100 mM NaCl, pH 8.6. Spectra were acquired using solution samples by excitation with a 413.1 nm line of a Kr+ laser with 10.5 mW power at the sample. All measurements were carried out with the sample immersed in a bath of ice water to reduce local heating. Peak positions were calibrated against a K2SO4 standard.
R266K Is Active but Exhibits Differential Behavior Towards Cofactors
The R266K variant, expressed as a soluble and tetrameric protein, was previously tested for cystathionine-synthesis activity (Scheme 1) (22). The results demonstrated that Fe(III) R266K hCBS displays maximal AdoMet- and PLP-responsive specific activity approximately 76% that of Fe(III) WT hCBS (Table 1). Interestingly, while the presence of exogenous PLP had no effect on WT cystathionine-synthesis activity, exogenous PLP increased basal R266K activity approximately 1.7-fold. wAdditionally, activation of the R266K variant by the hCBS allosteric activator AdoMet was significantly decreased compared to that of WT hCBS. Upon addition of AdoMet, R266K was activated approximately 2.3-fold without exogenous PLP and approximately 2.0-fold with exogenous PLP. Comparatively, upon addition of AdoMet, WT was activated approximately 3.5-fold without exogenous PLP and approximately 3.7-fold with exogenous PLP (22) (Table 1). The effect of PLP on R266K was consistent and greater than the error in the measurement.
Table 1.
Specific activity of Fe(III) WT and R266K hCBS. WT and R266K were each assayed for cysteine-synthesis activity (i.e. condensation of serine with H2S to form cysteine) and cystathionine-synthesis activity (i.e. condensation of serine with homocysteine to form cystathionine). Data are reported with and without the addition of exogenous PLP and AdoMet. Cystathionine-synthesis data are from reference (22), and are reported here for comparison. Error values represent the standard deviation from the mean value taken from three experimental repetitions on the same enzyme batch.
| hCBS Cysteine-Synthesis Activity (U)a | |||
|---|---|---|---|
| PLP | AdoMet | WT | R266K |
| − | − | 27.3 ± 0.1 | 10 ± 1 |
| − | + | 69 ± 2 | 20 ± 3 |
| + | − | 28 ± 1 | 15 ± 1 |
| + | + | 70 ± 4 | 23.4 ± 0.8 |
| hCBS Cystathionine-Synthesis Activity (U)a,b | |||
| PLP | AdoMet | WT | R266K |
| − | − | 128 ± 6 | 130 ± 10 |
| − | + | 444 ± 22 | 299 ± 35 |
| + | − | 137 ± 5 | 191 ± 17 |
| + | + | 507 ± 34 | 386 ± 26 |
1 Unit (U) = µmol product·(mg enzyme)−1·hr−1
Data from reference (22).
We tested R266K activity in an alternative cysteine-synthesis reaction (Scheme 1). In contrast with the cystathionine-synthesis results, Fe(III) R266K is significantly less active in cysteine synthesis and has maximal AdoMet- and PLP-responsive cysteine-synthesis activity approximately 34% that of Fe(III) WT (Table 1). In a manner similar to the cystathionine-synthesis results, the addition of PLP increases basal R266K cysteine-synthesis activity approximately 1.5-fold, whereas PLP has no effect on the cysteine-synthesis activity of WT Fe(III) hCBS. The presence of AdoMet increases cysteine-synthesis activity of R266K approximately 2.0-fold without exogenous PLP and approximately 2.3-fold with exogenous PLP (Table 1). Similar to the cystathionine-synthesis results, the effect of PLP on R266K in the cysteine-synthesis reaction was consistent and greater than the error in the measurement. When taken together, these results suggest that the R266K enzyme is functionally competent in the canonical and alternative reactions, but its differential behavior towards PLP and AdoMet suggests that 1) the PLP environment is disturbed by the R266K mutation; 2) the R266K enzyme may not be PLP replete; or 3) the conformation rearrangement of the R266K variant upon binding of AdoMet yields only partially activated enzyme.
Differences in the Fe(III) R266K Heme Are Transmitted to the PLP
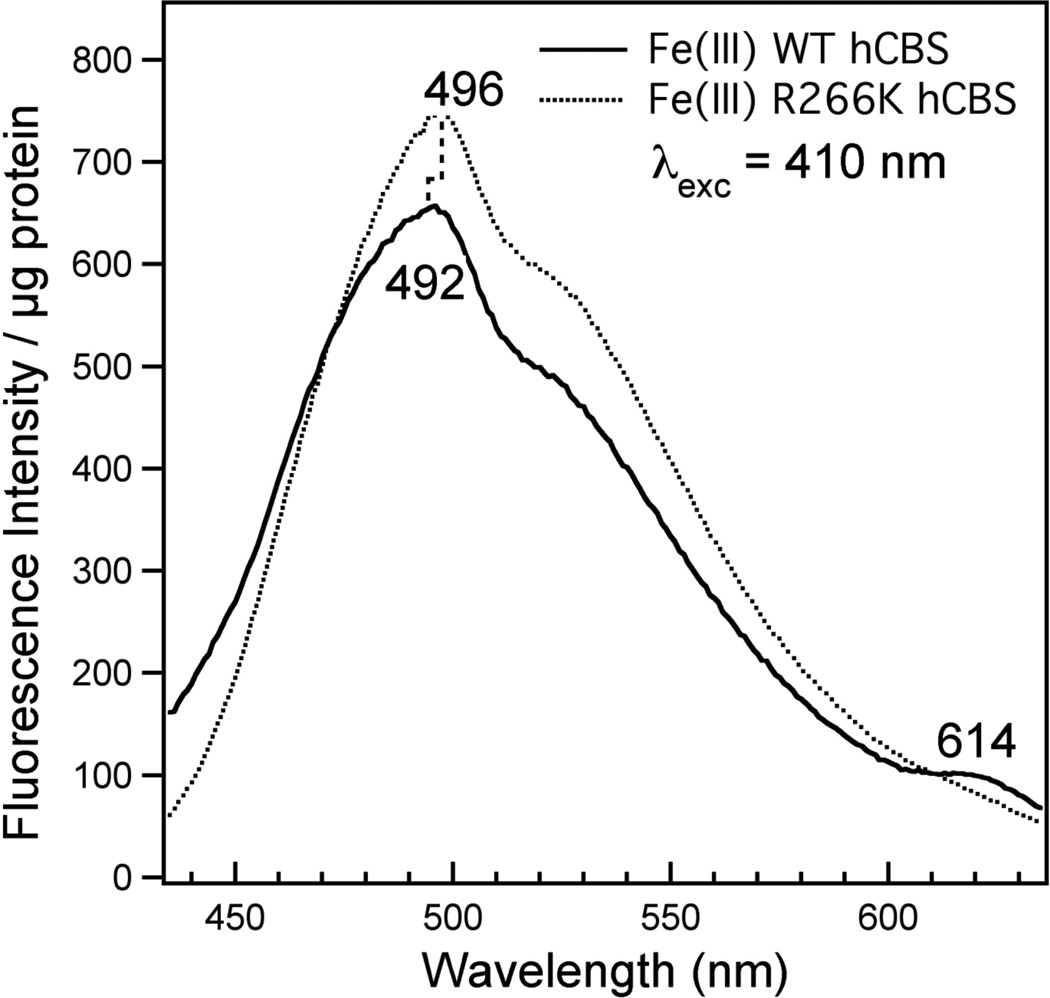
It appears that changes in the hydrogen-bonding stabilization of the Cys(thiolate) ligand in hCBS are transmitted to the PLP active site, as evidenced by a change in the PLP emission spectrum. The electronic absorption spectrum of the PLP active site is obscured by that of the heme; however, the presence and nature of the PLP may be interrogated using fluorescence spectroscopy. Figure 5 presents the fluorescence emission spectra (λexc = 410 nm) of full-length Fe(III) WT (solid) and R266K (dotted) hCBS at room temperature and pH 8.6. The PLP emission spectrum of Fe(III) WT hCBS is relatively weak and displays a broad emission envelope with maximal intensity at 492 nm as well as a smaller emission band at 614 nm; this emission spectrum is similar to, albeit slightly red-shifted from, that described previously for the ketoenamine tautomer of the PLP-Lys119 internal aldimine of hCBS-45 (36). Interestingly, the fluorescence spectrum of Fe(III) R266K hCBS is red-shifted by comparison to Fe(III) WT hCBS, and displays a new peak maximum at 496 nm as well as loss of the minor peak at 614 nm. Comparison of the WT and R266K emission spectra suggests that the internal aldimine ketoenamine tautomer is still present in R266K, consistent with the high canonical activity observed, but its environment is different. This observation implies that changes in the hydrogen-bonding network at the heme and/or the Cys(thiolate) axial ligand to the heme are transmitted to the PLP active site, a conclusion consistent with previous reports (21, 23, 25, 36–40).
