Abstract
The family of ubiquitin (Ub)-specific proteases (USP) removes Ub from Ub conjugates and regulates a variety of cellular processes. The human genome contains many putative USP-encoding genes, but little is known about USP tissue distribution, pattern of expression, activity, and substrate specificity. We have used a chemistry-based functional proteomics approach to identify active USPs in normal, virus-infected, and tumor-derived human cells. Depending on tissue origin and stage of activation/differentiation, different USP activity profiles were revealed. The activity of specific USPs, including USP5, -7, -9, -13, -15, and -22, was up-regulated by mitogen activation or virus infection in normal T and B lymphocytes. UCH-L1 was highly expressed in tumor cell lines of epithelial and hematopoietic cell origin but was not detected in freshly isolated and mitogen-activated cells. Up-regulation of this USP was a late event in the establishment of Epstein–Barr virus-immortalized lymphoblastoid cell lines and correlated with enhanced proliferation, suggesting a possible role in growth transformation.
Ubiquitination of cellular substrates, either at the N terminus or by way of isopeptide linkage to the ε-amino group of an internal lysine residue regulates many processes, from proteolysis to the control of intracellular trafficking (1). Although significant progress has been made in the characterization of enzymes that ligate ubiquitin (Ub) to target proteins, little is known about the removal of Ub from Ub conjugates. Yet, the activity of Ub-specific proteases (USPs), also known as deubiquitinating enzymes (DUBs), is likely to be central to the regulation of all processes in which Ub is involved, from the processing of poly-Ub precursors into Ub monomers, to the targeting or salvage of proteasomal substrates and the regulation of nonproteolytic functions of mono- and polyubiquitination (2, 3).
The human genome encodes 60–70 predicted members of the USP family, and four major classes have been identified. The best-studied classes, characterized by the presence of a catalytically active cysteine residue, are known as Ub-processing proteases (UBP) and Ub carboxyl-terminal hydrolases (UCH). UBPs can hydrolyze both linear and branched Ub modifications whereas the activity of UCH enzymes is restricted to the hydrolysis of small Ub C-terminal extensions (2, 3). A third USP family that contains an ovarian tumor (OTU) domain was recently discovered by using a proteomics approach (4), and several active members have been identified since (5, 6), confirming the USP activity of OTU domain-containing proteases. Finally, RPN11/POH-1, a proteasome 19S cap subunit belonging to the Jab1/MPN domain-associated metalloisopeptidase (JAMM) family that lacks the cysteine protease signature, was shown to cleave Ub from substrates in a Zn2+- and ATP-dependent manner (7, 8).
The large number of predicted USPs suggests that these enzymes may exhibit selectivity for the type of Ub linkage hydrolyzed or the protein substrates acted on and may thereby regulate specific cellular processes. Indeed, specific substrates have been identified for some USPs, the most recent example being the product of the cylindromatosis or turban tumor syndrome gene, CYLD (4), which was identified as a regulator of the NF-κB pathway (9–11). Despite the lack of information on their substrate specificity, it is clear that some USPs exert distinct growth regulatory activities by acting as oncoproteins (12–14) or tumor suppressor proteins (15–17). Furthermore, overexpression of certain USPs correlates with progression toward a more malignant phenotype in neuroblastoma (18) and carcinomas of lung, kidney, breast, and prostate (19–23), and the expression of some USPs is induced on growth factor stimulation (24–26). In addition, a strong increase of USP activity was observed on overexpression of the c-myc oncogene in cells of Burkitt's lymphoma (BL)-like phenotype, which correlated with resistance to apoptosis induced by pharmacological inhibition of the proteasome (27). Collectively, these findings identify USPs as important regulators of biological processes and potential targets for the treatment of a variety of human diseases, including several types of malignancies.
Studies aimed at a functional characterization of putative USPs and analysis of their expression and activity under different physiological and pathological conditions have been boosted by the development of influenza hemagglutinin (HA)-tagged Ub-derived active-site-directed probes that allow covalent modification of the active enzymes, followed by their isolation and identification by tandem mass spectrometry (MS/MS) (4). Here, we use this approach to characterize active USPs in tumor-derived and normal virus-infected or mitogen-activated human cells; we demonstrate considerably different USP activity profiles in tumor cells of various tissue origin and in normal cells depending on their state of activation/differentiation.
Materials and Methods
Synthesis of Suicide USP Substrates. HA-tagged Ub probes were synthesized as described (4). Freshly prepared HAUbBr2 (bromoethylamine functionalized probe) and HAUbVME (vinylmethyl ester functionalized probe) were stored in aliquots at –80°C until use. For details, see Figs. 5 and 6 and Table 3, which are published as supporting information on the PNAS web site.
Cell Lines and Primary Cells. For origin, cell type, and virus carrier status of the cell lines included in this investigation, see Table 3. Cell lines were cultured in RPMI medium 1640 supplemented with 2 mM l-glutamine, 100 international units/ml penicillin, 100 mg/ml streptomycin, and 10% heat-inactivated FCS. Peripheral blood lymphocytes were obtained from heparinized blood by Ficoll/Isopaque sedimentation, and macrophages were isolated by plastic adherence for 1 h at 37°C in medium containing 10% FCS. The nonadherent cells were further fractionated to yield T and B cell subpopulations by sheep red blood cell rosetting. The rosetting fraction contained >90% T lymphocytes as determined by surface staining with anti-CD3 antibodies and fluorescence-activated cell sorter (Becton Dickinson) analysis. T cells were activated with 1 μg/ml purified phyto-hemagglutinin (PHA-M, Sigma) for the indicated time, and B cells were activated with 1:100,000 dilution of formalin-fixed Staphylococcus aureus (Sigma). The purified B cells were also infected with spent supernatant from the Epstein–Barr virus (EBV) producer B95.8 cell line as described (28).
