Abstract
Escherichia coli TC44, a derivative of W3110, was engineered for the production of pyruvate from glucose by combining mutations to minimize ATP yield, cell growth, and CO2 production (ΔatpFH ΔadhE ΔsucA) with mutations that eliminate acetate production [poxB::FRT (FLP recognition target) ΔackA] and fermentation products (ΔfocA-pflB ΔfrdBC ΔldhA ΔadhE). In mineral salts medium containing glucose as the sole carbon source, strain TC44(ΔfocA-pflB ΔfrdBC ΔldhA ΔatpFH ΔadhE ΔsucA poxB::FRT ΔackA) converted glucose to pyruvate with a yield of 0.75 g of pyruvate per g of glucose (77.9% of theoretical yield; 1.2 g of pyruvate liters–1·h–1). A maximum of 749 mM pyruvate was produced with excess glucose. Glycolytic flux was >50% faster for TC44 producing pyruvate than for the wild-type W3110 during fully aerobic metabolism. The tolerance of E. coli to such drastic changes in metabolic flow and energy production implies considerable elasticity in permitted pool sizes for key metabolic intermediates such as pyruvate and acetyl-CoA. In strain TC44, pyruvate yield, pyruvate titer, and the rate of pyruvate production in mineral salts medium were equivalent or better than previously reported for other biocatalyts (yeast and bacteria) requiring complex vitamin feeding strategies and complex nutrients. TC44 offers the potential to improve the economics of pyruvate production by reducing the costs of materials, product purification, and waste disposal.
Keywords: metabolic engineering, glycolytic flux, fermentation, ATPase
Recent trends toward the production of “green” chemicals require the development of innovative synthesis techniques that are both efficient and cost-effective. Traditional chemical companies have begun to develop infrastructures for the production of compounds by using biocatalytic processes (1–3). Considerable progress has been reported toward new processes for commodity chemicals such as ethanol (4, 5), lactic acid (6–8), 1,3-propanediol (9, 10), and adipic acid (11). In addition, advances have been made in the genetic engineering of microbes for higher value specialty compounds, such as polyketides (12, 13) and carotenoids (14, 15). Each of these chemicals can be made directly or indirectly from the same central metabolite: pyruvic acid.
The metabolic fate of pyruvate, a key branch point in central carbon metabolism, is regulated in large part by the redox state of the cell. During the fermentative metabolism of glucose, NADH levels are high, and >95% of pyruvate is used as terminal electron acceptors for NADH oxidation; <5% is partitioned into biosynthesis. During oxidative metabolism, pyruvate is oxidatively decarboxylated by the pyruvate dehydrogenase complex to produce CO2 and acetyl-CoA. Oxygen or other external compounds serve as the terminal electron acceptor, and NADH levels remain relatively low. Up to half of the glucose carbon is used for biosynthesis, the remainder being fully oxidized by the tricarboxylic acid cycle to CO2.
Pyruvic acid is currently manufactured for use as a food additive, nutriceutical, and weight-control supplement (16, 17). Pyruvic acid can also be used as a starting material for the synthesis of amino acids, such as alanine, tyrosine, phenylalanine, and tryptophan, (16) and for acetaldehyde production (18). Pyruvate is produced commercially by both chemical and microbial processes (16). Chemical synthesis from tartrate involves toxic solvents and is energy-intensive (19, 20), with an estimated production cost of $8,650 per ton of pyruvate (16). Microbial pyruvate production is based primarily on two microorganisms, a multivitamin auxotroph of the yeast Torulopsis glabrata (21) and a lipoic acid auxotroph of Escherichia coli containing a mutation in the F1 ATPase component of (F1F0)H+-ATP synthase (22). Both strains require precise regulation of media composition during fermentation and complex supplements. Estimated production costs for pyruvate production with these strains is 14.5% ($1,255 per ton of pyruvate) that for chemical synthesis (16). Recently, Tomar et al. (23) have described a new E. coli mutant for pyruvate production from glucose and acetate in complex medium. This strain contains mutations in phosphoenolpyruvate carboxylase (ppc), pyruvate dehydrogenase (aceF), and alcohol dehydrogenase (adhE).
Nutrients in culture medium often represent a major cost associated with commercial fermentations. The use of a mineral salts medium and an inexpensive carbon source offers the potential to improve the economics of many biological processes by reducing the costs of materials, product purification, and waste disposal (24). In this article, we describe the development of a biocatalyst for the efficient production of pyruvate from glucose that requires only simple mineral salts as nutrients.
Materials and Methods
Microorganisms and Media. E. coli W3110 (ATCC 27325) derivatives and LY01 (25) (Table 1) were grown on mineral salts medium (26) containing glucose (2% in plates, 3% in broth). When needed for pH control, 3-[N-morpholino]propanesulfonic acid (0.1 M, pH 7.4) was added but was not included in pH-controlled fermentations. During plasmid and strain construction, cultures were grown in LB broth or on LB plates (1.5% agar) (27). Glucose (2%) was added to LB medium for all strains containing mutations in (F1F0)H+-ATP synthase. Antibiotics were included as appropriate (50 mg/liter kanamycin, 50 mg/liter ampicillin, 50 mg/liter apramycin, and 12.5 or 6.25 mg/liter tetracycline).
