Abstract
p53 promotes tumor suppression through its ability to function as a transcriptional factor and is activated by posttranslational modifications that include acetylation. Our earlier study demonstrated that p53 acetylation can enhance its sequence-specific DNA binding in vitro, and this notion was later confirmed in several other studies. However, a recent study has reported that in vitro acetylation of p53 fails to stimulate its DNA binding to large DNA fragments, raising an important issue that requires further investigation. Here, we show that unacetylated p53 is able to bind weakly to its consensus site within the context of large DNA fragments, although it completely fails to bind the same site within short oligonucleotide probes. Strikingly, by using highly purified and fully acetylated p53 proteins obtained from cells, we show that acetylation of the C-terminal domain can dramatically enhance site-specific DNA binding on both short oligonucleotide probes and long DNA fragments. Moreover, endogenous p53 apparently can be fully acetylated in response to DNA damage when both histone deacetylase complex 1 (HDAC1)- and Sir2-mediated deacetylation are inhibited, indicating dynamic p53 acetylation and deacetylation events during the DNA damage response. Finally, we also show that acetylation of endogenous p53 indeed significantly augments its ability to bind an endogenous target gene and that p53 acetylation levels correlate well with p53-mediated transcriptional activation in vivo. Thus, our results clarify some of the confusion surrounding acetylation-mediated effects on p53 binding to DNA and suggest that acetylation of p53 in vivo may contribute, at least in part, to its transcriptional activation functions.
Keywords: ChIP, transcription, CBP/p300
Often, p53 is referred to as the “guardian of the genome” (1). In response to DNA damage and other types of stress, p53 is able to induce cell growth arrest, apoptosis, and cell senescence (1–3). Mutations within the p53 tumor suppressor gene have been well documented in >50% of all human tumors (4). In cells that retain wild-type p53, multiple regulatory pathways play an important role in modulating its activities in vivo (2, 5, 6). p53 promotes tumor suppression through its ability to bind specific DNA sequences and to act as a transcription factor (7). The importance of p53-mediated transcriptional activation is underscored by the fact that the vast majority of tumor-associated p53 mutations occur within the domain responsible for sequence-specific DNA binding (4). A number of genes that are critically involved in either cell growth arrest or apoptosis have been identified as p53 direct targets. These include p21CIP1/WAF1, Mdm2, GADD45, Cyclin G, 14–3-3σ, Noxa, p53AIP1, and PUMA, among others (7–15).
p53 is tightly regulated, with the protein often found in latent form and the protein levels being very low in unstressed cells (16). This regulation is essential for its effect on tumorigenesis, as well as for maintaining normal cell growth. It is generally thought that the mechanism of p53 activation by cellular stress involves mainly posttranslational modifications, including phosphorylation and acetylation (17, 18). The discovery that CREB-binding protein (CBP)/p300 could acetylate p53 and increase its DNA binding activity established a new paradigm for transcriptional regulation (19), and subsequent studies have shown that many transcriptional factors are functionally modified by acetylation (20–22).
p53 is acetylated by CBP/p300 at multiple lysine residues (positions 370, 372, 373, 381, and 382) within the C-terminal regulatory domain. The sequence-specific DNA binding activity of p53 in electrophoretic mobility-shift assays (EMSA) was shown to be dramatically stimulated by acetylation, possibly as a result of an acetylation-induced conformational change (19, 23, 24). The physiological relevance of p53 acetylation became more evident with the finding that p53 can be acetylated in vivo in response to a variety of cellular stresses (25). In addition, another histone acetyltransferase, p300/CBP-associated factor (PCAF), was shown to acetylate p53 at Lys-320, although functional consequences in vivo need to be further elucidated (23, 24, 26). The level of acetylated p53 is up-regulated in primary fibroblasts by the Ras oncogene and by promyelocytic leukemia (PML), which also induce premature senescence of these cells in a p53-dependent manner (27, 28). Interestingly, several studies have indicated that p53 C-terminal domain lysine residues, five of which are acetylation sites, play a critical role in Mdm2-mediated ubiquitination and subsequent degradation (29–32). In addition, increasing the levels of p53 acetylation with deacetylase inhibitors was found to prevent p53 from degradation in vivo (41, 46), indicating that acetylation of p53 may directly regulate its stability. Furthermore, Barlev et al. (33) have reported that acetylation of p53 is important for efficient in vivo recruitment of CBP and PCAF complexes to promoter regions and for activation of p53-targeted gene expression. These recent findings thus expand the in vivo role of p53 to include protein stability and protein–protein interactions, in addition to DNA binding.