Labeling of Cell Extracts and Western Blot Analysis. Cells were harvested, washed three times with PBS, and lysed with glass beads in twice the pellet volume of buffer containing 50 mM Tris (pH 7.4), 5 mM MgCl2, 250 mM sucrose, 1 mM DTT, and 1 mM ATP. Nuclei, membranes, and intact cells were removed by centrifugation, and 20 μg of clarified protein extract corresponding to 2–4 × 106 cells was incubated for 1 h at 37°C with HAUb derivatives. After boiling in reducing sample buffer, the cell lysates were fractionated in precast SDS/PAGE 4–12% gradient gels (Invitrogen), blotted to poly(vinylidene difluoride) membranes and probed with the anti-HA monoclonal antibody 12CA5.
Immunoprecipitation and MS/MS Analysis. Procedures for experiments using HAUb-derived probes, immunoprecipitation, and MS-based identification of target enzymes and associated proteins were performed essentially as described (4). MS/MS data were processed and subjected to database searches by using mascot (Matrix Science, London) against the National Center for Biotechnology Information nonredundant (nr) or human EST databases or by using proteinlynx global server 1.1 software (Micromass, Manchester, U.K.) against swiss-prot trembl/new (www.expasy.ch).
Results
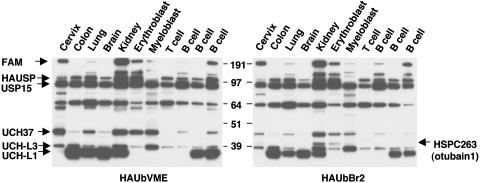
Identification of Active Human USPs. To obtain an overview of USP activity in human cells, cell extracts from a panel of tumor cell lines were labeled with HA-tagged Ub probes that allow simultaneous detection of multiple USPs and their identification by MS/MS (4). These probes measure activity levels of the target enzymes rather than protein levels. Distinct patterns of activity were detected in cell lines of different histological origin (Fig. 1). The most prominent differences were observed in the activity of a USP migrating at ≈35 kDa, which, based on comparison with the migration of previously characterized USP in mouse cells, was tentatively identified as UCH-L1 (PGP9.5), and confirmed as such by MS/MS (Table 1). This USP was highly active in four of five carcinoma lines and in two of three lymphoma lines of B cell origin whereas other cell lines of hematopoietic cell origin and one cervical carcinoma line were negative. Variations were also detected in the activity of USPs migrating at ≈38 kDa and 45 kDa, which we tentatively identified as UCH-L3 and UCH37 (UCH-L5), again confirmed by MS/MS, and in several USPs migrating between 55 and 290 kDa. Increased activity of a 290-kDa USP was observed in carcinoma lines from cervix and kidney, the erythroleukemia line K562, and one of three B cell lymphoma lines. The only known USP with this predicted molecular mass is USP9X (FAM), the human ortholog of Drosophila fat-facets that is known to exert growth regulatory activity. As previously observed in mouse cells (29), a slightly different pattern of USPs was detected by using the HAUbVME and HAUbBr2 probes. In particular, a USP of ≈60 kDa was detected only by the former in nearly all cell lines whereas only the latter detected a USP of ≈42 kDa in cell lines from cervix, kidney, erythroblastoid, and myeloid origin (compare Fig. 1 Left with Fig. 1 Right). The 42-kDa polypeptide detected by HAUbBr2 was identified as HSPC263 (otubain-1; refs. 4 and 5), a member of the ovarian tumor (OTU) domain-containing family of USPs.
Fig. 1.
USP activity profile in human cells. Cell extracts from a panel of tumor cell lines were labeled with HA-tagged Ub probes. Major variations in USP activity profiles were demonstrated in the epithelial cell lines HeLa, CoLo, U1906, SH-SY-5Y, and HEK293 (lanes 1–5), and the hematopoietic cell lines K562, U937.1, Molt-3, HDLM-2, Namalwa, and SU.DHL-4 (lane 6–11). Different USP activities were detected by using the HAUbVME (Left) and HAUbBr2 (Right) probes.