Table 1. Sources and characteristics of strains and plasmids.
| Strains/plasmids | Relevant characteristics | Ref. |
|---|---|---|
| Strains | ||
| W3110 | K12 wild type | ATCC 27325 |
| TOP10F′ | laclq (episome) | Invitrogen |
| LY01 | E. coli B, frd pfl::pdoZm adhEZm cat | 25 |
| LY74 | W3110, ΔpoxB::FRT-tet-FRT | This study |
| SZ61 | W3110, ΔackA::FRT-tet-FRT | 8 |
| TC36 | W3110, (Succ+), ΔfocA-pflB::FRT ΔfrdBC ΔldhA Δatp(FH)::FRT ΔadhE::FRT ΔsucA::FRT | 26 |
| TC37 | W3110, (Succ+), ΔfocA-pflB::FRT ΔfrdBC ΔldhA Δatp(FH)::FRT ΔadhE::FRT ΔsucA::FRT ΔackA::FRT-tet-FRT | This study |
| TC38 | W3110, (Succ+), ΔfocA-pflB::FRT ΔfrdBC ΔldhA Δatp(FH)::FRT ΔadhE::FRT ΔsucA::FRT ΔackA::FRT | This study |
| TC41 | W3110, (Succ+), ΔfocA-pflB::FRT ΔfrdBC ΔldhA Δatp(FH)::FRT ΔadhE::FRT ΔsucA::FRT ΔpoxB::FRT-tet-FRT | This study |
| TC42 | W3110, (Succ+), ΔfocA-pflB::FRT ΔfrdBC ΔldhA Δatp(FH)::FRT ΔadhE::FRT ΔsucA::FRT ΔpoxB::FRT | This study |
| TC43 | W3110, (Succ+), ΔfocA-pflB::FRT ΔfrdBC ΔldhA Δatp(FH)::FRT ΔadhE::FRT ΔsucA::FRT ΔpoxB::FRT ΔackA::FRT-tet-FRT | This study |
| TC44 | W3110, (Succ+), ΔfocA-pflB::FRT ΔfrdBC ΔldhA Δatp(FH)::FRT ΔadhE::FRT ΔsucA::FRT ΔpoxB::FRT ΔackA::FRT | This study |
| Plasmids | ||
| pCR2.1-TOPO | bla kan, TOPO TA cloning vector | Invitrogen |
| pFT-A | bla flp low-copy vector containing recombinase and temperature-conditional pSC101 replicon | 27 |
| pKD46 | bla γ β exo low-copy vector containing red recombinase and temperature-conditional pSC101 replicon | 28 |
| pLO12065 | bla, Smal fragment with FRT flanked tet gene | 5 |
| pLO12075 | bla kan poxB | This study |
| pLO12078 | bla poxB | This study |
| pLO12080 | bla poxB::FRT-tet-FRT | This study |
| pLO12403 | bla | 29 |
Genetic Methods. Standard methods were used for plasmid construction, phage P1 transduction, electroporation, and PCR (27, 28). Coding regions for ackA and poxB were amplified by using ORFmer primers (Sigma-Genosys, The Woodlands, TX) and initially cloned into pCR2.1-TOPO (Invitrogen). Chromosomal integration of mutated genes was facilitated by pKD46 containing an arabinose-inducible Red recombinase (29). Integrants were selected by using antibiotic resistance, screened for appropriate antibiotic resistance markers and phenotypic traits, and verified by analyses of PCR products and fermentation profiles. FRT (FLP recognition target)-flanked antibiotic resistance genes were deleted by using FLP recombinase (30, 31).
Disruption of Pyruvate Oxidase (poxB). A single derivative of pCR2.1-TOPO was selected in which the amplified poxB gene was oriented in the same direction as the lac promoter (pLOI2075). To eliminate extraneous BsaBI sites in the vector, the small EcoRI fragment containing poxB was ligated into the corresponding site of pLOI2403 to produce pLOI2078. The SmaI fragment from pLOI2065 containing a tet gene flanked by FRT sites was ligated into the unique BsaBI site in poxB to produce pLOI2080. Plasmid pLOI2080 served as a template for the amplification of poxB::FRT-tet-FRT (3.4 kbp) with poxB primers. Amplified DNA was integrated into E. coli W3110(pKD46) as described (26) to produce LY74. The poxB::FRT-tet-FRT mutation in LY74 was transduced into TC36 to produce TC41. The tet gene was removed by using FLP recombinase, and the resulting strain was designated TC42 (ΔfocA-pflB::FRT ΔfrdBC::FRT Δ ldhA ΔatpFH::FRT ΔadhE::FRT ΔsucA::FRT poxB::FRT).
Deletion of ackA (Acetate Kinase). The ΔackA::FRT-tet-FRT mutation was transduced from SZ61 (8) into TC36 and TC42 to produce strains TC37 (ΔfocA-pflB::FRT ΔfrdBC::FRT Δ ldhA ΔatpFH::FRT ΔadhE::FRT ΔsucA::FRT ΔackA::FRT-tet-FRT) and TC43 (ΔfocA-pflB::FRT ΔfrdBC::FRT Δ ldhA ΔatpFH::FRT ΔadhE::FRT ΔsucA::FRT poxB::FRT ΔackA::FRT-tet-FRT), respectively. After removal of the tet gene by using FLP recombinase, resulting strains were designated TC38 (ΔfocA-pflB::FRT ΔfrdBC::FRT ΔldhA ΔatpFH::FRT ΔadhE::FRT ΔsucA::FRT ΔackA::FRT) and TC44 (ΔfocA-pflB::FRT ΔfrdBC::FRT ΔldhA ΔatpFH::FRT ΔadhE::FRT ΔsucA::FRT poxB::FRT ΔackA::FRT), respectively.