The ability of an mAb (PAb421) that recognizes the C terminus of p53 (34) or of p53 C-terminal peptides (35) to activate DNA binding and transcription functions of p53, both in vitro and in vivo, underscores the importance of sequence-specific DNA binding in p53 activation. These findings also suggest the presence of a cellular pool of latent p53 that can be activated posttranslationally. According to the allosteric model that has been proposed in many studies (34, 36, 37), the C-terminal tail of p53 acts as a negative regulator and may interact with the core DNA binding domain to lock the DNA binding domain into an inactive conformation. However, single-strand DNA interactions, antibody binding, and posttranslational modifications, such as acetylation or phosphorylation, could disrupt interactions between the C-terminal domain and the core domain, thus allowing the DNA binding domain to adopt an active conformation (19, 34, 36, 37).
Recently, Espinosa and Emerson (38) proposed a contradictory model based on their finding that, in the context of an artificially reconstituted chromatin-assembled p21 promoter, p53 can activate transcription equally well regardless of its acetylation status (38). Further, a C-terminal deletion mutant of p53 was reported to be transcriptionally inactive in the in vitro system, in contrast to reports that C-terminal deletion mutants exhibit very strong p21 promoter activation and cell growth repression (39, 40). Espinosa and Emerson also failed to observe a significant enhancement of p53 binding to DNA, in response either to antibody binding or to in vitro acetylation, when using either a larger DNA fragment or an artificially reconstituted chromatin template. To reevaluate the allosteric model, as well as the role of acetylation in the regulation of p53-mediated function, we have obtained highly purified and fully acetylated p53 from cells and demonstrated that acetylation of p53 strongly enhances its sequence-specific DNA binding both in vitro and in vivo.
Experimental Procedures
p53 Protein Expression and Purification. pET-Flag-p53 and pET-Flag-p53 (K-Q) mutant plasmids were constructed as previously (41). Both plasmids were transformed into Escherichia coli BL21 competent cells. The proteins were induced by 1 mM isopropyl β-d-thiogalactoside at 25°C overnight and extracted with buffer BC500 (20 mM Tris·HCl, pH 8.0/500 mM KCl/0.5 mM EDTA/20% glycerol/1 mM DTT/0.5 mM PMSF) containing 1% Nonidet P-40, and purified by anti-Flag M2-agarose affinity gel (Sigma). Total p53 and acetylated p53 were expressed in and purified from H1299 cells as described (41) with some modifications. To purify the acetylated p53 proteins, H1299 cells were cotransfected with Flag-p53 and p300. Twenty hours after the transfection, the cells were treated with 1 mM trichostatin A (TSA) and 5 mM nicotinamide for 6 h, then lysed in Flag–lysis buffer (50 mM Tris·HCl, pH 7.8/137 mM NaCl/10 mM NaF/1 mM EDTA/1% Triton X-100/0.2% Sarkosyl/1 mM DTT/10% glycerol and fresh proteinase inhibitors) with mild sonication, and the cell extracts were immunoprecipitated with anti-Flag mAb beads (M2, Sigma). After elution with the Flag peptide, the total p53 proteins were loaded onto a PAb421 antibody column. The unacetylated portion of the p53 protein was depleted by the PAb421 column after repeated passage. Unbound proteins consisted of purified acetylated p53 and were further tested for purity by Western blot analysis with anti-p53 antibodies (DO-1, PAb421, or acetylated p53-specific antibody). To avoid p53 deacetylation by both HDAC1- and Sir2-mediated deacetylases (23, 24), 1 mM TSA and 5 mM nicotinamide were added in each step.