Table 1. USPs identified in human cells.
| Molecular mass, kDa
|
Present in
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Accession no. swissprot/trembl | Expected | Observed | BL | LCL | Sequence coverage, % | No. of matches | Remarks | |
| USP9X | Q8WWT3/Q8WX12/Q93008 | 292.8 | 300 | + | ++ | 15 | 46 | FAFX, FAM |
| USP7 | Q93009 | 129.3 | 140 | + | ++ | 44 | 80 | HAUSP |
| USP22 | Q9UPT9 | 66.6 | 47 | + | - | 2 | 1 | |
| USP5 | P45974 | 96.6 | 140,100 | + | ++ | 45 | 47 | IsoT1 |
| USP13 | Q92995 | 80 | 100 | + | - | 7 | 9 | IsoT3 |
| USP15 | Q9Y4E8 | 113.6 | 140,100 | + | + | 21 | 23 | |
| USP15i | Q9R085 | 109.2 | 100 | + | - | 2 | 1 | |
| USP8 | P40818 | 127.5 | 120 | + | - | 2 | 1 | Ubp-Y |
| UCH-L5/UCH37 | Q9XSJ0/Q9Y5K5/Q9WUP7 | 37.4 | 48 | + | + | 53 | 23 | 19S subunit |
| UCH-L3 | P15374 | 26.2 | 35,38 | + | + | 16 | 5 | |
| UCH-L1 | P9936 (AAH06305) | 24.8 | 35-37 | ++ | + | 56 | 60 | |
Cell lines exhibiting different USP profiles were then chosen for identification of the HAUbVME and HAUbBr2 reactive polypeptides by anti-HA immunoprecipitation followed by SDS/PAGE, tryptic digestion of excised bands, MS/MS, and bioinformatics (Tables 1 and 2 and Fig. 6). Eleven USPs were identified by using this approach, many of these at different activity levels in various cell types. USP-8, USP-13 (IsoT3), and USP-22 were detected in the BL cell line Raji but not in the EBV-transformed lymphoblastoid cell line (LCL) LCL-R whereas the activities of USP-5 (IsoT), -7 (HAUSP), -9, and -15i seemed to be higher in the virus-transformed cells. Polypeptides corresponding to IsoT and IsoT3 were found throughout the 110- to 150-kDa molecular mass region whereas HAUSP and USP15 were consistently found at 140 and 100 kDa, respectively. Higher levels of UCH-L1, and to a minor extent UCH-L3, were observed in Raji as compared with LCL-R.
Table 2. USP-associated proteins identified in human cells.
| Molecular mass, kDa
|
Present in
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Accession no. swissprot/trembl | Expected | Observed | BL | LCL | Sequence coverage, % | No. of matches | Remarks | |
| MVP | Q14764 | 99.6 | 100 | + | - | 8 | 4 | Drug resistance |
| EF1α | P04720 | 50.4 | 45 | + | + | 10 | 4 | |
| Drebrin | Q16643 | 71.8 | 47 | + | + | 9 | 2 | |
| S-acetyltransferase | Q86YI5 | 69 | 70 | + | + | 18 | 6 | |
| Damage-specific DNA bdg protein | Q16531 | 127 | 140 | + | - | 7 | 3 | |
| 3′-5′ RNA exonuclease | Q8IWX1 | 86 | 90 | + | - | 10 | 3 | |
| Ku70 | P12956 | 69.7 | 70 | + | + | 18 | 6 | |
| Retinoblastoma A-associated | P06400 | 106.2 | 65 | + | - | 8 | 2 | |
| 26S-associated Pad1 (POH1) | O00487 | 34.7 | 35 | + | - | 31 | 10 | Drug resistance |
| 26S S12 (p40) | P51665 | 37 | 37 | + | - | 18 | 6 | |
| 26S MSS1 | P35998 | 48.6 | 47 | + | - | 61 | 45 | |
| 26S 6A | P17980 | 49.5 | 45 | + | - | 17 | 7 | |
| 26S 6B | P43686 | 47.3 | 47 | + | + | 12 | 4 | |
| 26S S2 | Q9DBA1 | 49 | 47 | + | + | 42 | 22 | |
| 26S S4 | Q03527 | 47.5 | 47 | + | - | 12 | 4 | |
| 26S S45/S8 | O43208/P47210 | 31.3 | 47 | + | - | 15 | 3 | |
| 26S S1 | Q99460/O88761 | 105.8 | 120 | + | + | 27 | 25 | |
Differences in proteins that coprecipitate with USPs were also detected. Most notably, the major vault protein (MVP) and POH1 (RPN11) were observed in precipitates from Raji but not LCL-R. MVP has been implicated in multidrug resistance (30) whereas POH1 is a proteasome subunit known to be up-regulated in several forms of cancer and likewise implicated in multidrug resistance (31). Consistent with previous results obtained with mouse thymoma cells (4), several subunits of the 19S cap coprecipitated with USPs from both BL cells and LCLs (Tables 1 and 2).