Figure 5.
Emission spectra of Fe(III) WT (solid line) and R266K (dotted line) hCBS. Proteins (7.4 µM WT and 10.4 µM R266K) were in 100 mM CHES buffer and 100 mM NaCl, pH 8.6 at room temperature. Emission spectra were recorded as follows: excitation at 410 nm with a 4 mm excitation slit width; output recorded from 425 nm to 635 nm with a 2 mm emission slit width. Total emission counts were normalized to protein concentration for each sample.
Fe(II) R266K hCBS is Less Thermally Stable Than Fe(II) WT hCBS and Undergoes a More Facile Ligand-Switch
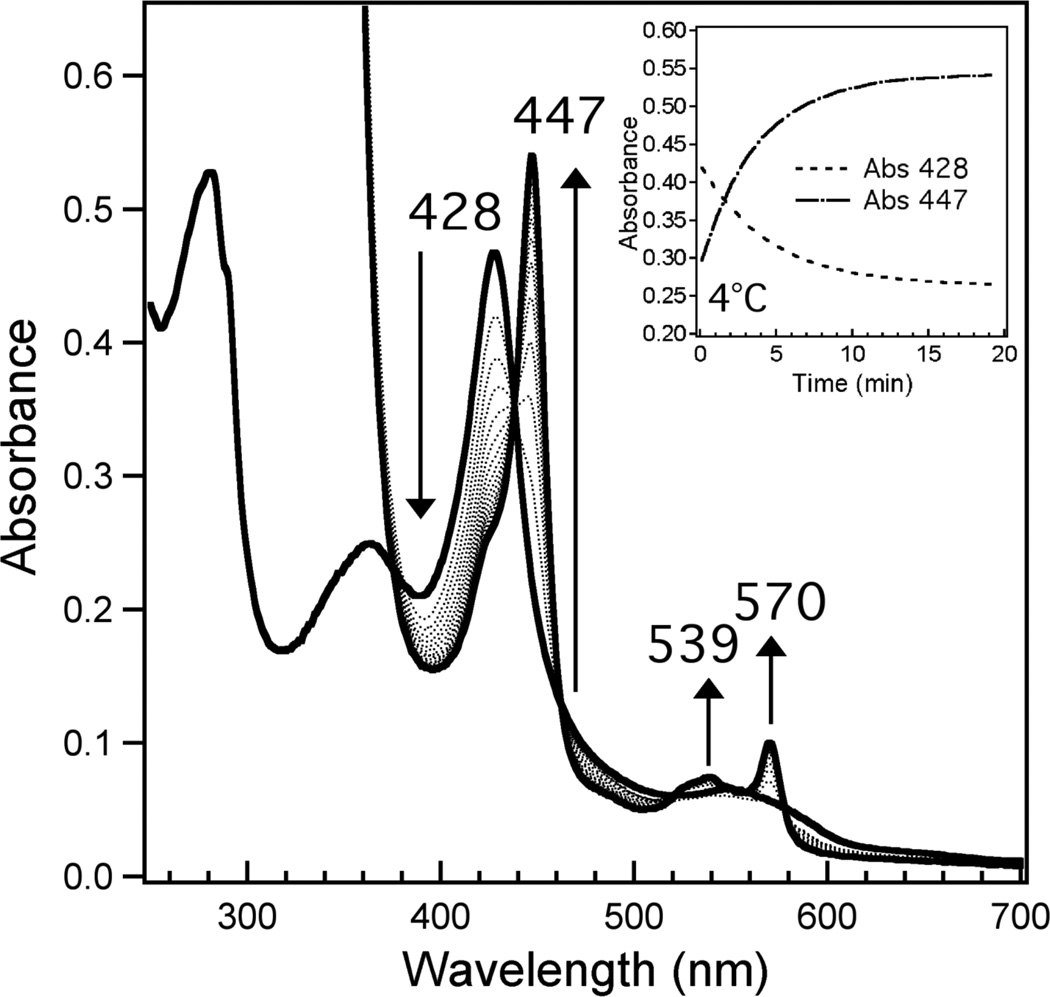
Low-temperature reduction of Fe(III) R266K hCBS is accompanied by retention of the native Cys(thiolate)/His ligation motif. Addition of a solution of the reducing agent sodium dithionite to the Fe(III) R266K variant, each equilibrated at 4°C, induces changes in the electronic absorption spectrum; the Soret band sharpens and is shifted to 447 nm with transformation of the broad α/β absorption envelope into two discrete bands at 539 nm and 570 nm (α and β, respectively, Figure 6), indicative of a six-coordinate, low-spin Fe(II) heme. This process is isosbestic and appears to follow first-order kinetics (Figure 6, inset); additionally, the positions of the highly red-shifted Soret (~450 nm) and α and β bands are almost identical to that of WT Fe(II) hCBS when it is initially reduced (16, 18), suggesting that Fe(II) R266K hCBS retains the Cys(thiolate)/His ligation motif (like that of WT) when reduced at low temperature (17).
Figure 6.