EMSA. EMSA was carried out essentially as described (19) with some modifications. The short probe (25 bp) was generated by annealing the single-stranded oligonucleotides 5′-caggaacatgtcccaagatgttgaa-3′ and its complementary sequence. The long probe (220 bp) was generated by PCR with 5′-tgctgcctgcttcccaggaaca-3′ (sense) and 5′-ccatccccttcctcacctgaaa-3′ (antisense) primers. Both probes were 32P end-labeled by using T4 polynucleotide kinase and further purified by QIAquick nucleotide removal kit (Qiagen, Valencia, CA). The DNA binding reactions (20 μl) contained 20 mM Tris·HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, 10% glycerol, 0.5 mg/ml BSA, and 100 ng of poly(dI-dC) and proteins as indicated. Reaction mixtures were preincubated at room temperature for 30 min before a 32P-labeled DNA probe was added and further incubated at room temperature for 30 min. The mixtures were then resolved on a native 4% polyacrylamide gel at 4°C followed by autoradiography.
Chromatin Immunoprecipitation (ChIP) Assay. The ChIP assay was performed essentially as described (42) with some modifications. A total of 107 H460 cells (treated with 20 μm of etoposide for 4 h or untreated) were crosslinked with 1% formaldehyde for 10 min at room temperature. Cells were harvested in SDS buffer (0.5% SDS/100 mM NaCl/50 mM Tris·HCl, pH 8.1/5 mM EDTA). After centrifugation, the cell pellet was resuspended in 2.5 ml of IP buffer (0.3% SDS/100 mM NaCl/50 mM Tris·HCl, pH 8.1/5 mM EDTA/2% Triton X-100), followed by sonication to an average DNA length of 500–1,000 bp. The sample was cleared by centrifugation and divided into five aliquots of 0.5 ml, with 25 μl being saved for input control. Antibodies were added to each of the sample tubes, which were then rotated at 4°C overnight. Thirty microliters of protein A beads was then added to each of the tubes, which then were further rotated for 2 h at 4°C. The beads were washed three times with 1 ml of mixed Micelle wash buffer (150 mM NaCl/20 mM Tris·HCl, pH 8.1/5 mM EDTA/5% sucrose/1% Triton X-100/0.2% SDS), two times with 1 ml of buffer 500 (0.1% deoxycholic acid/1 mM EDTA/50 mM Hepes, pH 7.5/500 mM NaCl/1% Triton X-100), two times with 1 ml of LiCl/detergent solution (0.5% deoxycholic acid/1 mM EDTA/250 mM LiCl/0.5% Nonidet P-40/10 mM Tris·HCl, pH 8.0), and once with 1 ml of TE buffer. The beads were eluted with 300 μl of elution buffer (1% SDS/0.1 M NaHCO3) and incubated overnight at 65°C to reverse crosslinks. The eluates were treated with proteinase K and extracted with phenol/chloroform followed by ethanol precipitation. The DNAs were dissolved in TE buffer and analyzed by PCR. The following antibodies were used: anti-p53 FL (Santa Cruz Biotechnology), antiacetylated p53 (43), and antiosteoprotegerin (Santa Cruz Biotechnology). Primers used in PCR were from p21 promoter sequences: 5′-ctcacatcctccttcttcag-3′ (sense) and 5′-cacacacagaatctgactccc-3′ (antisense). Input DNA was diluted 1:10 for PCR. The PCR products were amplified for 27 cycles.
Measuring the Proportion of Acetylated p53 Proteins. H460 cells were harvested after treatment with 20 μm of etoposide (Sigma), 1 μM TSA (Sigma), and 5 mM nicotinamide (Sigma), or all three drugs for 6 h. Cell pellets were lysed in RIPA buffer (2 mM Tris·HCl, pH 7.4/5 mM EDTA/150 mM NaCl/1% Nonidet P-40/1% deoxycholic acid/0.025% SDS/1 mM PMSF/1 mM TSA/5 mM nicotinamide) with mild sonication. The cell extracts were immunoprecipitated with anti-p53 (FL) and antiacetylated p53 antibodies. The beads were eluted by SDS sample buffer after being washed with 0.5 ml of RIPA buffer five times. The samples were further tested by Western blot analysis with anti-p53 (DO-1).