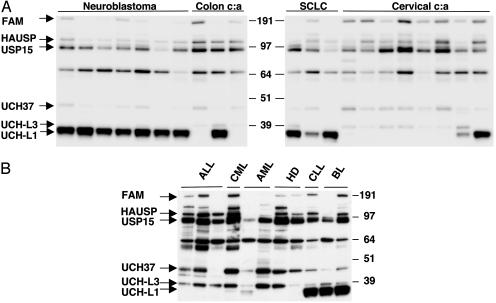
USP Profiling in Tumor Cell Lines. To investigate whether the extensive differences observed in the first screen reflects cell-type or tumor-specific features, USP activity profiles were compared in a large panel of tumor cell lines of epithelial and hematopoietic origin. Both unique and tumor-specific patterns of activity were observed. The greatest differences were seen in the activity of UCH-L1, which was highly active in all seven neuroblastoma lines and all three small-cell lung carcinoma lines tested, but in only one of three colon carcinoma lines and two of eight human papillomavirus-positive cervical carcinoma lines (Fig. 2A). Among cell lines of hematopoietic origin, only some B cell lymphoma lines were strongly positive whereas three T cell lymphomas, three myeloid, and two Hodgkin's lymphoma lines were negative (Fig. 2B). The high UCH-L1 activity found in neuroblastoma lines could reflect the tissue origin of this tumor because UCH-L1 is highly expressed in neurons and accounts for 1–2% of the total protein content in brain (32). Similar information is lacking for other cell types. Consistent differences were also observed in the activity of UCH-L3, which was not detected in neuroblastoma lines but was present in all other cell types, although at different levels (Fig. 2 A and B). UCH37 (UCH-L5), a known subunit of the proteasome 19S regulatory particle (33), was regularly detected although in varying amounts. The activity levels of UCH37 seemed to be inversely proportional to the levels of UCH-L1 in several cases (compare Fig. 2 A and B), raising the question whether the two enzymes may complement each other's activity. The activity profiles of other USPs varied significantly between individual cell lines without distinguishable tumor or cell type-specific pattern.
Fig. 2.
USP activity in tumor cell lines of different cell origin. Shown is USP profiling in tumor cell lines of different tissue origin illustrating the expression of unique and tumor-specific patterns of activity. (A) USP activity of cell lines derived from neuroblastoma [FL-II, SK-SY-5Y, SK-N-AS, SHEP-1, Lan-5, SK-N-BE (2), and IMR-32; lanes 1–7]; colon carcinoma (colon c:a) (HCTC, CoLo, and LIM; lanes 8–10); small cell lung carcinoma (SCLC) (H69, U1906, and U1285; lanes 11–13); and cervical carcinoma (cervical c:a) (HeLa, SiHa, HT3, SW756, ME180, MS751, CasKi, and C33A; lanes 14–21) is shown. Both unique and tumor-specific patterns of activity were observed. (B) USP activity in cell lines from T cell acute lymphocytic leukemia (ALL) (Jurkat, Molt-4, and Molt-3; lane 1–3); chronic myeloid leukemia (CML) in blast crisis (K562; lane 4); acute myeloid leukemia (AML) (U937.1 and MAC-6; lanes 5 and 6); B cell-derived Hodgkin's disease lymphoma (HD) (HDLM-2 and L1236; lanes 7 and 8); chronic lymphocytic leukemia (CLL) (SU.DHL-4; lane 9); and BL (Namalwa and Ramos; lanes 10 and 11).
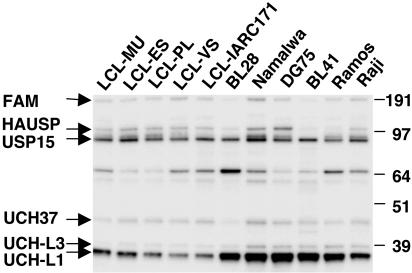
USP Profiling in Freshly Isolated, Mitogen-Stimulated, and Virus-Infected Cells. To gain some insight on whether the activity of various USPs is a tumor- or cell type-associated characteristic, we examined a large panel of BL-derived cell lines and in vitro EBV-transformed LCLs of normal B cell origin. In several cases, the malignant and virus-transformed cells were derived from the same patient, which eliminates the possible influence of genetic variations between individuals. Previous observations indicated that USP activity is significantly higher in BL compared with LCL cells, and higher levels of UCH-L1 were detected in Western blots using a specific antibody (27). This finding was confirmed and extended by labeling with the HAUbVME probe, which detected significantly higher levels of UCH-L1 in all BL lines tested compared with LCLs (Fig. 3). A regular although less prominent increase was also observed for UCH-L3 levels whereas the reactivity of other USPs did not differ consistently in the two cell types. A great heterogeneity in the activity levels of the high molecular mass USPs, including FAM and USP7/HAUSP, was evident in BL cells.
Fig. 3.
Comparison of USP activity in BL vs. LCL cells. Higher UCH-L1/UCH-L3 activity and heterogeneous expression of the high MW USPs are observed in BL cells compared with LCLs.
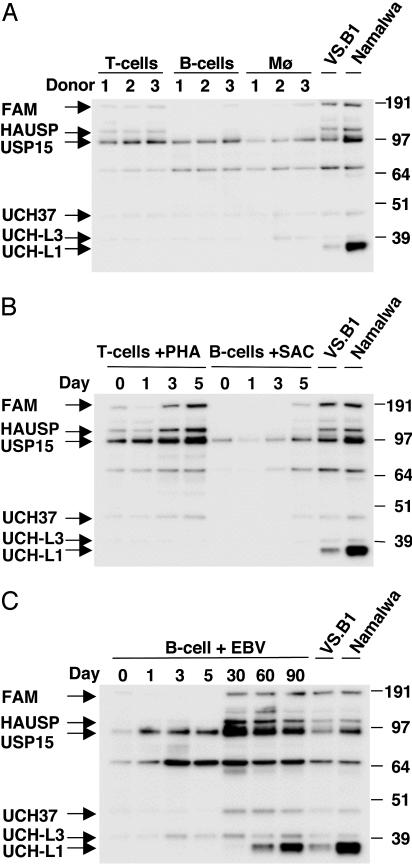
The differing USP activities between BLs and LCLs, and the absence of active UCH-L1 in HD cell lines that share a common germinal center cell origin as BL cells, suggest that high activity levels of UCH-L1, and possibly UCH-L3, may be part of the malignant phenotype of this tumor. To explore this further, USP profiling was performed in freshly isolated subpopulations of blood mononuclear cells and in cells activated by mitogen stimulation and EBV infection (Fig. 4). Very low overall USP activity was detected in freshly isolated T, B, and monocyte-enriched mononuclear cell subpopulations, which seemed to have virtually no UCH-L1 or UCH-L3 activity (Fig. 4A). Stimulation of T cells with PHA led to blast transformation and proliferation, accompanied by increased activity of several high molecular mass USPs, including USP7/HAUSP, USP9X (FAM), and USP15, whereas there was no significantly increased USP activity in B cells stimulated with formalin-fixed S. aureus, despite comparable blast transformation and proliferation (Fig. 4B and data not shown). Importantly, mitogen stimulation did not induce any detectable UCH-L1 activity, confirming that up-regulation of this USP is not simply a marker of cell proliferation.