Fermentation. Fermentations (5 and 10 liters) were conducted at 37°C (dual Rushton impellers, 350–450 rpm; pH 7.0) in BioFlo 3000 fermentors (New Brunswick Scientific). Unless stated otherwise, dissolved oxygen levels were 100% of air saturation at the time of inoculation and allowed to fall to 5% of air saturation during cell growth with continuous air sparging [0.2 vvm (vessel volume per minute)]. This level of oxygen was maintained by mixing O2 with air at a constant flow rate of 1.0 liter/min. Broth pH was controlled with 11.4 M KOH. For fed-batch studies, glucose was added from a sterile 4-M stock. Two fed batch regimes were investigated: (i) 3% initial glucose followed by the addition of 3% glucose after 15 h (6% total glucose) and (ii) 3% initial glucose with the addition of 590 ml of 4 M glucose at a constant rate over a 20-h period (9.8% total glucose).
Seed cultures prepared as described (26) were used to provide an inoculum of 16.5 mg of dry cell weight per liter. Broth samples were removed to measure organic acids, residual glucose, and cell mass. Volumetric and specific rates were estimated from measured values for glucose and acetate by using prism (Graph-Pad, San Diego). A smooth curve was generated with 10 points per min (Lowess method) to fit measured results. The first derivative (acetate or glucose versus time) of each curve served as an estimate of volumetric rate. Specific rates (mM·liter–1·h–1·mg–1 dry cell weight) were calculated by dividing volumetric rates by respective values for cell mass.
Analyses. Organic acids and glucose were measured with a Hewlett–Packard HPLC (HP 1090 series II) equipped with a UV monitor (210 nm) and refractive index detector (26). Cell mass was estimated with a Bausch & Lomb Spectronic 70 spectrophotometer (1.0 OD550 nm is equivalent to 0.33 g/liter dry cell weight).
Results
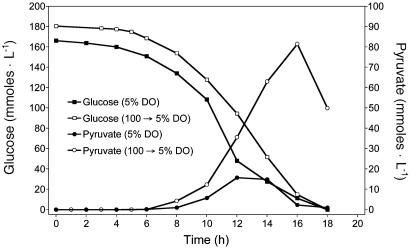
Pyruvate As a Coproduct During Acetate Fermentations. E. coli TC36 (ΔfocA-pflB ΔfrdBC ΔldhA ΔatpFH ΔadhE ΔsucA) was previously engineered from W3110 (wild type) for the production of acetate (Fig. 1A) by combining chromosomal deletions that minimize cell yield, oxygen consumption, CO2 evolution, and reduced fermentation products (26). In this strain, glycolytic flux was higher than the parent because of a deletion of genes (ΔatpFH) encoding two membrane proteins that couple the F1 and F0 components of the (F1F0)H+-ATP synthase complex. This mutation eliminated ATP production by oxidative phosphorylation and created an active, cytoplasmic F1-ATPase (Fig. 1 B and C). Glycolytic flux in TC36 exceeded the capacity for acetate production under our initial test conditions (5% air saturation at inoculation and during fermentation), resulting in the transient accumulation of ≈16 mM pyruvate near the end of exponential growth (Fig. 2). The peak level of pyruvate was increased to 81 mM (Fig. 2) by inoculating the fermentor at an initial dissolved oxygen level of 100% air saturation (rather than 5% of saturation) with continuous air sparging until the oxygen level declined to 5% air saturation, then adding oxygen to maintain 5% of air saturation. Under this condition, pyruvate yield at the peak was 25% of the maximum theoretical yield (Fig. 2).
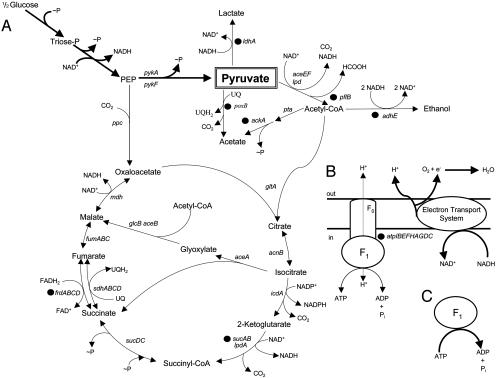
Fig. 1.
Summary of central metabolism in E. coli. (A) Carbon metabolism. (B) Oxidative phosphorylation. (C) Cytoplasmic F1ATPase subunit (active). Filled circles indicate metabolic steps that have been blocked by mutation or deletion of genes encoding respective enzymes.
Fig. 2.
Effect of oxygen level on pyruvate production by TC36. Cells were inoculated into fermentation broth at 100% air saturation and continuously sparged with air until the oxygen levels declined to 5% saturation. At this time, oxygen was blended to maintain 5% saturation during the remaining period of incubation (open symbols). Alternatively, media was sparged with a mixture of air and nitrogen to provide 5% air saturation before inoculation. Sparging was switched to air and oxygen as needed to maintain 5% air saturation (filled symbols).
We have explored additional genetic modifications of TC36 to further increase the efficiency of pyruvate production from glucose. Although many metabolic routes lead to acetate, two primary routes are present in E. coli (Fig. 1A): (i) conversion of acetyl-CoA to acetate by phosphotransacetylase (pta) and acetate kinase (ackA) and (ii) direct oxidation of pyruvate to acetate by pyruvate oxidase (poxB) (Fig. 1A). Derivatives of TC36 were constructed with mutations in both pathways.