Results
Acetylation, which modifies the lysine residues of both histone and nonhistone target proteins, is now recognized as an important regulatory step in transcriptional regulation (19–21). Many transcription factors other than p53, including GATA-1, MyoD, HMG-1, E2F-1, ACTR, EKLF, and Smad7, have been demonstrated to be bona fide substrates of acetyltransferases (44, 45). The functional consequences of acetylation are diverse and include increased DNA binding, increased stability, and altered protein–protein interactions. However, most of the functional studies on acetylated transcription factors, including those with p53, used preparations of acetylated proteins that lacked quantitative analyses of the acetylation levels because of an inability to resolve acetylated and unacetylated forms of the nonhistone proteins on regular SDS/PAGE gels (19–21, 23, 24, 26, 46). Thus, functional differences between the true acetylated and unacetylated forms could be hard to distinguish if only a minor fraction of the molecules in an acetylated preparation actually were acetylated.
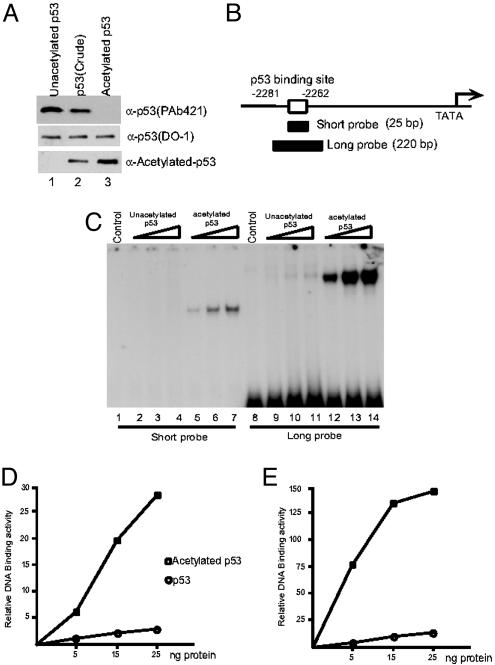
Recently, we developed a method to purify CBP/p300-acetylated p53 proteins from mammalian cells on the basis of their inability to be recognized by the anti-p53 mAb PAb421, raised specifically against the C-terminal domain of p53 (41). Thus, a highly purified form of acetylated p53 was successfully obtained through affinity chromatography procedures. To detect acetylated p53, we used an antibody that specifically recognizes acetylated p53 polypeptides at all different p300-mediated acetylation sites (43). As shown in Fig. 1A, the purified acetylated p53 reacted strongly with the acetylated p53-specific antibody but poorly with the PAb421 antibody, whereas the opposite was true for the purified unacetylated p53 (Fig. 1 A Bottom, lane 3 vs. 1). Equal reactivity with the DO-1 antibody, which recognizes the N-terminal domain of p53, confirmed equal molar inputs of acetylated vs. unacetylated p53. To examine the effect of acetylation on site-specific DNA binding, we tested the DNA binding activities of acetylated p53 on a short oligonucleotide probe (25 bp) and a 220-bp-long DNA fragment, both of which are derived from the p21 promoter region (Fig. 1B, ref. 7). As expected (19), the unmodified p53 was virtually inactive in site-specific DNA binding to the 25-bp probe in an EMSA (Fig. 1C, lanes 2–4), whereas significantly enhanced binding was observed with acetylated p53 (Fig. 1C, lanes 5–7, and D). A weak, but rather significant binding was observed by the same amount of unacetylated p53 proteins on a 220-bp probe (Fig. 1C, lanes 9–11), indicating that unacetylated forms of p53 isolated from mammalian cells are capable of binding the target site. This observation is consistent with our earlier results (19) and those of Espinosa and Emerson (38). However, in contrast to Espinosa and Emerson (38) and by using a highly purified acetylated p53, we found that acetylation of p53 strongly augments its DNA binding on a long DNA fragment comparable with that used by these investigators (Fig. 1C, lanes 12–14, and E).