Reduction process of Fe(III) to Fe(II) R266K hCBS at 4°C. Protein (7.8 µM) was in 100 mM CHES buffer, 100 mM NaCl, pH 8.6; the reduction process was initiated by addition of a stock solution of sodium dithionite to a final concentration of 1.5 mM. Solid lines indicate the initial (428 nm Soret) and final (447 nm Soret) spectra; dotted spectra were taken at 1 min intervals after addition of reductant. Inset: time course plots showing the loss of the Fe(III) Soret (428 nm, dashed) and the growth of the Fe(II) Soret (447 nm, dashed-dot) upon introduction of sodium dithionite at 4°C.
Similar to that of WT Fe(II) hCBS, the Cys(thiolate)-ligated Fe(II) R266K species is not stable to heat treatment; in contrast, the Cys(thiolate)-ligated Fe(II) R266K variant is significantly less thermally stable. We reported previously that WT Cys(thiolate)-ligated Fe(II) hCBS undergoes an irreversible ligand-switch process at 37°C and pH 8.6 that is accompanied by a loss of the WT Fe(II) Soret at 449 nm and growth of a new Soret at 424 nm (CBS424); however, this process took nearly 48 h to reach completion at physiological temperature (25, 39). The Soret shift from 449 nm to 424 nm results from loss of the Cys(thiolate) as an Fe(II) axial ligand (25). While the identity of the new ligand is unknown, EXAFS data have confirmed that Cys52 is replaced by a neutral heme ligand that is either a nitrogenous or oxygen-containing Lewis base (41).
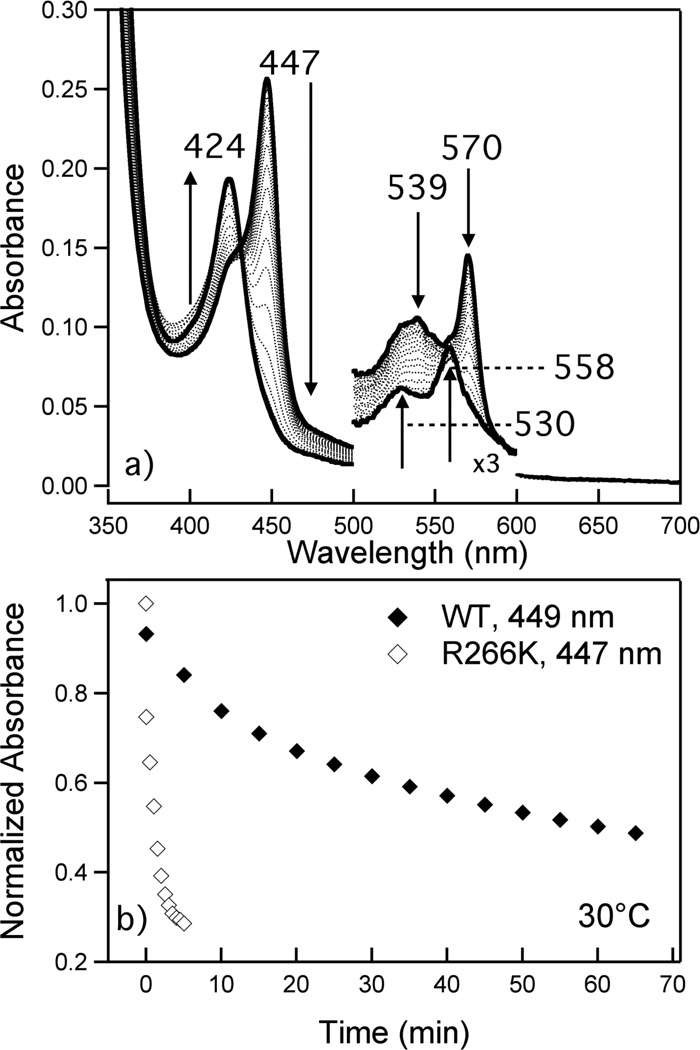
Upon elevation of temperature, Cys(thiolate)-ligated Fe(II) R266K hCBS undergoes a thermally-induced ligand switch with a more rapid apparent rate than Cys(thiolate)-ligated WT Fe(II) hCBS. Thermally treated Cys(thiolate)-ligated Fe(II) R266K loses its Soret at 447 nm with concomitant formation of new Soret, β and α positions at 424 nm, 530 nm and 558 nm, respectively (Figure 7A). Ligand switching begins spontaneously in the R266K variant at 4°C and, although this process is slow at 4°C (apparent rate not measured), the rate is more rapid near physiological temperatures (Figure 7B, Table 2). In stark contrast with Cys(thiolate)-ligated Fe(II) WT hCBS, the R266K variant converts to CBS424 almost instantaneously upon reduction at 37°C (i.e. upon equilibration of reducing agent and protein), prohibiting the direct comparison of ligand switching rates between WT and R266K at physiological temperature (39). Rate constants for the R266K ligand-switching processes were measured at 10, 20 and 30°C (Table 2), and using an Arrhenius plot, rate constants were extrapolated to 37°C (Figure S2; Table 2). Unlike the WT enzyme, in which the loss of 449 nm appears to follow a three-state triexponential decay, the R266K variant exhibits different kinetics. Loss of 447 nm peak is best modeled as a biexponential decay, suggesting that the ligand-switch process may be different in the R266K compared to WT. Alternatively, the first step of the ligand-switch process may be so facile in the R266K variant that it is not measured, and thus the apparent rate of ligand-switching is biexponential. At 37°C there is a nearly 300-fold increase in the slowest observed rate constant of the ligand-switch process for Fe(II) R266K hCBS (k = 9 ± 1.1 × 10−2 min−1) compared to that of Fe(II) WT hCBS (k = 3.0 × 10−4 min−1) (39). These data suggest that the weaker Fe-S bond in R266K significantly destabilizes Cys(thiolate) ligation in the Fe(II) form, leading to more facile ligand switching.
Figure 7.
Ligand switch process of Fe(II) R266K hCBS at 30°C. Protein (3.8 µM) was in 100 mM CHES buffer, 100 mM NaCl, pH 8.6; sodium dithionite was added to a final concentration of 1.5 mM. (A) Solid lines indicate the initial (447 nm Soret) and final (424 nm Soret) spectra; dotted spectra were taken at 1 min intervals after addition of reductant. (B) Time course plots showing the loss of the Fe(II) WT hCBS Cys(thiolate)-ligated heme Soret at 449 nm (♦) and the loss of the Fe(II) R266K hCBS Cys(thiolate)-ligated heme Soret at 447 nm (◊).
Table 2.