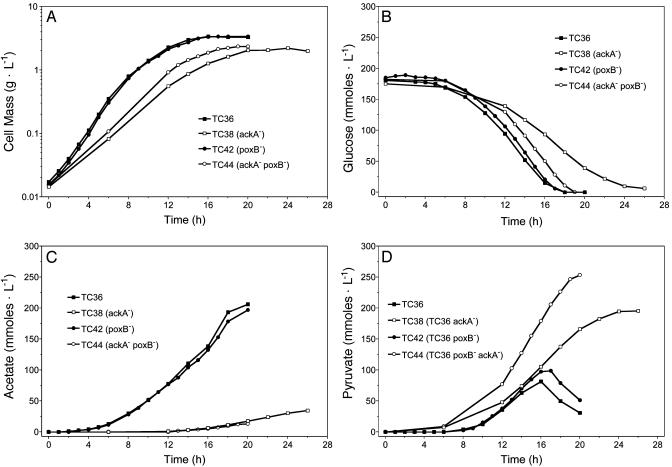
Acetate Kinase (ΔackA) Enhances Pyruvate Production but Inhibits Glycolysis. To block acetate production by the acetate kinase pathway, the central region of the ackA gene was deleted in TC36 to produce TC38. Acetate production was reduced by 85% (Fig. 3C and Table 2), consistent with the acetate kinase pathway being the dominant route for acetate production in TC36. However, this deletion also reduced net ATP production (estimated) by 30% (Fig. 1A), reduced cell yield by 36% (Fig. 3A and Table 2), and reduced the maximum specific growth rate by 45% (Table 3). Glycolytic flux was reduced by 45% (Table 3), increasing the time required to complete fermentations from 18 h for TC36 to 24 h for TC38 (Fig. 3B). Although both volumetric and specific rates of glucose metabolism were lower for TC38 (Table 3), pyruvate yield was 5.5-fold higher (Table 2 and Fig. 3D), and the specific rate of pyruvate production was 4-fold higher (Table 3) than for TC36. Small amounts of 2-oxoglutarate, succinate, and fumarate were produced by both strains. Ten percent to 15% of the carbon was not recovered as cell mass or acidic fermentation products and is presumed to be lost as CO2 because of metabolite cycling. With strain TC38, the pyruvate yield (195 mM) was 58% of the theoretical maximum. Acetate (28.9 mM) remained the second most abundant product.
Fig. 3.
Batch fermentation of glucose by mutant strains of E. coli.(A) Cell growth. (B) Glucose utilization. (C) Acetate production. (D) Pyruvate production.
Table 2. Products formed from glucose catabolism by E. coli strains used in this study.
| Product concentrations, mM*
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Condition | Replicates | Cell mass, g/liter | Carbon balance, % | Pyruvate yield, % theoretical† | Pyruvate | Acetate | 2-oxoglutarate | Succinate | Fumarate |
| W3110 | 3% glucose | 2 | 4.13 | 80.6 | 5.9 | 20.8 | 180.0 | 8.3 | 13.7 | 0.9 |
| TC36 | 3% glucose | 3 | 3.47 ± 0.23 | 89.0 ± 2.7 | 10.5 ± 7.9 | 38.1 ± 27.2‡ | 197.7 ± 21.1 | 16.6 ± 16.2 | 13.7 ± 13.2 | 1.4 ± 0.2 |
| TC38 | 3% glucose | 3 | 2.21 ± 0.09 | 84.3 ± 5.2 | 57.5 ± 2.6 | 194.5 ± 9.1 | 28.9 ± 16.7 | 10.5 ± 1.9 | 8.1 ± 9.1 | 0.8 ± 0.7 |
| TC38 | 3% glucose§ | 2 | 2.40 | 84.7 | 68.8 | 241.9 | 7.0 | 7.9 | <2.0 | <0.2 |
| TC42 | 3% glucose | 2 | 3.40 | 86.8 | 29.1 | 79.0 | 178.4 | 76.2 | 24.3 | 1.7 |
| TC44 | 3% glucose | 3 | 2.36 ± 0.10 | 88.5 ± 0.6 | 69.3 ± 1.5 | 252.5 ± 6.2 | 11.6 ± 1.2 | 3.6 ± 1.2 | 16.8 ± 0.7 | 1.1 ± 0.2 |
| TC44 | 3% glucose, ½ nitrogen | 2 | 2.02 | 73.6 | 38.8 | 125.2 | 50.3 | 30.0 | 7.7 | 2.9 |
| TC44 | 3% + 3% glucose¶ | 2 | 2.63 | 86.7 | 72.3 | 479.8 | 39.8 | 31.7 | 10.9 | 0.7 |
| TC44 | 6% glucose | 2 | 1.95 | 94.8 | 588.9 | 46.0 | 26.1 | <2.0 | 0.7 | |
| TC44 | Excess glucose∥ | 2 | 2.51 | - | - | 749.0 | 62.4 | 45.3 | 14.7 | 3.3 |
Unless stated otherwise the concentrations represent measurements at the time of glucose exhaustion.
Maximum theoretical yield is 2 mol of pyruvate per mol of glucose (0.978 g of pyruvate per g of glucose).
Pyruvate concentration during glucose fermentations ranged from 14.88 to 111.89 mM and was very sensitive to small differences in dissolved oxygen.
Dissolved oxygen allowed to fall from 100% to 50% of air saturation.
Three percent initial glucose followed by the addition of 3% glucose after 15 h.
Three percent initial glucose with the addition of 590 ml of 4 M glucose over a period of 20 h.