Fig. 4.
Ub isopeptidase activity in freshly isolated, mitogen-activated, and virus-infected cells. (A) Freshly isolated subpopulation of T cells, B cells, and adherent monocytes do not express UCH-L1/UCH-L3. Relatively higher activity of the high molecular mass USPs is observed in T cells compared with B cells and monocytes. (B) Changes in USP activity profiles are observed on phytohemagglutinin (PHA) stimulation and Staphylococcus aureus Cowan strain (SAC) activation of T and B cells, respectively. (C) EBV infection of B cells resembles mitogen stimulation at early times postinfection whereas different activity patterns are observed after long-term culture. UCH-L1/UCH-L3 activity is not observed in primary cells and is detected in EBV-infected cells only after long-term culture. The EBV-transformed LCL VS.B1 and BL line Namalwa were used as reference for USP labeling. Results shown are from one representative experiment of three.
EBV has been implicated in the pathogenesis of human malignancies including BL, Hodgkin's lymphoma, and nasopharyngeal carcinoma (reviewed in ref. 34). To investigate the effect of EBV on USP activity, the reactivity with the HAUbVME probe was monitored over a period of 90 days after infection of freshly isolated B cells. As illustrated by the representative experiment shown in Fig. 4C, some increase in the activity of UCH-L3, USP-15, and an unknown USP comigrating with an anti-HA antibody cross-reactive polypeptide of 68 kDa was detected already at early times postinfection whereas a significant increase in UCH-L1, UCH37, USP15, USP7/HAUSP, and USP9X (FAM) activity became apparent only at later time points. In particular, UCH-L1 activity was first detected at day 30 and increased progressively until day 90 when it reached the level of established LCLs. This progressive increase correlated with a switch of growth pattern from slow to rapid proliferation. Similar results were obtained in repeated experiments.
Discussion
Increasing evidence places USPs at the core of a multitude of physiological and pathological processes. The role of this enzyme family in cancer is highlighted by the presence of members with oncogenic or tumor suppressor activity, which underscores the importance of assessing the expression and activity of USPs in different cell types and in cells corresponding to discrete stages of differentiation, activation, and malignant transformation. Using previously described Ub-based probes, we have found dramatic differences in the USP profile of a large panel of human primary and tumor-derived cells. Some of these differences are likely to reflect tissue-specific expression/activity patterns, as previously observed in mouse primary tissues (29), whereas others seem to be specific for individual tumors or tumor types.
The most striking differences were observed in the expression of a 35-kDa USP that was identified as UCH-L1. All cell lines derived from neuroblastoma, lung carcinoma, and various types of B cell lymphomas were strongly positive whereas cell lines derived from other hematopoietic precursors, including T cell blasts, myeloblasts, erythroblasts, two EBV-negative Hodgkin's disease lymphoma lines, and the majority of cell lines from carcinomas of cervix and colon were clearly negative. The high UCH-L1 enzyme activity in neuroblastoma, lung carcinoma, colon carcinoma, and BL lines is in agreement with previous studies demonstrating high expression levels in these and other malignant tumors by immunohistochemical methods (19, 20, 23, 27). The significance of this finding in the context of tumor pathogenesis and progression is unclear. UCH-L1 is highly expressed in brain (32) where its physiological role is underscored by the neurodegenerative phenotype of a naturally occurring null mutation. The Gad mice show a gracile axonal dystrophy phenotype and exhibit progressive sensory and motor ataxia associated with axonal degeneration (35). The contribution of UCH-L1 to this phenotype is unknown, but the low amounts of Ub detected in cells derived from these animals suggest a possible role in the maintenance of Ub steady-state levels, perhaps through direct binding to monoubiquitin (36).
The mechanism(s) by which UCH-L1 may contribute to the growth phenotype of certain cell types remains unknown. The enzyme cleaves pro-Ub cotranslationally (37) and may therefore participate in the generation of free Ub from polyubiquitin precursors and small Ub adducts. The expression of Ub genes is rapidly up-regulated under stress condition (38), suggesting a possible role of UCH-L1 in stress responses. In addition, recent findings suggest that UCH-L1 may have the unique capacity to change its enzymatic activity and serve as a ligase under conditions that favor dimerization (39). It remains to be seen whether a switch to this type of activity may be an important consequence of UCH-L1 overexpression in vivo.