Experimentally fit rate constants (min−1) for loss of the Cys(thiolate)-ligated heme Soret of Fe(II) R266K hCBS. The loss of the Cys(thiolate)-ligated heme Soret was monitored at at 447 nm for Fe(II) R266K hCBS and was fit to a biexponential decay.
| Rate of Cys52 Ligand Loss (min−1) | ||||
|---|---|---|---|---|
| 37°Ca | 30°C | 20°C | 10°C | |
| k1 | (9 ± 1.1) × 10−2 | (5 ± 1.4) × 10−2 | (1.4 ± 0.1) × 10−2 | (9.1 ± 0.5) × 10−3 |
| k2 | (1.2 ± 0.5) × 100 | (6.6 ± 0.8) × 10−1 | (2.5 ± 0.3) × 10−1 | (1.0 ± 0.3) × 10−1 |
Upon equilibration at physiological temperature (37°C), Fe(II) R266K hCBS exists almost exclusively as the ligand-switched form (424 nm Soret), which prevented the measurement of k1 and k2 at this temperature; reported values were extrapolated from Arrhenius plots of k1 and k2 (Figure S3).
Discussion
The influence and importance of the hydrogen-bonding network associated with the heme iron-Cys(thiolate) interaction has been extensively explored for cytochrome P450 (Cyt P450) and nitric oxide synthase (NOS). Both enzymes possess a heme cofactor ligated by a strongly-donating mercaptide Lewis base in the axial position (42). A hydrogen-bonding network to the Cys(thiolate) heme ligand is found in a number of Cyt P450 active sites consisting of Leu (backbone amide), Gly (backbone amide), and Gln (sidechain amide) (43–46). Similarly, a number of NOS enzymes have a highly conserved Trp residue that hydrogen bonds to the Cys(thiolate) via the indole nitrogen (47–51). While a definitive consensus on the role of this hydrogen-bonding network has yet to be achieved, studies (both in vitro and in silico) of hydrogen-bonding variants at the Cys(thiolate) heme ligand of Cyt P450CAM (44, 52–54) and NOS (55–58) have demonstrated changes in the redox potential, stability, and reactivity of these proteins relative to the WT proteins.
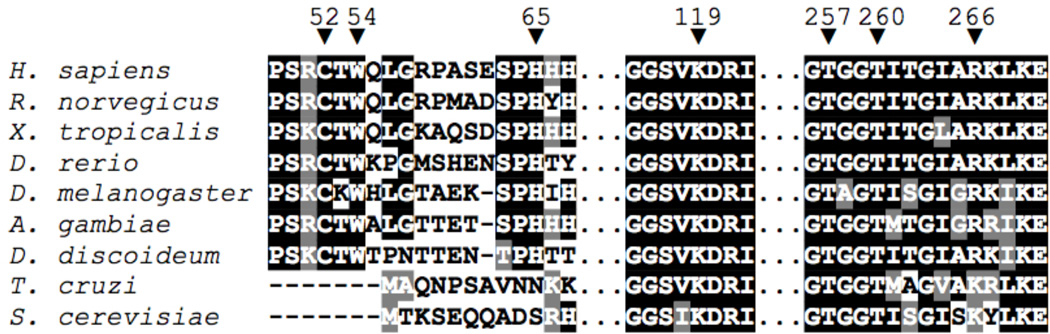
CBS exhibits similarities to and differences from Cyt P450 and NOS. Whereas Cyt P450 and NOS are monooxygenases and use heme as their active site (42), CBS is a PLP-dependent enzyme in which the PLP cofactor is spatially removed from the heme macrocycle, which is proposed to be a regulatory site (14, 15, 59, 60). The heme cofactors of Cyt P450 and NOS are each ligated by a single Cys(thiolate) ligand with labile sixth coordination site (occupied sometimes by H2O) (42, 61), whereas the CBS heme is coordinatively-saturated and is ligated by both a Cys(thiolate) and His residue (14, 15). Similar to Cyt P450 and NOS, the hCBS and DmCBS crystal structures reveal residues within hydrogen-bonding distance of the Cys(thiolate) ligand (Figure 1): the amide backbone of a Trp residue (3.59 Å distance) and the guanidinium group of an Arg side chain (3.54 Å distance) (14, 15). Additionally, sequence alignments of CBS enzymes from a number of different organisms illustrate strong conservation of both the Arg and Trp residues (Figure 8), similar to the strong conservation of hydrogen-bonding residues in the Cyt P450 and NOS enzymes (vide supra). In CBS enzymes that do not possess heme, such as those from T. cruzi and S. cerevisiae, a Lys residue is found in a position analogous to Arg266, maintaining a locus of positive charge in this area. Unlike the NOS enzymes, which use the Trp indole N-H moiety to hydrogen bond directly to the Cys(thiolate), CBS enzymes use the Trp backbone amide; however, the strong conservation of this residue (Figure 8) suggests that the sidechain bulk and aromaticity may play an important role in the heme binding pocket. Interestingly, the Arg residue that hydrogen bonds to the Cys(thiolate) heme ligand is a relatively frequent site for human mutation, and the loss or alteration of this hydrogen-bonding residue is correlated with increased levels of Hcy in human patients (62–64).
Figure 8.
Partial sequence alignment of CBS enzymes from selected species, with human enzyme numbering. Heme ligands Cys52 and His65, PLP-internal-aldimine forming Lys119, Cys52(thiolate)-contacts Trp54 and Arg266, and PLP-contacts Thr257 and Thr260 are all labeled with an inverted arrow (▼). Human (H. sapiens accession no. P35520), rat (R. norvegicus, P32232), frog (X. tropicalis, Q640V0), zebrafish (D. rerio, B7ZV69), fruit fly (D. melanogaster, Q9VRD9), mosquito (A. gambiae, Q7QEV0) and slime mold (D. discoideum, P46794) sequences all contain the necessary ligands for heme binding and have Arg266 completely conserved. Trypanosoma (T. cruzi, Q9BH24) and yeast (S. cerevisiae, P32582) CBS, which do not contain heme or its associated ligands, have a Lys residue at the position that is analogous to human Arg266. Sequence alignments were visualized using the MEGA5 program (70).
CBS missense mutations represent the most common cause of CBS deficiency and hyperhomocysteinemia (5, 62, 65), and Arg266 is the location of two missense mutations, R266G and R266K, which were originally identified in Japanese and Norwegian patients, respectively (63, 64). Interestingly, while the patient with the R266G pathogenic mutation was unresponsive to vitamin B6 (a PLP precursor) supplementation (63), the patients carrying the R266K mutation were B6-responsive (64). Previous studies on the variants R266G, R266A and R266E have clearly demonstrated that these proteins are either unstable and do not accumulate (R266G) or create soluble but heme- and PLP-deplete protein (R266A/E) (41, 66). These results showed that mutation of Arg266 to either a hydrophobic or anionic residue dramatically affected the overall stability of the polypeptide. Analysis of R266M (an isosteric but not pathogenic variation) demonstrated that retention of the salt bridge between Cys52 and Arg266 appears to be necessary for maximal AdoMet- and PLP-responsive activity. Furthermore, electronic absorption spectroscopy of the R266M variant demonstrated complete conversion to the catalytically inactive CBS-424 species upon heme reduction (41); fluorescence and rR data correlated the loss of the Arg-Cys salt bridge in the R266M variant with a shift to the inactive enolimine PLP tautomer (36). These results demonstrated that complete loss of one of the hydrogen-bonding partners to the Cys(thiolate) heme ligand disrupts CBS function.