Table 3. Comparison of metabolic rates.
| Glucose consumption rate
|
Pyruvate production rate
|
||||
|---|---|---|---|---|---|
| Strain | μmax, h-1 | Volumetric,* mmol·liter-1·h-1 | Specific,† mmol·liter-1·h-1 g-1 dcw | Volumetric,* mmol·liter-1·h-1 | Specific,† mmol·liter-1·h-1 g-1 dcw |
| E. coli W3110 (wild type) | 0.69 | 11.9 | 17.35 | nd | nd |
| TC36‡ (focA-pflB frdBC ldhA atpFH adhE sucA) | 0.49 ± 0.03 | 10.1 ± 2.6 | 17.6 ± 1.5 | nd | nd |
| TC36§ (focA-pflB frdBC ldhA atpFH adhE sucA) | 0.51 ± 0.01 | 10.7 ± 0.9 | 29.7 ± 3.5 | 3.8 ± 3.0 | 5.3 ± 3.1 |
| TC38§ (focA-pflB frdBC ldhA atpFH adhE sucA ackA) | 0.28 ± 0.01 | 6.7 ± 0.6 | 16.3 ± 2.2 | 8.3 ± 0.7 | 21.1 ± 3.7 |
| TC38¶ (focA-pflB frdBC ldhA atpFH adhE sucA ackA) | 0.21 | 6 | 28 | 8 | 28 |
| TC42§ (focA-pflB frdBC ldhA atpFH adhE sucA poxB) | 0.55 | 10 | 17 | 6 | 10 |
| TC44§ (focA-pflB frdBC ldhA atpFH adhE sucA poxB ackA) | 0.34 ± 0.02 | 9.7 ± 0.7 | 27.2 ± 4.1 | 13.1 ± 0.3 | 40.4 ± 7.4 |
nd, not determined.
Standard deviations are included for data from three or more fermentations; others represent an average of two fermentations.
Maximum specific rates per g of dry cell weight (dcw).
Dissolved oxygen controlled at 5% of air saturation by adjusting the ratio of O2 and N2.
Dissolved oxygen allowed to fall from 100% to 5% of air saturation during growth with air sparging; O2 added to maintain 5% air saturation.
Dissolved oxygen allowed to fall from 100% to 50% of air saturation during growth with air sparging; O2 added to maintain 50% air saturation.
Pyruvate Oxidase (poxB) Mutation Causes a Small Reduction in Acetate Production. Pyruvate can be metabolized to acetate by the membrane-bound protein pyruvate oxidase by using the electron transport system to couple oxygen as a terminal electron acceptor. The poxB gene is typically repressed during exponential growth but is induced by stress or entry into stationary phase (32, 33). The poxB gene in TC36 was disrupted by constructing stop codons in the central region to produce TC42. In contrast to the ackA deletion (TC38), the poxB mutation (TC42) caused a relatively small decease in acetate and an increase in pyruvate (Table 2 and Fig. 3 C and D), consistent with a minor role for the PoxB pathway. Pyruvate yield with TC42 was <30% of the theoretical maximum. Unlike the mutation in ackA (TC38), inactivation of poxB (TC42) did not reduce the rate of growth or glucose metabolism (Table 3).
Combining Mutations in Pyruvate Oxidase (poxB) and Acetate Kinase (ackA) Increased Pyruvate Production. Strain TC44 (ΔfocA-pflB ΔfrdBC ΔldhA ΔatpFH ΔadhE ΔsucA poxB::FRT ΔackA) was constructed in which both the acetate kinase and pyruvate oxidase mutations were combined in a TC36 background. Inactivation of poxB was beneficial for growth and pyruvate production (Fig. 3A and Tables 2 and 3) in comparison to TC38 (an isogenic strain containing functional poxB). Both volumetric and specific rates of glucose metabolism were higher for TC44 than for TC36 or TC38 (Table 3). Acetate production by TC44 was half that of TC38, and the pyruvate yield was 17% higher. The specific rate of pyruvate production by TC44 was 8-fold that of TC36 and twice that of TC38 (Table 3). The time required to complete fermentation with TC44 was 30% shorter than with TC38 (Fig. 3B). Broth containing 3% glucose (167 mM) was metabolized to 2.2% pyruvate (252 mM) after 18 h in mineral salts medium (Fig. 3D). Although acetate levels were substantially reduced by combining the poxB and ackA mutations (Fig. 3C), acetate (12 mM) and dicarboxylic acids (20 mM) remained as minor products.
The Beneficial Role of a poxB Mutation for Pyruvate Production. Although eliminating the primary route for acetyl-CoA dissimilation (ΔackA) in TC38 increased pyruvate production, this was accompanied by detrimental effects on growth and fermentation rates (Fig. 2 and Tables 2 and 3) that were substantially reduced by inactivation of PoxB (TC44). Pyruvate oxidase competes with NADH oxidation for oxygen as a terminal electron acceptor and represents a potential mechanism for this action. Oxygen transfer rates are frequently limiting during aerobic fermentations at relatively high levels of saturation (34) and may be even more problematic under our fermentation conditions (5% of air saturation). Increased PoxB activity resulting from larger pyruvate pools in the acetate kinase mutant (TC38) would be expected to decrease the availability of NAD+ for glycolysis and to increase the levels of NADH, an allosteric inhibitor of key biosynthetic enzymes, such as citrate synthase (35) and phosphoenolpyruvate carboxylase (36). This hypothesis was tested in part by examining the effect of increasing the oxygen level during fermentation with TC38.