Although the high expression of UCH-L1 in neuroblastoma may reflect the phenotype of the normal precursor, this is clearly not the case in other tumor types. There was virtually no detectable UCH-L1 activity in resting T and B lymphocytes, and activity was not induced on stimulation with mitogens that induced blast transformation and proliferation (Fig. 3). Thus, the high levels of UCH-L1 detected in some B cell lymphomas are unlikely to reflect the phenotype of the normal precursor and are not directly associated with B cell proliferation. It is noteworthy that, whereas BL and Hodgkin's disease lymphoma share a common germinal center cell precursor, only the former regularly expressed high levels of UCH-L1 (Fig. 2B), suggesting a complex relationship between UCH-L1 expression/activity and B cell malignancies. This result was further substantiated by the observation that LCLs obtained by in vitro EBV infection of normal B cells regularly expressed UCH-L1 although at significantly lower levels as compared with BL cells. UCH-L1 up-regulation was clearly not a direct consequence of viral gene expression because it became evident only after >30 days postinfection whereas virtually all cells in the infected cultures expressed the latency-associated EBV nuclear antigens within 6–10 days (not shown). The late appearance of UCH-L1 coincides with the time when EBV-infected cultures become oligoclonal due to overgrowth of cells that are better suited to in vitro culture conditions. Such transition from slow to rapid proliferation was indeed observed in our experiments, suggesting that UCH-L1 may play a role in, or be a consequence of, this adaptation.
Several interesting differences were observed on direct comparison of USP activity in tumor-derived BL lines and in vitro EBV-transformed LCLs of normal B cell origin. In addition to the strong overexpression of UCH-L1, USP8, -13, -15, and -22 were detected in BL cells only (Tables 1 and 2). The activity of USP8 (also called Ubp-y or HUMORF8) increases in proliferating cells (26), suggesting that it may contribute to the malignant phenotype. This finding is supported by the identification of an oncogenic fusion protein involving USP8 and the p85 β-subunit of phophatidylinositol-3 kinase in one case of chronic myeloproliferative disorder (13). The putative USP-22 was identified in this study as an active USP in BL although its specific function remains unknown. USP9X (FAM) was found to be active in both BL and LCLs but was not expressed in resting B lymphocytes and several tumor cell lines of different origin. USP9X is the human homolog of the Drosphila protein fat-facets, an enzyme required for eye and embryo development (40). In mammalian cells, FAM binds to and stabilizes β-catenin (41). β-Catenin serves a dual role as structural member of the adhesion junction, and member of a transcription activation complex (reviewed in ref. 42). The product of the adenomatous polyposis coli gene (APC) is a negative regulator of β-catenin levels, with cancer-associated mutations deficient in this function. The inability of APC to down-regulate β-catenin leads to enhanced transcriptional activation and enhanced growth. Increased levels of β-catenin occur in human leukemias (43), suggesting a possible growth-enhancing effect in these cancers.
In addition to the previously reported coisolation of USP with 19S proteasome subunits (4), a number of potentially interacting partners copurified with USPs in BL and LCL cells. Among these were several RNA and DNA binding proteins, including the DNA repair enzyme Ku70 (44), and the protein synthesis elongation factor EF-1α, which was shown to be required for the degradation of certain N-alpha-acetylated substrates (45). Of note, the drug resistance gene product MVP, originally isolated from a drug-resistant lung cancer cell line (46), was detected in immunoprecipitates from BLs, but not LCLs. Increased expression of MVP has been reported in multidrug-resistant tumor cells (47) and seems to be predictive of a poor prognosis in patients with multiple myeloma and diffuse large B cell lymphomas (48, 49). It is tempting to speculate that the increase in USP activity seen in BL might stabilize MVP by rescuing it from proteolysis and thus contribute to drug resistance.
Supplementary Material
Acknowledgments
This work was supported by grants awarded by the Swedish Cancer Society, the Swedish Foundation for Strategic Research, and the Karolinska Institute (to M.G.M.); the National Institutes of Health and Astra Zeneca (to H.L.P.); an Innovational Research Incentives Scheme Veni grant from the Netherlands Organization for Scientific Research (to H.O.); and the Multiple Myeloma Research Foundation and the Harvard Center for Neurodegeneration and Repair (to B.M.K.). P.J.G. was supported by a National Institutes of Health training grant, and M.G.M. was supported by a short-term travel grant awarded by the Yamagiwa-Yoshida Memorial Union Internationale Contre le Cancer International Cancer Study Grants program.
Abbreviations: Ub, ubiquitin; UCH, Ub carboxyl-terminal hydrolase; USP, Ub-specific protease; MS/MS, tandem MS; BL, Burkitt's lymphoma; LCL, lymphoblastoid cell line; EBV, Epstein–Barr virus; HA, hemagglutinin; MVP, major vault protein.