Recently, we have shown that the position and nature of the affinity tag for expression and purification of R266K hCBS plays an important role in enabling isolation of this variant (22). Moreover, biochemical characterization of purified R266K suggested that this pathogenic mutation negatively impacts the enzyme’s saturation with PLP, its response to AdoMet, and its thermal stability, despite having similar heme saturation as compared to that of WT hCBS (22). However, none of the previous studies addressed the mode in which the structurally modest and charge invariant R266K substitution elicits changes at the heme and the PLP cofactors that are associated with diminished enzyme activity.
In this work, we have used spectroscopic methods to demonstrate that R266K hCBS bears a geometric distortion at the heme iron center, resulting in a weakening and a lengthening of the Fe(III)-Cys(thiolate) bond. The longer Fe(III)-S(Cys) bond present in the R266K variant is likely due to the closer match between the free amino acid pKa values of Cys (8.2) and Lys (10.5) versus Arg (12.5) (67). This closer pKa match between Cys and Lys would be expected to form a stronger hydrogen bond and a more fully protonated thiolate residue, thus destabilizing the Coulombic attraction between the ferric and mercaptide ions and leading to a longer Fe-S(Cys) bond. Destabilization of the Fe-Cys(thiolate) interaction results in rapid displacement of Cys52 upon reduction of the heme iron, as evidenced by the increased rate of ligand switching in Fe(II) R266K. A similar facile ligand switch was seen in the Fe(II) R266M variant (41). These in vitro findings suggest that the R266K hCBS heme would be unstable if reduced in vivo. Surprisingly, spectroelectrochemical titration showed that the redox potential of the hCBS heme (approximately −350 mV) remained virtually unchanged in the R266K variant (41); however, the facile ligand switching that we observed for this variant may call into question the accuracy of the prior measurement. A weaker Fe-thiolate interaction, with a more fully compensated charge neutralization through stronger hydrogen bonding to Lys, would be expected to make the reduction potential of the heme iron in R266K more thermodynamically favorable. Whether redox chemistry is relevant to CBS function is unclear; the unfavorable reduction potential may imply that ferric is the only relevant oxidation state of the CBS heme in vivo.
The spectral and activity data suggest that changes at the PLP active site induced by alteration of the hydrogen-bonding partner to the Cys(thiolate) heme ligand may be the source of diminished enzymatic activity in R266K hCBS. The shift in the PLP emission spectrum suggests an alteration of the PLP environment in R266K. Interestingly, this variant is more impaired in cysteine synthesis than in cystathionine synthesis. While the first substrate (Ser) is identical in each enzymatic reaction, the second substrate differs (Scheme 1). Sulfide (Km= 3.1 mM, T. cruzi CBS; 16.8 mM S. cerevisiae CBS) has been shown to be a poorer second substrate than Hcy (Km 0.9 mM, T. cruzi CBS; 2.25 mM S. cerevisiae CBS) (68, 69). We speculate that a looser PLP environment in R266K, coupled with the inefficiency of sulfide as a substrate, may facilitate loss of the PLP-Ser external aldimine from the enzyme during the cysteine-synthesis reaction. This speculation is supported by the observation that saturation of the enzyme’s active site with Ser, followed by extensive dialysis, has been used as an effective method to generate PLP-free, and thus inactive, hCBS (36). Furthermore, the greater sensitivity of R266K toward partial rescue of activity by exogenous PLP in both the cystathionine- and cysteine-synthesis reactions is consistent with a more facile loss of the PLP-Ser external aldimine.
This study directly demonstrates that even modest changes at the heme Cys(thiolate) ligand may be communicated to the PLP active site, suggesting that the heme environment modulates CBS activity. Indeed, numerous examples exist in which CBS activity is abrogated upon loss of the metal-thiolate bond (21, 23, 25, 37–39, 60), strongly implying that communication exists between the heme and PLP active site, consistent with recent proposals (23, 36, 39, 41, 60). While a consensus on the function of heme in CBS has yet to be achieved, these data, in combination with previous results, demonstrate a clear necessity for the presence of a metal-thiolate bond and an intact hydrogen-bonding network for optimal activity in heme-containing CBS.
Conclusion
In this study, we have demonstrated that the charge invariant R266K mutation generates a CBS protein that exhibits subtle spectroscopic and enzymatic changes consistent with alteration of the environment of the ligating cysteine residue. We have used electronic absorption and EPR spectroscopies to show that a change in a hydrogen bonding residue (R→K) elicits minor geometric perturbations at the Fe(III)-Cys(thiolate) bond, as evidenced by red shifts in LMCT transitions of the visible spectrum, as well as alterations of the rhombic (V) and axial distortions (Δ) of the EPR spectrum. Using rR spectroscopy, we have demonstrated that the R266K variant exhibits a longer and weaker Fe-S(Cys) bond; upon heme reduction, this weaker interaction causes a more facile rate of ligand-switching compared to WT. Finally, we have used fluorescence spectroscopy to demonstrate that even minor changes in hydrogen bonding at the heme site may be transmitted to the PLP active site; the alteration of the PLP environment is correlated with diminished canonical and alternative activities in CBS. Taken together, these data illustrate that the heme and PLP active sites are able to communicate to one another via hydrogen bonding at the heme Cys(thiolate) axial ligand.
Supplementary Material
Acknowledgements
The authors of this manuscript express thanks to Professor Thomas C. Brunold (University of Wisconsin—Madison) and his research group for their knowledge and the generous use of their instrumentation. We also thank Katherine M. Freeman for her helpful suggestions and careful reading of the manuscript. Sequence searches utilized both database and analysis functions of the Universal Protein Resource (UniProt) Knowledgebase and Reference Clusters (http://www.unprot.org) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
This work was supported by N.I.H. grant HL-065217 to J.P.K and J.N.B, by American Heart Association Grant In-Aid 09GRNT2110159 to J.P.K and by Postdoctoral Fellowship 0920079G from the American Heart Association to T. M.
Abbreviations
- Hcy
homocysteine
- AdoMet
S-adenosylmethionine
- AdoHcy
S-adenosylhomocysteine
- hCBS
human cystathionine β-synthase
- CBSDH
CBS-deficient homocystinuria
- PLP
pyridoxal-5’-phosphate
- DmCBS
cystathionine β-synthase from Drosophila melanogaster
- WT
wild-type
- EDTA
ethylenediaminetetraacetic acid
- BSA
bovine serum albumin
- DTT
dithiothreitol
- CHES
N-cyclohexyl-2-aminoethanesulfonic acid
- TCEP
tris(2-carboxyethyl)phospine
- Tris
tris(hydroxymethyl)aminomethane
- MCD
magnetic circular dichroism
- EPR
electron paramagnetic resonance
- rR
resonance Raman
- LMCT
ligand-to-metal charge-transfer
- P450CAM
camphor-binding cytochrome P450
- ImH
imidazole
- BxRcoM
regulator of CO-metabolism protein from Burkholderia xenovorans
- hRev-Erbβ
human heme-containing nuclear receptor protein
- DmE75
heme-containing nuclear receptor protein from Drosophila melanogaster
- RrCooA
heme-containing CO-sensing transcriptional regulator protein from Rhodospirillum rubrum
- CBS424
Fe(II) ligand-switched form of cystathionine β-synthase
- NOS
nitric oxide synthase
Footnotes
Supporting Information Available
Figures showing the mid-frequency window of the Fe(III) WT and R266K hCBS rR spectra (S1), and Arrhenius plots of the fit rate constants (min−1) for loss of the Cys(thiolate)-ligated heme Soret of Fe(II) R266K hCBS (S2) are available free of charge via the internet at http://pubs.acs.org.