Increasing oxygen saturation from 5% to 50% during TC38 fermentations (Tables 2 and 3) was beneficial, consistent with our hypothesis. Cell yield, pyruvate yield, and the specific rate of glucose metabolism were 8–41% higher for TC38 at 50% air saturation than at 5% air saturation. These results were very similar to those observed for the isogenic poxB mutant TC44 during fermentation at 5% air saturation. Increasing the oxygen saturation during TC38 fermentations also decreased the final concentrations of acetate to a level equivalent to TC44 at 5% air saturation and decreased the production of dicarboxylic acids (Table 2).
Prior experience with the acetate-producing strain TC36 demonstrated that yields could be improved by simple changes in fermentation conditions (26). Unlike TC36, decreasing the ammonia level by half did not increase product yields for TC44 (Table 2). However, doubling of the initial concentration of glucose or providing a second addition of glucose (3% plus 3%) increased pyruvate yields by 11% and doubled the final pyruvate titer. With excess glucose, 749 mM pyruvate was produced. This may represent the limit for pyruvate tolerance. Addition of 600 mM pyruvate to mineral salts medium inhibited the growth of wild-type W3110 (data not shown).
Discussion
Construction of E. coli TC44 for pyruvate excretion during the oxidative metabolism of glucose required mutations to reduce the utilization of pyruvate for cell growth (ΔatpFH ΔsucA) and the elimination of all major nonessential pathways (ΔfocA-pflB ΔfrdBC ΔldhA adhE poxB::FRT ΔackA) that can consume pyruvate. In this strain, glycolytic flux exceeded that of the unmodified parent, W3110, by >50%. This increased rate of glycolysis is attributed to the ATPase (ΔatpFH) mutation that provided a cytoplasmic F1-ATPase, increasing the availability of ADP for glycolysis and minimizing allosteric inhibition by ATP accumulation. A similarly high flux was previously reported for an analogous strain designed for the efficient production of acetate (26). Strain TC44 converted glucose to pyruvate with a yield of 0.75 g of pyruvate per g of glucose (77.9% of theoretical yield) under optimal conditions in minimal medium. With excess glucose, a maximum of 749 mM pyruvate was produced.
Addition of a pyruvate oxidase mutation was surprisingly beneficial for growth and pyruvate production. Introduction of the poxB deletion into the acetate-producing strain (TC36) resulted in a small improvement in growth rate, in contrast to the ackA deletion that reduced growth rate by half (Table 3). The decreased growth rate in the acetate kinase mutant of TC36 cannot be explained by the predicted 30% reduction in ATP yield, because the growth rate was substantially restored by adding a subsequent mutation in pyruvate oxidase, which should not affect ATP production. These results suggested that the reduction in growth rate by the acetate kinase mutation may result from an increase in PoxB activity associated with a larger pyruvate pool.
Pyruvate oxidase transfers electrons from pyruvate to ubiquinone on the decarboxylation of pyruvate. An increase in pyruvate metabolized by pyruvate oxidase would increase oxygen demand and reduce the oxygen available for NADH oxidation (37). Because the total nicotinamide adenine dinucleotide pool is relatively constant (38) and the NAD+/NADH ratio is responsive to changes in oxygen availability (38), increased pyruvate oxidase activity would also reduce the level of NAD+ available for glycolysis. High NADH levels also serve as an important allosteric regulator of key enzymes such as citrate synthase (35) and phosphoenolpyruvate carboxykinase (36). Allosteric inhibition of these enzymes in minimal medium would limit both glycolytic flux and the partitioning of carbon into biosynthesis. Oxidation of NADH is also essential to provide the stoichiometric levels of NAD+ required for glycolysis. Thus, inactivating pyruvate oxidase may be expected to improve growth by eliminating the increase in oxygen demand, restoring the NAD+ pools for glycolysis, and decreasing the level of NADH. This hypothesis was supported by further tests in which the availability of oxygen was increased from 5% of air saturation to 50% of air saturation during fermentation with the acetate kinase mutant TC38. The increase in oxygen (50% air saturation) substantially reversed the negative impact of the acetate kinase mutation on growth rate and metabolism, analogous to the effect of adding a pyruvate oxidase mutation. A similar increase in oxygen was of no benefit for TC44 in which both acetate pathways had been mutated (data not shown).
Pyruvate can be produced by a variety of microorganisms, including mutants of yeasts and bacteria (Table 4). Typical production rates for these biocatalysts are ≈1 g·liter–1·h–1, with yields exceeding half the weight of substrate. T. glabrata, a yeast strain currently used for the commercial production of pyruvate, can achieve pyruvate titers of 69 g/liter, although yields are somewhat lower than for TC44 and more elaborate process controls are required. T. glabrata strains used in the commercial process are multivitamin auxotrophs requiring tight regulation of vitamin concentrations that result in complex vitamin feeding strategies during fermentation (16). Previous E. coli strains constructed for pyruvate production were cultured in complex media and achieved low titers (22, 23). In contrast, E. coli TC44 produced pyruvate at high yields and high titers when used with only mineral salts, glucose, and simple process controls.