References
- 1.Glickman, M. H. & Ciechanover, A. (2002) Physiol. Rev. 82, 373–428. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson, K. D. (2000) Semin. Cell Dev. Biol. 11, 141–148. [DOI] [PubMed] [Google Scholar]
- 3.Wing, S. S. (2003) Int. J. Biochem. Cell Biol. 35, 590–605. [DOI] [PubMed] [Google Scholar]
- 4.Borodovsky, A., Ovaa, H., Kolli, N., Gan-Erdene, T., Wilkinson, K. D., Ploegh, H. L. & Kessler, B. M. (2002) Chem. Biol. 9, 1149–1159. [DOI] [PubMed] [Google Scholar]
- 5.Balakirev, M. Y., Tcherniuk, S. O., Jaquinod, M. & Chroboczek, J. (2003) EMBO Rep. 4, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans, P. C., Smith, T. S., Lai, M. J., Williams, M. G., Burke, D. F., Heyninck, K., Kreike, M. M., Beyaert, R., Blundell, T. L. & Kilshaw, P. J. (2003) J. Biol. Chem. 278, 23180–23186. [DOI] [PubMed] [Google Scholar]
- 7.Yao, T. & Cohen, R. E. (2002) Nature 419, 403–407. [DOI] [PubMed] [Google Scholar]
- 8.Verma, R., Chen, S., Feldman, R., Schieltz, D., Yates, J., Dohmen, J. & Deshaies, R. J. (2000) Mol. Biol. Cell 11, 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovalenko, A., Chable-Bessia, C., Cantarella, G., Israel, A., Wallach, D. & Courtois, G. (2003) Nature 424, 801–805. [DOI] [PubMed] [Google Scholar]
- 10.Brummelkamp, T. R., Nijman, S. M., Dirac, A. M. & Bernards, R. (2003) Nature 424, 797–801. [DOI] [PubMed] [Google Scholar]
- 11.Trompouki, E., Hatzivassiliou, E., Tsichritzis, T., Farmer, H., Ashworth, A. & Mosialos, G. (2003) Nature 424, 793–796. [DOI] [PubMed] [Google Scholar]
- 12.Gilchrist, C. A. & Baker, R. T. (2000) Biochim. Biophys. Acta 1481, 297–309. [DOI] [PubMed] [Google Scholar]
- 13.Janssen, J. W., Schleithoff, L., Bartram, C. R. & Schulz, A. S. (1998) Oncogene 16, 1767–1772. [DOI] [PubMed] [Google Scholar]
- 14.Gray, D. A., Inazawa, J., Gupta, K., Wong, A., Ueda, R. & Takahashi, T. (1995) Oncogene 10, 2179–2183. [PubMed] [Google Scholar]
- 15.Li, M., Chen, D., Shiloh, A., Luo, J., Nikolaev, A. Y., Qin, J. & Gu, W. (2002) Nature 416, 648–653. [DOI] [PubMed] [Google Scholar]
- 16.Li, Z., Wang, D., Na, X., Schoen, S. R., Messing, E. M. & Wu, G. (2002) Biochem. Biophys. Res. Commun. 294, 700–709. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, D. E., Proctor, M., Marquis, S. T., Gardner, H. P., Ha, S. I., Chodosh, L. A., Ishov, A. M., Tommerup, N., Vissing, H., Sekido, Y., et al. (1998) Oncogene 16, 1097–1112. [DOI] [PubMed] [Google Scholar]
- 18.Yanagisawa, T. Y., Sasahara, Y., Fujie, H., Ohashi, Y., Minegishi, M., Itano, M., Morita, S., Tsuchiya, S., Hayashi, Y., Ohi, R. & Konno, T. (1998) Tohoku J. Exp. Med. 184, 229–240. [DOI] [PubMed] [Google Scholar]
- 19.D'Andrea, V., Malinovsky, L., Berni, A., Biancari, F., Biassoni, L., Di Matteo, F. M., Corbellini, L., Falvo, L., Santoni, F., Spyrou, M. & De Antoni, E. (1997) G Chir 18, 521–524. [PubMed] [Google Scholar]
- 20.Schumacher, U., Mitchell, B. S. & Kaiserling, E. (1994) DNA Cell Biol. 13, 839–843. [DOI] [PubMed] [Google Scholar]
- 21.Tezel, E., Hibi, K., Nagasaka, T. & Nakao, A. (2000) Clin. Cancer Res. 6, 4764–4767. [PubMed] [Google Scholar]
- 22.Brichory, F., Beer, D., Le Naour, F., Giordano, T. & Hanash, S. (2001) Cancer Res. 61, 7908–7912. [PubMed] [Google Scholar]
- 23.Sasaki, H., Yukiue, H., Moriyama, S., Kobayashi, Y., Nakashima, Y., Kaji, M., Fukai, I., Kiriyama, M., Yamakawa, Y. & Fujii, Y. (2001) Jpn J. Clin. Oncol. 31, 532–535. [DOI] [PubMed] [Google Scholar]
- 24.Zhu, Y., Lambert, K., Corless, C., Copeland, N. G., Gilbert, D. J., Jenkins, N. A. & D'Andrea, A. D. (1997) J. Biol. Chem. 272, 51–57. [DOI] [PubMed] [Google Scholar]