References
- 1.Trudinger PA, Loughlin RE. Metabolism of Simple Sulphur Compounds. In: Neuberger A, editor. Comprehensive Biochemistry: Amino Acid Metabolism and Sulphur Metabolism. Amsterdam: Elsevier; 1981. [Google Scholar]
- 2.Jakubowski H. Homocysteine Thiolactone: Metabolic Origin and Protein Homocysteinylation in Humans. J. Nutr. 2000;130:377S–381S. doi: 10.1093/jn/130.2.377S. [DOI] [PubMed] [Google Scholar]
- 3.Jakubowski H. The Pathopysiological Hypothesis Of Homocysteine Thiolactone-Mediated Vascular Disease. J. Physiol. Pharmacol. 2008;59:155–167. [PubMed] [Google Scholar]
- 4.de Koning ABL, Werstuck GH, Zhou J, Austin RC. Hyperhomocysteinemia and its role in the development of atherosclerosis. Clin. Biochem. 2003;36:431–441. doi: 10.1016/s0009-9120(03)00062-6. [DOI] [PubMed] [Google Scholar]
- 5.Mudd SH, Levy HL, Kraus JP, editors. Disorders of Transsulfuration. New York: McGraw-Hill; 2001. [Google Scholar]
- 6.Clarke R, Smith AD, Jobst KA, Refsum H, Sutton L, Ueland PM. Folate, vitamin B12, and serum total homocysteine levels in confirmed Alzheimer disease. Arch. Neurol. 1998;55:1449–1455. doi: 10.1001/archneur.55.11.1449. [DOI] [PubMed] [Google Scholar]
- 7.Mattson MP, Shea TB. Folate and homocysteine metabolism in neural plasticity and neurodegenerative disorders. Trends Neurosci. 2003;26:137–146. doi: 10.1016/S0166-2236(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 8.Refsum H, Ueland PM, Nygard O, Vollset SE. Homocysteine and cardiovascular disease. Annu. Rev. Med. 1998;49:31–62. doi: 10.1146/annurev.med.49.1.31. [DOI] [PubMed] [Google Scholar]
- 9.Mills JL, McPartlin JM, Kirke PN, Lee YJ, Conley MR, Weir DG, Scott JM. Homocysteine metabolism in pregnancies complicated by neural-tube defects. Lancet. 1995;345:149–151. doi: 10.1016/s0140-6736(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 10.Kery V, Poneleit L, Meyer JD, Manning MC, Kraus JP. Binding of Pyridoxal 5'-Phosphate to the Heme Protein Human Cystathionine β-Synthase. Biochemistry. 1999;38:2716–2724. doi: 10.1021/bi981808n. [DOI] [PubMed] [Google Scholar]
- 11.Oliveriusová J, Kery V, Maclean KN, Kraus JP. Deletion Mutagenesis of Human Cystathionine beta-synthase. Impact on Activity, Oligomeric Status, and S-Adenosylmethionine Regulation. J. Biol. Chem. 2002;277:48386–48394. doi: 10.1074/jbc.M207087200. [DOI] [PubMed] [Google Scholar]
- 12.Janosik M, Kery V, Gaustadnes M, MacLean KN, Kraus JP. Regulation of human cystathionine β-synthase by S-adenosyl-L-methionine: Evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry. 2001;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- 13.Kery V, Poneleit L, Kraus JP. Trypsin Cleavage of Human Cystathionine β-Synthase into an Evolutionarily Conserved Active Core: Structural and Functional Consequences. Arch. Biochem. Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 14.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine β-synthase: A unique pyridoxal 5'-phosphate dependent heme protein. EMBO J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koutmos M, Kabil O, Smith JL, Banerjee R. Structural basis for substrated activation and regulation by cystathionine beta-synthase (CBS) domains in cystathionine β-synthase. Proc. Natl. Acad. Sci. USA. 2010;107:20958–20963. doi: 10.1073/pnas.1011448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omura T, Sadano H, Hasegawa T, Yoshida Y, Kominami S. Hemoprotein H-450 Identified as a Form of Cytochrome P-450 Having an Endogenous Ligand at the 6th Coordination Position of the Heme. J. Biochem. 1984;96:1491–1500. doi: 10.1093/oxfordjournals.jbchem.a134978. [DOI] [PubMed] [Google Scholar]
- 17.Svastits EW, Alberta JA, Kim I-C, Dawson JH. Magnetic Circular Dichroism Studies of the Active Site Structure of Hemoprotein H-450: Comparison to Cytochrome P-450 and Sensitiviy to pH Effects. Biochem. Biophys. Res. Commun. 1989;165:1170–1176. doi: 10.1016/0006-291x(89)92725-3. [DOI] [PubMed] [Google Scholar]
- 18.Kery V, Bukovska G, Kraus JP. Transsulfuration depends on heme in addition to pyridoxal 5'-phosphate. Cystathionine β-synthase is a heme protein. J. Biol. Chem. 1994;269:25283–25288. [PubMed] [Google Scholar]
- 19.Evande R, Ojha S, Banerjee R. Visualization of PLP-bound intermediates in hemeless variants of human cystathionine β-synthase: evidence that lysine 119 is a general base. Arch. Biochem. Biophys. 2004;427:188–196. doi: 10.1016/j.abb.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 20.Bruno S, Schiaretti F, Burkhard P, Kraus JP, Janosik M, Mozzarelli A. Functional Properties of the Active Core of Human Cystathionine β -Synthase Crystals. J. Biol. Chem. 2001;276:16–19. doi: 10.1074/jbc.C000588200. [DOI] [PubMed] [Google Scholar]
- 21.Taoka S, West M, Banerjee R. Characterization of the Heme and Pyridoxal Phosphate Cofactors of Human Cystathionine β-Synthase Reveals Nonequivalent Active Sites. Biochemistry. 1999;38:2738–2744. doi: 10.1021/bi9826052. [DOI] [PubMed] [Google Scholar]
- 22.Majtan T, Kraus JP. Folding and activity of mutant cystathionine β-synthase depends on the position and nature of the purification tag: Characterization of the R266K CBS mutant. Protein Expres. Purif. 2012;82:317–324. doi: 10.1016/j.pep.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith AT, Majtan T, Freeman KM, Su Y, Kraus JP, Burstyn JN. Cobalt Cystathionine ß-Synthase: A Cobalt-Substituted Heme Protein with a Unique Thiolate Ligation Motif. Inorg. Chem. 2011;50:4417–4427. doi: 10.1021/ic102586b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaitonde MK. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occuring amino acids. Biochem. J. 1967;104:627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pazicni S, Cherney MM, Lukat-Rogers GS, Oliveriusová J, Rodgers KR, Kraus JP, Burstyn JN. The Heme of Cystathionine β-synthase Likely Undergoes a Thermally Induced Redox-Mediated Ligand Switch. Biochemistry. 2005;44:16785–16795. doi: 10.1021/bi051305z. [DOI] [PubMed] [Google Scholar]
