Abstract
Cockayne syndrome (CS) is a rare autosomal recessive disorder characterized by progressive multisystem degeneration and segmental premature aging. Mutations in the DNA repair gene ERCC6 are responsible for the majority of CS cases reported. In this study, we identified 4 patients presenting with CS from 2 Old Order Amish families. Sequence analysis of the ERCC6 gene revealed 2 novel mutations associated with the disorder in these patients. The patients from family 1 were homozygous for a splice-site mutation, c.2709 + 1G>T, in intron 14 of ERCC6, whereas the patients from family 2 were compound heterozygous for c.2709 + 1G>T and a short deletion in exon 5 (c.1293_1320del). Our findings provide evidence of allelic heterogeneity in Old Order Amish, which is extremely uncommon for a rare condition in an isolated founder population.
Key Words: Cockayne syndrome, ERCC6, Old Order Amish, Sequence analysis
Cockayne syndrome (CS; OMIM 133540) is a rare autosomal recessive multisystem disorder characterized by growth failure, mental retardation, progressive neurological dysfunction, premature aging, and photosensitivity [Nance and Berry, 1992]. The underlying cause of the disease is a defect in transcription-coupled DNA repair, specifically in the nucleotide excision repair pathway [Venema et al., 1990; van Hoffen et al., 1993; Evans and Bohr, 1994]. Mutations in 2 excision-repair cross-complementing genes, ERCC6 and ERCC8, have been associated with the majority of CS cases [Troelstra et al., 1992, 1993; Henning et al., 1995]. Among the reported CS cases with mutations in ERCC6 or ERCC8 genes, about two-thirds have mutations in the ERCC6 gene [Laugel et al., 2010].
Here, we report the identification of 2 novel mutations in the ERCC6 gene in the Old Order Amish segregating CS.
Case Report
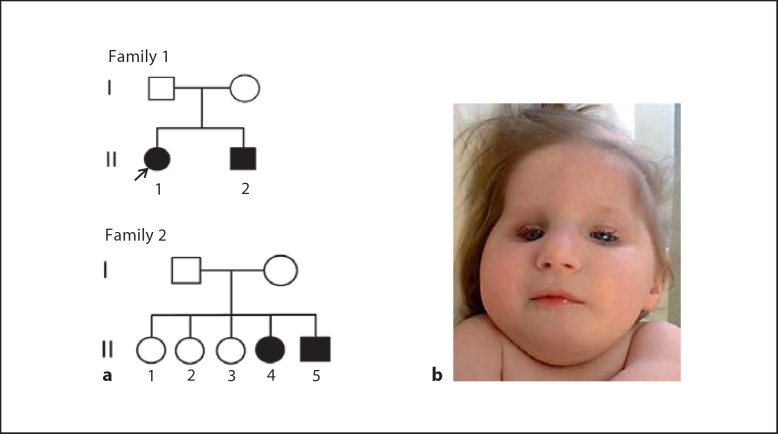
A total of 4 patients from 2 sibships were identified at our clinic (fig. 1a). All the patients were Old Order Amish from the State of Ohio. The proband (II-1 in family 1, fig. 1a) was a 2-year-old female who presented at our clinic with developmental delay and failure to thrive. She was a term newborn with a weight of 2,722 g (8th percentile), length of 47 cm (13th percentile) and a head circumference of 32.5 cm (6th percentile) at birth after an uneventful pregnancy and delivery. Poor feeding, fussiness, low-pitched and weak cry, floppiness, and lack of eye contact were noted by the parents soon after the birth. Other clinical features found during early infancy included hypotonia, nystagmus, cataracts, cutaneous photosensitivity, and significant delays in growth and development. Upon examination at presentation, the patient weighed 7.1 kg (<1st percentile) with a length of 70.5 cm (<1st percentile) and head circumference of 39.5 cm (<1st percentile) with obvious facial dysmorphism including microcephaly, micrognathia, high palate, hypoplastic teeth, and sunken eyes, consistent with classic facial features of CS as shown in figure 1b. Her upper arms and legs appeared ‘chubby’ while the forearms and lower legs were thin with significant muscle atrophy. Deep tendon reflexes were absent. The other 3 patients showed similar signs and symptoms as the proband and were clinically diagnosed as CS.
Fig. 1.
The Old Order Amish families with CS. a Pedigrees of families 1 and 2. Filled symbols represent affected individuals, open symbols represent unaffected individuals. An arrow indicates the proband. b Facial view of the proband showing the typical facial features of CS.
Materials and Methods
The study was approved by the Institutional Review Board of DDC Clinic for Special Needs Children. Peripheral blood samples were obtained from all participants after obtaining written informed consent. Genomic DNA was isolated from white blood cells using standard methods. PCR primers were designed to amplify each of the 21 protein-coding exons of the ERCC6 gene and their flanking intronic sequences. Direct sequencing of the PCR products was performed using an ABI 310 Sequencer. The identified mutation was verified with repeat PCR amplification and sequencing in both orientations. The base-pair number of the mutation site was determined according to the GenBank mRNA reference sequence NM_000124.