- 25.Zhu, Y., Carroll, M., Papa, F. R., Hochstrasser, M. & D'Andrea, A. D. (1996) Proc. Natl. Acad. Sci. USA 93, 3275–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naviglio, S., Mattecucci, C., Matoskova, B., Nagase, T., Nomura, N., Di Fiore, P. P. & Draetta, G. F. (1998) EMBO J. 17, 3241–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavioli, R., Frisan, T., Vertuani, S., Bornkamm, G. W. & Masucci, M. G. (2001) Nat. Cell Biol. 3, 283–288. [DOI] [PubMed] [Google Scholar]
- 28.Miller, G. (1974) J. Infect. Dis. 130, 187–205. [DOI] [PubMed] [Google Scholar]
- 29.Borodovsky, A., Kessler, B. M., Casagrande, R., Overkleeft, H. S., Wilkinson, K. D. & Ploegh, H. L. (2001) EMBO J. 20, 5187–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein, U., Jurchott, K., Schlafke, M. & Hohenberger, P. (2002) J. Clin. Oncol. 20, 3282–3292. [DOI] [PubMed] [Google Scholar]
- 31.Spataro, V., Toda, T., Craig, R., Seeger, M., Dubiel, W., Harris, A. L. & Norbury, C. (1997) J. Biol. Chem. 272, 30470–30475. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson, K. D., Lee, K. M., Deshpande, S., Duerksen-Hughes, P., Boss, J. M. & Pohl, J. (1989) Science 246, 670–673. [DOI] [PubMed] [Google Scholar]
- 33.Li, T., Duan, W., Yang, H., Lee, M. K., Bte Mustafa, F., Lee, B. H. & Teo, T. S. (2001) FEBS Lett. 488, 201–205. [DOI] [PubMed] [Google Scholar]
- 34.Kieff, E. (1996) in Fields Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott, Raven, Philadelphia), Vol. 2, 3rd Ed., pp. 2343–2396. [Google Scholar]
- 35.Saigoh, K., Wang, Y. L., Suh, J. G., Yamanishi, T., Sakai, Y., Kiyosawa, H., Harada, T., Ichihara, N., Wakana, S., Kikuchi, T. & Wada, K. (1999) Nat. Genet. 23, 47–51. [DOI] [PubMed] [Google Scholar]
- 36.Osaka, H., Wang, Y. L., Takada, K., Takizawa, S., Setsuie, R., Li, H., Sato, Y., Nishikawa, K., Sun, Y. J., Sakurai, M., et al. (2003) Hum. Mol. Genet. 12, 1945–1958. [DOI] [PubMed] [Google Scholar]
- 37.Larsen, C. N., Krantz, B. A. & Wilkinson, K. D. (1998) Biochemistry 37, 3358–3368. [DOI] [PubMed] [Google Scholar]
- 38.Finch, J. S., St. John, T., Krieg, P., Bonham, K., Smith, H. T., Fried, V. A. & Bowden, G. T. (1992) Cell Growth Differ. 3, 269–278. [PubMed] [Google Scholar]
- 39.Liu, Y., Fallon, L., Lashuel, H. A., Liu, Z. & Lansbury, P. T., Jr. (2002) Cell 111, 209–218. [DOI] [PubMed] [Google Scholar]
- 40.Fischer-Vize, J. A., Rubin, G. M. & Lehmann, R. (1992) Development (Cambridge, U.K.) 116, 985–1000. [DOI] [PubMed] [Google Scholar]
- 41.Taya, S., Yamamoto, T., Kanai-Azuma, M., Wood, S. A. & Kaibuchi, K. (1999) Genes Cells 4, 757–767. [DOI] [PubMed] [Google Scholar]
- 42.Henderson, B. R. & Fagotto, F. (2002) EMBO Rep. 3, 834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung, E. J., Hwang, S. G., Nguyen, P., Lee, S., Kim, J. S., Kim, J. W., Henkart, P. A., Bottaro, D. P., Soon, L., Bonvini, P., et al. (2002) Blood 100, 982–990. [DOI] [PubMed] [Google Scholar]
- 44.Song, J. Y., Lim, J. W., Kim, H., Morio, T. & Kim, K. H. (2003) J. Biol. Chem. 278, 36676–36687. [DOI] [PubMed] [Google Scholar]
- 45.Gonen, H., Smith, C. E., Siegel, N. R., Kahana, C., Merrick, W. C., Chakraburtty, K., Schwartz, A. L. & Ciechanover, A. (1994) Proc. Natl. Acad. Sci. USA 91, 7648–7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheper, R. J., Broxterman, H. J., Scheffer, G. L., Kaaijk, P., Dalton, W. S., van Heijningen, T. H., van Kalken, C. K., Slovak, M. L., de Vries, E. G., van der Valk, P., et al. (1993) Cancer Res. 53, 1475–1479. [PubMed] [Google Scholar]
- 47.Kickhoefer, V. A., Rajavel, K. S., Scheffer, G. L., Dalton, W. S., Scheper, R. J. & Rome, L. H. (1998) J. Biol. Chem. 273, 8971–8974. [DOI] [PubMed] [Google Scholar]
- 48.Filipits, M., Jaeger, U., Simonitsch, I., Chizzali-Bonfadin, C., Heinzl, H. & Pirker, R. (2000) Clin. Cancer Res. 6, 3417–3423. [PubMed] [Google Scholar]
- 49.Filipits, M., Drach, J., Pohl, G., Schuster, J., Stranzl, T., Ackermann, J., Konigsberg, R., Kaufmann, H., Gisslinger, H., Huber, H., et al. (1999) Clin. Cancer Res. 5, 2426–2430. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.