- 26.Spiro TG. Resonance Raman spectroscopic studies of heme proteins. Biochim. Biophys. Acta. 1975;416:169–189. doi: 10.1016/0304-4173(75)90006-3. [DOI] [PubMed] [Google Scholar]
- 27.Hu S, Smith KM, Spiro TG. Assignment of Protoheme Resonance Raman Spectrum by Heme Labeling in Myoglobin. J. Am. Chem. Soc. 1996;118:12638–12646. [Google Scholar]
- 28.Green EL, Taoka S, Banerjee R, Loehr TM. Resonance Raman Characterization of the Heme Cofactor in Cystathionine β-Synthase. Identification of the Fe-S(Cys) Vibration in the Six-Coordinate Low-Spin Heme. Biochemistry. 2000;40:459–463. doi: 10.1021/bi0010874. [DOI] [PubMed] [Google Scholar]
- 29.Pazicni S, Lukat-Rodgers GS, Oliveriusová J, Rees KA, Parks RB, Clark RW, Kraus JP, Rodgers KR, Burstyn JN. The redox behavior of the heme in cystathionine β-synthase is sensitive to pH. Biochemistry. 2004;43:14684–14695. doi: 10.1021/bi0488496. [DOI] [PubMed] [Google Scholar]
- 30.Shimizu T, Iizuka T, Shimada H, Ishimura Y, Nozawa T, Hatano M. Magnetic circular dichroism studies of P-450CAM. Characterization of axial ligands of ferric and ferrous low-spin complexes. Biochim. Biophys. Acta. 1981;670:341–354. doi: 10.1016/0005-2795(81)90106-9. [DOI] [PubMed] [Google Scholar]
- 31.Marvin KA, Kerby RL, Youn H, Roberts GP, Burstyn JN. The Transcription Regulator RcoM-2 from Burkholderia xenovorans Is a Cysteine-Ligated Hemoprotein That Undergoes a Redox-Mediated Ligand Switch. Biochemistry. 2008;47:9016–9028. doi: 10.1021/bi800486x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marvin KA, Reinking JL, Lee AJ, Pardee K, Krause HM, Burstyn JN. Nuclear Receptors Homo sapiens Rev-erbβ and Drosophila melanogaster E75 Are Thiolate-Ligated Heme Proteins Which Undergo Redox-Mediated Ligand Switching and Bind CO and NO. Biochemistry. 2009;48:7056–7071. doi: 10.1021/bi900697c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhawan IK, Shelver D, Thorsteinsson MV, Roberts GP, Johnson MK. Probing the heme axial ligation in the CO-sensing CooA protein with magnetic circular dichroism spectroscopy. Biochemistry. 1999;38:12805–12813. doi: 10.1021/bi991303c. [DOI] [PubMed] [Google Scholar]
- 34.Palmer G. Electron Paramagnetic Resonance of Hemoproteins. In: Lever ABP, Gray HB, editors. Iron Porphyrins, Part II. New York: VCH Publishers; 1983. pp. 45–88. [Google Scholar]
- 35.Blumberg WE, Peisach J. Low-spin compounds of heme proteins. Adv. Chem. Ser. 1971;100:271–291. [Google Scholar]
- 36.Weeks CL, Singh S, Madzelan P, Banerjee R, Spiro TG. Heme Regulation of Human Cystathionine β-Synthase Activity: Insights from Fluorescence and Raman Spectroscopy. J. Am. Chem. Soc. 2009;131:12809–12816. doi: 10.1021/ja904468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taoka S, Green EL, Loehr TM, Banerjee R. Mercuric chloride-induced spin or ligation state changes in ferric or ferrous human cystathionine β-synthase inhibit enzyme activity. J. Inorg. Biochem. 2001;87:253–259. doi: 10.1016/s0162-0134(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 38.Taoka S, Banerjee R. Characterization of NO binding to human cystathionine β-synthase: Possible implications of the efects of CO and NO binding to the human enzyme. J. Inorg. Biochem. 2001;87:245–251. doi: 10.1016/s0162-0134(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 39.Cherney MM, Pazicni S, Frank N, Marvin KA, Kraus JP, Burstyn JN. Ferrous Human Cystathionine β-Synthase Loses Activity during Enzyme Assay Due to a Ligand Switch Process. Biochemistry. 2007;46:13199–13210. doi: 10.1021/bi701159y. [DOI] [PubMed] [Google Scholar]
- 40.Puranik M, Weeks CL, Lahaye D, Kabil Ö, Shinichi T, Nielsen SB, Groves JT, Banerjee R, Spiro TG. Dynamics of Carbon Monoxide Binding to Cystathionine β-Synthase. J. Biol. Chem. 2006;281:13433–13438. doi: 10.1074/jbc.M600246200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh S, Madzelan P, Stasser J, Weeks CL, Becker D, Spiro TG, Penner-Hahn J, Banerjee R. Modulation of the heme electronic structure and cystathionine β-synthase activity by second coordination sphere ligands: The role of heme ligand switching in redox regulation. J. Inorg. Biochem. 2009;103:689–697. doi: 10.1016/j.jinorgbio.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sono M, Roach MP, Coulter ED, Dawson JH. Heme-Containing Oxygenases. Chem. Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 43.Cupp-Vickery J, Poulos TL. Structure of cytochrome P450 eryF: an enzyme involved in erythromycin biosynthesis. Nat. Struct. Biol. 1995;2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 44.Galinato MGI, Spolitak T, Ballou DP, Lehnert N. Elucidating the Role of the Proximal Cysteine Hydrogen-Bonding Network in Ferric Cytochrome P450cam and Corresponding Mutants Using Magnetic Circular Dichroism Spectroscopy. Biochemistry. 2011;50:1053–1069. doi: 10.1021/bi101911y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravichandran KG, Boddupalli SS, Hasemann CA, Peterson J, Deisenhofer J. Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450's. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 46.Hasemann CA, Ravichandran KG, Boddupalli SS, Peterson J, Deisenhofer J. Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 47.Bird LE, Ren J, Zhang J, Foxwell N, Hawkins AR, Charles IG, Stammers DK. Crystal Structure of SANOS, a Bacterial Nitric Oxide Synthase Oxygenase Protein from Staphylococcus aureus. Structure. 2002;10:1687–1696. doi: 10.1016/s0969-2126(02)00911-5. [DOI] [PubMed] [Google Scholar]