Results and Discussion
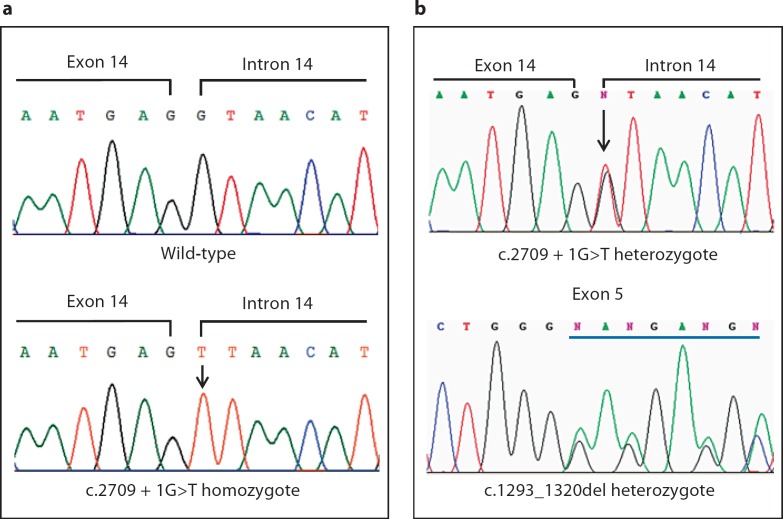
We first carried out whole gene sequencing analysis of the ERCC6 gene using genomic DNA from the proband. Direct sequencing revealed a homozygous splice-donor site mutation, c.2709 + 1G>T, in intron 14 of the ERCC6 gene (fig. 2a). As illustrated by the effects of a similar mutation (c.3330 + 1G>T) in the MYBPC3 gene in our previous report [Xin et al., 2007], abolition of the donor splice site results in skipping of the upstream exon, in this case exon 14, which leads to a frameshift in translation. Targeted c.2709 + 1G>T mutation analysis in family 1 showed that the affected sibling of the proband was homozygous and both parents were heterozygous for the c.2709 + 1G>T mutation.
Fig. 2.
Identification of 2 novel mutations in the ERCC6 gene. Sequence chromatograms showing the homozygous c.2709 + 1G>T mutation identified in the patients from family 1 (a) and the compound heterozygous mutations of c.2709 + 1G>T and c.1293_1320del in affected individuals from family 2 (b). Affected residuals are underlined or indicated by an arrow.
Given the high degree of consanguinity in the Old Order Amish, we next performed the same c.2709 + 1G>T targeted mutation test for the 2 affected individuals from family 2. Surprisingly, the sequencing results showed that both of them carried only 1 mutated allele of c.2709 + 1G>T (fig. 2b). Further sequencing analysis of the full-length ERCC6 gene revealed a novel heterozygous mutation, a 28-bp deletion in exon 5 (c.1293_1320del), in both patients (fig. 2b). The 28-bp deletion led to a frameshift and creation of a premature termination in translation (p.Glu432Lysfs*24). Targeted mutation analysis in both parents showed that the father was heterozygous for the 28-bp deletion while the mother carried the heterozygous c.2709 + 1G>T mutation, indicating the biparental origin of the described mutations. Of the 3 unaffected siblings, 2 were wild-type normal and 1 was heterozygous for the c.2709 + 1G>T variant.
Genetically, the identification of 2 distinct mutations in the Old Order Amish was unexpected. The Old Order Amish are descended from a very small number of 18th-century founders who migrated from Europe. They live in many small endogamous demes, which represent a collection of genetically closed communities across North America. Their population has experienced very rapid growth and has had very small degrees of immigration. Under these circumstances, particularly in view of the high inbreeding rates and restricted founder pool, the chance that any rare variant or mutation will become homozygous is extremely large in comparison with outbred populations. Our finding in this study challenges the classical view of a single founder mutation segregating in such an isolate. Since the Amish have been subjects of diverse genetic studies for many years, an important implication of our study is that in such small isolated populations, homozygosity mapping studies may be compromised unless mutational diversity is taken into account.
In conclusion, we identified 2 novel mutations, homozygosity for c.2709 + 1G>T and compound heterozygosity for c.2709 + 1G>T and c.1293_1320del, in the ERCC6 gene associated with CS in Old Order Amish. Our findings indicate the possibility of allelic heterogeneity in the Old Order Amish, which is highly uncommon for a rare condition in an isolated founder population.
Acknowledgements
We thank the family for their participation and helpful cooperation in this study. We acknowledge the help of Alicia Bright in data collection. This work was supported in part by The Elisabeth Severance Prentiss Foundation, The Reinberger Foundation, and the Leonard Krieger Fund of the Cleveland Foundation (L2009-0078).
References
- Evans MK, Bohr VA. Gene-specific DNA repair of UV-induced cyclobutane pyrimidine dimers in some cancer-prone and premature-aging human syndromes. Mutat Res. 1994;314:221–231. doi: 10.1016/0921-8777(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, et al. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995;82:555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- Laugel V, Dalloz C, Durand M, Sauvanaud F, Kristensen U, et al. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31:113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- Troelstra C, Hesen W, Bootsma D, Hoeijmakers JH. Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne's syndrome group B. Nucleic Acids Res. 1993;21:419–426. doi: 10.1093/nar/21.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoffen A, Natarajan AT, Mayne LV, van Zeeland AA, Mullenders LH, Venema J. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin B, Puffenberger E, Tumbush J, Bockoven JR, Wang H. Homozygosity for a novel splice site mutation in the cardiac myosin-binding protein C gene causes severe neonatal hypertrophic cardiomyopathy. Am J Med Genet Part A. 2007;143A:2662–2667. doi: 10.1002/ajmg.a.31981. [DOI] [PubMed] [Google Scholar]