Table 4. Comparison of biocatalysts for pyruvate production.
| Strain | Relevant genotype/phenotype | Carbon source | Nitrogen source | Fermentation time, h | [Pyruvate], g/liter | Volumetric production, g·liter-1·h-1 | Pyruvate yield, g/g | Ref. |
|---|---|---|---|---|---|---|---|---|
| Candida lipolytica AJ 14353 | B1- Met- | Glucose | NH4NO3 | 72 | 44 | 0.61 | 0.44 | 16 |
| Debaryomyces hansenii Y-256 | B1- Bio- | Glucose | Peptone | 96 | 42 | 0.44 | 0.42 | 16 |
| T. glabrata ACII-3 | B1- Bio- B6- NA- acetate leaky | Glucose | Soy hydrolysate (NH4)2SO4 | 47 | 60 | 1.28 | 0.68 | 16 |
| T. glabrata WSH-IP 303 | B1- Bio- B6- NA- | Glucose | NH4Cl | 56 | 69 | 1.23 | 0.62 | 16 |
| E. coli TBLA-1 | lipA2 bgl+ atpA401 | Glucose | Polypeptone | 24 | 30 | 1.25 | 0.60 | 22 |
| E. coli CGSC7916 | aceF fadR adhE ppc | Glucose acetate | Tryptone (NH4)2HPO4 | 36 | 35 | 0.97 | 0.65 | 23 |
| E. coli TC44 | pflB frdBC ldhA atpFH adhE sucA ackA poxB | Glucose | (NH4)2HPO4 | 43 | 52 | 1.21 | 0.76 | This study |
The remarkable tolerance of E. coli to drastic changes in metabolic fluxes that allows the production of acetate or pyruvate as alternative products of metabolism implies considerable elasticity in permitted pool sizes for key metabolic intermediates, such as pyruvate and acetyl-CoA. It is interesting to note that pyruvate transiently accumulated in broth during fermentations with TC36 but was rapidly cometabolized with glucose during the latter stages of fermentation. Similar transient accumulation and reutilization was previously observed in minimal medium for other central metabolites (carboxylic acids and acetate) in other derivatives of W3110 (26). Exogenously added pyruvate has been shown to be rapidly mixed and metabolized with intracellular pyruvate (39). Carboxylic acids (40, 41) and acetate (5, 26) are also cometabolized with sugars. Active transport systems have been identified in E. coli for pyruvate (42), acetate (43), and dicarboxylic acids (44–46). Thus, for pyruvate and perhaps many other compounds, the extracellular milieu may be reasonably regarded as a reservoir for the temporary expansion of metabolic pools. A similar argument can be made for CO2 (47) and for acetaldehyde during ethanol production (48–50). The transient accumulation of metabolites in the aqueous milieu may be of evolutionary advantage. This accumulation could arguably be regarded as a form of metabolic conditioning of the environment, as a storage reservoir, and as an expanded metabolic pool to relieve a temporary imbalance in metabolic flux. Enzymes are relatively stable in E. coli. Substantial remodeling of metabolism is primarily regulated at the level of transcription with dilution during subsequent growth. Transient storage of products from imbalanced metabolism in the extracellular milieu could increase the metabolic flexibility of E. coli during adaptation to a changing environment.
Acknowledgments
This work was supported by U.S. Department of Agriculture Grants 01-35504-10669 and 00-52104-9704 and U.S. Department of Energy Grant FG02–96ER20222 (FAES No. R09934).
Abbreviation: FRT, FLP recognition target.
References
- 1.Schmid, A., Dordick, J. S., Hauer, B., Kiener, A., Wubbolts, M. & Witholt, B. (2001) Nature 409, 258–268. [DOI] [PubMed] [Google Scholar]
- 2.Stringer, J. (1996) Chem. Week 158, 52. [Google Scholar]
- 3.Strohl, W. R. (2001) Metab. Eng. 3, 4–14. [DOI] [PubMed] [Google Scholar]
- 4.Ingram, L. O., Aldrich, H. C., Borges, A. C. C., Causey, T. B., Martinez, A., Morales, F., Saleh, A., Underwood, S. A., Yomano, L. P. & York, S. W. (1999) Biotechnol. Prog. 15, 855–866. [DOI] [PubMed] [Google Scholar]
- 5.Underwood, S. A., Zhou, S., Causey, T. B., Yomano, L. P., Shanmugam, K. T. & Ingram, L. O. (2002) Appl. Environ. Microbiol. 68, 6263–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, D., Shin, S., Rhee, J. & Pan, J. (1999) Appl. Environ. Microbiol. 65, 1384–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dien, B. S., Nichols, N. N. & Bothast, R. J. (2001) J. Ind. Microbiol. Biotechnol. 27, 259–264. [DOI] [PubMed] [Google Scholar]
- 8.Zhou, S., Causey, T. B., Hasona, A., Shanmugam, K. T. & Ingram, L. O. (2003) Appl. Environ. Microbiol. 69, 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura, C. E., Gatenby, A. A., Hsu, A. K.-H., La Reau, R. D., Haynie S. L., Diaz-Torres, M., Trimbur, D. E., Whited, G. M., Nagarajan, V., Payne, M. S., et al. (2000) U.S. Patent 6,013,494.