- 48.Sughamsu J, Crane BR. Structure and Reactivity of a Thermostable Prokaryotic Nitric-oxide Synthase That Forms a Long-lived Oxy-Heme Complex. J. Biol. Chem. 2006;281:9623–9632. doi: 10.1074/jbc.M510062200. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Igarashi J, Jamal J, Yang W, Poulos TL. Structural studies of constitutive nitric oxide synthases with diatomic ligands bound. J. Biol. Inorg. Chem. 2006;11:753–768. doi: 10.1007/s00775-006-0123-8. [DOI] [PubMed] [Google Scholar]
- 50.Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Tainer JA. Structure of Nitric Oxide Synthase Oxygenase Dimer with Pterin and Substrate. Science. 1998;279:2121–2126. doi: 10.1126/science.279.5359.2121. [DOI] [PubMed] [Google Scholar]
- 51.Pant K, Bilwes AM, Adak S, Stuehr DJ, Crane BR. Structure of a Nitric Oxide Synthase Heme Protein from Bacillus subtilis. Biochemistry. 2002;41:11071–11079. doi: 10.1021/bi0263715. [DOI] [PubMed] [Google Scholar]
- 52.Yoshioka S, Tosha T, Takahashi S, Ishimori K, Hori H, Morishima I. Roles of the Proximal Hydrogen Bonding Network in Cytochrome P450cam-Catalyzed Oxidation. J. Am. Chem. Soc. 2002;124:14571–14579. doi: 10.1021/ja0265409. [DOI] [PubMed] [Google Scholar]
- 53.Yoshioka S, Takahashi S, Ishimori K, Morishima I. Roles of the axial push effect in cytochrome P450cam studied with site-directed mutagenesis at the heme proximal site. J. Inorg. Biochem. 2000;81:141–151. doi: 10.1016/s0162-0134(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 54.Usharani D, Zazza C, Lai W, Chourasia M, Waskell L, Shaik S. A Single-Site Mutation (F429H) Converts the Enzyme CYP 2B4 into a Heme Oxygenase: A QM/MM Study. J. Am. Chem. Soc. 2012;134:4053–4056. doi: 10.1021/ja211905e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lang J, Driscoll D, Gélinas S, Rafferty SP, Couture M. Trp180 of endothelial NOS and Trp56 of bacterial saNOS modulate sigma bonding of the axial cysteine to the heme. J. Inorg. Biochem. 2009;103:1102–1112. doi: 10.1016/j.jinorgbio.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Brunel A, Wilson A, Henry L, Dorlet P, Santolini J. The Proximal Hydrogen Bond Network Modulates Bacillus subtilis Nitric-oxide Synthase Electronic and Structural Properties. J. Biol. Chem. 2011;286:11997–12005. doi: 10.1074/jbc.M110.195446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Couture M, Adak S, Stuehr DJ, Rousseau DL. Regulation of the Properties of the Heme-NO Complexes in Nitric-oxide Synthase by Hydrogen Bonding to the Proximal Cysteine. J. Biol. Chem. 2001;276:38280–38288. doi: 10.1074/jbc.M105341200. [DOI] [PubMed] [Google Scholar]
- 58.Hannibal L, Somasundaram R, Tejero J, Wilson A, Stuehr DJ. Influence of Heme-Thiolate in Shaping the Catalytic Properties of a Bacterial Nitric-Oxide Synthase. J. Biol. Chem. 2011;286:39224–39235. doi: 10.1074/jbc.M111.286351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banerjee R, Zou C-G. Redox regulation and reaction mechanism of human cystathionine-β-synthase: a PLP-dependent hemesensor protein. Arch. Biochem. Biophys. 2005;433:144–156. doi: 10.1016/j.abb.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 60.Kabil O, Weeks CL, Carballal S, Gherasim C, Alvarez B, Spiro TG, Banerjee R. Reversible Heme-Dependent Regulation of Human Cystathionine β-Synthase by a Flavoprotein Oxidoreductase. Biochemistry. 2011;50:8261–8263. doi: 10.1021/bi201270q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Omura T. Heme-thiolate proteins. Biochem. Biophys. Res. Commun. 2005;338:404–409. doi: 10.1016/j.bbrc.2005.08.267. [DOI] [PubMed] [Google Scholar]
- 62.Mudd SH. Hypermethioninemias of Genetic and Non-Genetic Origin: A Review. Am. J. Med. Genet. Part C Semin. Med. Genet. 2011;157:3–32. doi: 10.1002/ajmg.c.30293. [DOI] [PubMed] [Google Scholar]
- 63.Katsushima F, Oliveriusova J, Sakamoto O, Ohura T, Kondo Y, Iinuma K, Kraus E, Stouracova R, Kraus JP. Expression study of mutant cystathionine β-synthase found in Japanese patients with homocystinuria. Mol. Genet. Metab. 2006;87:323–328. doi: 10.1016/j.ymgme.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 64.Kim CE, Gallagher PM, Guttormsen AB, Refsum H, Ueland PM, Ose L, Følling I, Whitehead AS, Tsai MY, Kruger WD. Functional Modeling of Vitamin Responsiveness in Yeast: A Common Pyridoxine-Responsive Cystathionine β-Synthase Mutation in Homocystinuria. Hum. Mol. Genet. 1997;6:2213–2221. doi: 10.1093/hmg/6.13.2213. [DOI] [PubMed] [Google Scholar]
- 65.Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine β-synthase. Biochim. Biophys. Acta. 2003;1647:206–213. doi: 10.1016/s1570-9639(03)00048-7. [DOI] [PubMed] [Google Scholar]
- 66.Ozaki S-i, Sakaguchi C, Nakahara A, Yoshiya M. Mutagenesis Studies of Human Cystathionine β-Synthase: Residues Important for Heme Binding and Substrate Interaction. Protein Peptide Lett. 2010;17:351–355. doi: 10.2174/092986610790780233. [DOI] [PubMed] [Google Scholar]
- 67.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 5th ed. New York: W. H. Freeman and Company; 2008. [Google Scholar]
- 68.Nozaki T, Shigeta Y, Saito-Nakano Y, Imada M, Kruger WD. Characterization of Transsulfuration and Cysteine Biosynthetic Pathways in the Protozoan Hemoflagellate Trypanosoma cruzi. J. Biol. Chem. 2001;276:6516–6523. doi: 10.1074/jbc.M009774200. [DOI] [PubMed] [Google Scholar]
- 69.Ono B-I, Kijima K, Inoue T, Miyoshi S-I, Matsuda A, Shinoda S. Purification of properties of Saccaromyces cerevisiae cystathionine β-synthase. Yeast. 1994;10:333–339. doi: 10.1002/yea.320100306. [DOI] [PubMed] [Google Scholar]
- 70.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.