Fig. 1.
p53 acetylation strongly enhances its DNA binding activity in vitro.(A) Western blot analysis of purified acetylated p53 (lane 3), unacetylated p53 (lane 1), and a preparation of unresolved acetylated and unacetylated p53 (lane 2) proteins with the antiacetylated p53-specific antibody (Bottom), the DO-1 antibody (Middle), or the PAb421 antibody (Top). (B) Schematic representation of the short and long probes in the p21 promoter region used for EMSA. (C) EMSA assay of unacetylated p53 (lanes 2–4 and 9–11) and acetylated p53 (lanes 5–7 and 12–14) with short (lanes 1–7) and long (lanes 8–14) probes. (D and E) The DNA/protein complexes in C were quantitated by phosphorimaging by using imagequant software (Amersham Biosciences). (D) Results with the short probe. (E) Results with the long probe.
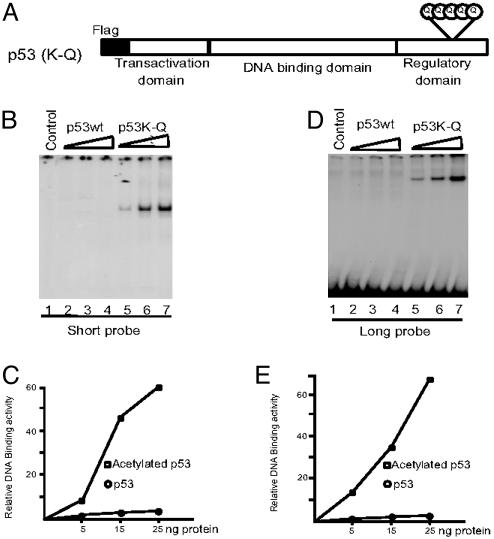
Lysine residues (Lys-370, -372, -373, -381, and -382) of the C-terminal regulatory domain of p53 are specifically acetylated by CBP/p300. Several studies have suggested that neutralization of the positive charges on these lysine residues is critical for disrupting the negative regulation of the p53 C terminus and that replacing them with either alanine or histidine residues significantly enhances the DNA binding activity of p53 on a short nucleotide probe (19–21, 23, 24, 38). Interestingly, it was previously reported that a histone H4 protein with a lysine to glutamine mutation at the H4 acetylation site was functionally comparable with the acetylated form in preventing Sir3p from binding H4 and spreading to form heterochromatin in yeast (47), consistent with the notion that this mutation mimicked the acetylated state of lysine through neutralization of the positive charges (41). Thus, to further assess whether neutralization of positive charges on p53 by acetylation might augment its DNA binding on a long DNA fragment, we tested the DNA binding activity of a p53 (K-Q) mutant in which all five known p300-dependent acetylation sites (Lys-370, -372, -373, -381, and -382) were replaced by glutamine residues (Fig. 2A).
Fig. 2.
Lysine to glutamine mutations in the p53 C terminus enhance DNA binding activity in vitro.(A) Schematic representation of the p53 (K-Q) mutant with lysine to glutamine changes at residues 370, 372, 373, 381, and 382. (B and C) EMSA assay of p53 wild type (wt) (lanes 2–4) and p53 (K-Q) mutant (lanes 5–7) with short probe (B). The resulting DNA-protein complexes in B were quantitated by phosphorimaging by using imagequant software (C). (D and F) EMSA assay of p53 wt (lanes 2–4) and p53 (K-Q) mutant (lanes 5–7) with long probe (D). The resulting DNA–protein complexes in D were quantitated by phosphorimaging by using imagequant software (F).
Both wild-type p53 and p53 (K-Q) proteins were expressed in bacteria as Flag-tagged proteins, purified to near homogeneity on M2 mAb affinity columns, and tested for DNA binding to both types of probes. As expected, wild-type p53 showed almost no binding to a 25-bp probe containing the p53 binding site from the p21 promoter. In contrast, p53 (K-Q) showed strong DNA binding activity (Fig. 2 B and C). Significantly, when a 220-bp DNA fragment was used as a probe, we observed the same enhancement of DNA binding by the p53 (K-Q) mutant protein (Fig. 2 D and E). These results demonstrate that neutralization of the positively charged lysine residues at the C terminus and/or a structural change is sufficient to render p53 able to bind strongly to a target site within both a short probe and a large DNA fragment.