- 10.Tong, I., Liao, H. H. & Cameron, D. C. (1991) Appl. Env. Microbiol. 57, 3541–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu, W., Draths, K. M. & Frost, J. W. (2002) Biotechnol. Prog. 18, 201–211. [DOI] [PubMed] [Google Scholar]
- 12.Beck, B. J., Aldrich, C. C., Fecik, R. A., Reynolds, K. A. & Sherman, D. H. (2003) J. Am. Chem. Soc. 125, 4682–4683. [DOI] [PubMed] [Google Scholar]
- 13.Dayem, L. C., Carney, J. R., Santi, D. V., Pfeifer, B. A., Khosla C. & Kealey, J. T. (2002) Biochemistry 41, 5193–5201. [DOI] [PubMed] [Google Scholar]
- 14.Lee, P. C. & Shmidt-Dannert, C. (2002) Appl. Microbiol. Biotechnol. 60, 1–11. [DOI] [PubMed] [Google Scholar]
- 15.Wang, C.-W., Oh, M.-K. & Liao, J. C. (2000) Biotechnol. Prog. 16, 922–926. [DOI] [PubMed] [Google Scholar]
- 16.Li, Y., Chen, J. & Lun, S.-Y. (2001) Appl. Microbiol. Biotechnol. 57, 451–459. [DOI] [PubMed] [Google Scholar]
- 17.Anonymous (2002) China Chem. Rep. 13, 28. [Google Scholar]
- 18.Ingram, L. O., Conway, T., Clark, D. P., Sewell, G. W. & Preston, J. F. (1987) Appl. Environ. Microbiol. 53, 2420–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blair, G. T. (1999) in Concise Encyclopedia of Chemical Technology, eds. Kroschwitz, J. I., Howe-Grant, M., Siegel, P. M., Richman, B., Zayatz, E., Altieri, L., Bickford, M. & Humphreys, L. (Wiley, New York), p. 1101.
- 20.Ondrey, G. (2001) Chem. Eng. 108, 21. [Google Scholar]
- 21.Li, Y., Chen, J. & Lun, S.-Y. & Rui, X. S. (2001) Appl. Microbiol. Biotechnol. 55, 680–685. [DOI] [PubMed] [Google Scholar]
- 22.Yokota, A., Terasawa, Y., Takaoka, N., Shimizu, H. & Tomita, F. (1994) Biosci. Biotechnol. Biochem. 58, 2164–2167. [DOI] [PubMed] [Google Scholar]
- 23.Tomar, A., Eiteman, M. A. & Altman, E. (2003) Appl. Microbiol. Biotechnol. 62, 76–82. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, J. & Greasham, R. (1999) Appl. Microbiol. Biotechnol. 51, 407–421. [DOI] [PubMed] [Google Scholar]
- 25.Yomano, L. P., York S. W. & Ingram, L. O. (1998) J. Ind. Microbiol. Biotechnol. 20, 132–138. [DOI] [PubMed] [Google Scholar]
- 26.Causey, T. B., Zhou, S., Shanmugam, K. T. & Ingram, L. O. (2003) Proc. Natl. Acad. Sci. USA 100, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria (Cold Spring Harbor Lab. Press, Plainview, NY).
- 28.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 29.Datsenko, K. A. & Wanner, B. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posfai, G., Koob, M. D., Kirkpatrick, H. A. & Blattner, F. C. (1997) J. Bacteriol. 179, 4426–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Morales, F., Borges, A. G., Martinez, A., Shanmugam, K. T. & Ingram, L. O. (1999) J. Bacteriol. 181, 7143–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang, Y.-Y. & Cronan, J. E., Jr. (1983) J. Bacteriol. 154, 756–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang, Y.-Y., Wang, A.-Y. & Cronan, J. E., Jr. (1994) Mol. Microbiol. 11, 1019–1028. [DOI] [PubMed] [Google Scholar]
- 34.Varma, A., Boesch, B. W. & Palsson, B. O. (1993) Appl. Environ. Microbiol. 59, 2465–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weitzman, P. D. J. (1966) Biochim. Biophys. Acta 128, 213–215. [DOI] [PubMed] [Google Scholar]
- 36.Krebs, A. & Bridger, W. A. (1976) Can. J. Biochem. 54, 22–26. [DOI] [PubMed] [Google Scholar]
- 37.Cunnungham, C. C. & Hager, L. P. (1975) J. Biol. Chem. 250, 7139–7146. [PubMed] [Google Scholar]
- 38.Wimpenny, J. W. T. & Firth, A. (1972) J. Bacteriol. 111, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, Y.-T., Bennett, G. N. & San, K.-Y. (2001) Metab. Eng. 3, 115–123. [DOI] [PubMed] [Google Scholar]
- 40.Lutgens, M. & Gottschalk, G. (1980) J. Gen. Microbiol. 119, 63–70. [DOI] [PubMed] [Google Scholar]
- 41.Underwood, S. A., Buszko, M. L., Shanmugam, K. T. & Ingram, L. O. (2004) Appl. Environ. Microbiol. 70, in press. [DOI] [PMC free article] [PubMed]
- 42.Lang, V. J., Leystra-Lantz, C. & Cook, R. A. (1987) J. Bacteriol. 169, 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gimenez, R., Nuñez, M. F., Badia, J., Aguilar, J. & Baldoma, J. (2003) J. Bacteriol. 185, 6448–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo, T. C. Y., Rayman, M. K. & Sanwal, B. D. (1972) J. Biol. Chem. 247, 6323–6331. [PubMed] [Google Scholar]
- 45.Pos, K. M., Dimroth, P. & Bott, M. (1998) J. Bacteriol. 180, 4160–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seol, W. & Shatkin, A. J. (1991) Proc. Natl. Acad. Sci. USA 88, 3802–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merlin, C., Masters, M., McAteer, S. & Coulson, A. (2003) J. Bacteriol. 185, 6415–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dombek, K. M. & Ingram, L. O. (1984) J. Bacteriol. 157, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanley, G. A. & Pamment, N. B. (1993) Biotechnol. Bioeng. 42, 24–29. [DOI] [PubMed] [Google Scholar]
- 50.Stanley, G. A., Douglas, N. G., Every, E. J., Tzanatos, T. & Pamment, N. B. (1993) Biotechnol. Lett. 15, 1199–1204. [Google Scholar]