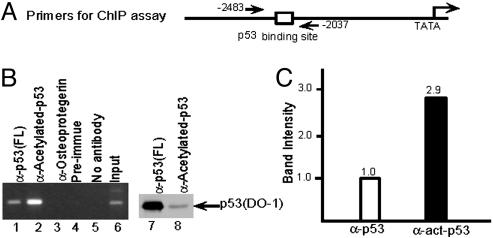
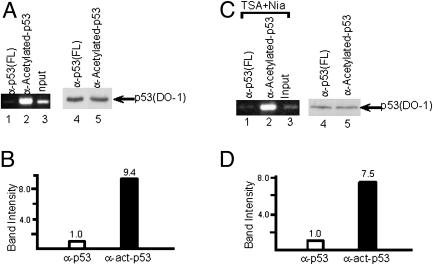
To examine the role of acetylation in the regulation of p53 binding to DNA in vivo, we tested the effect of acetylation on the ability of endogenous p53 to bind to DNA sequences within a natural target gene (the 5′ p53 binding site in the p21 promoter; Fig. 3A and ref. 7). ChIP assays were performed in human lung carcinoma H460 cells by using anti-p53 antibodies. To more accurately compare the DNA binding activity of acetylated p53 vs. total p53 proteins, we used a rabbit polyclonal antibody directed against the full-length p53 (Santa Cruz Biotechnology) and a rabbit polyclonal antibody that is specific for CBP/p300-acetylated p53 (43) to immunoprecipitate chromatin from cells after the treatment by DNA damage agents. As indicated in Fig. 3B (lanes 1 and 2), a high level of p53 binding to the p21 promoter was detected by immunoprecipitation with both anti-full-length p53 and antiacetylated p53 antibodies. Interestingly, although the antiacetylated p53 antibody only immunoprecipitated a small fraction of the total crosslinked p53 protein (Fig. 3B, lane 8 vs. 7), the p53 bound to the p21 promoter was enriched about 3-fold in acetylated p53 relative to total p53 (Fig. 3C). To quantitatively analyze differences in p53 binding to the endogenous p21 promoter, the amounts of the p53 proteins immunoprecipitated by these two different antibodies were normalized on the basis of immunoreactivity with the DO-1 antibody. As shown in Fig. 4, ChIP analysis using similar amounts of immunoprecipitated p53 proteins in the PCR assay (Fig. 4A, lanes 4 and 5) showed an almost 10-fold enhancement of binding to p21 promoter DNA by acetylated p53 (Fig. 4A, lanes 1–3, and B). Thus, our results also indicate that it is critical to normalize the p53 protein amount when two different antibodies are used to immunoprecipitate the endogenous p53 in the ChIP assay, which may explain the failure of others to detect an effect of acetylation on p53 binding to DNA in vivo (33).
Fig. 3.
p53 acetylation strongly enhances its DNA binding activity in vivo.(A) Schematic representation of the primers in p21 promoter region used for ChIP assay. (B) ChIP assay by using anti-p53 (FL) (lane 1), antiacetylated p53 (lane 2), antiosteoprotegerin (lane 3), preimmune (lane 4), and no antibody (lane 5). Western blot shows p53 protein amount in ChIP assay by anti-p53 (FL) (lane 7) and by antiacetylated p53 (lane 8). Cells were treated by etoposide before the ChIP assay. (C) The resulting DNA amounts in ChIP assay by anti-p53 (FL) and antiacetylated p53 (B, lanes 1 and 2) were quantitated by phosphorimaging using imagequant software.
Fig. 4.
Acetylation of p53 enhances its DNA binding on endogenous promoters. (A) ChIP assay with equal amounts of immunoprecipitated acetylated and total p53 proteins (lanes 1 and 2). A Western blot with the DO-1 antibody shows the amounts of immunoprecipitated p53 proteins, either by anti-p53 (FL) (lane 4) or by antiacetylated-p53 (lane 5) antibodies, used in the ChIP assays in lanes 1 and 2. Cells were treated by etoposide before the ChIP assay. (B) The resulting DNA amounts in A were quantitated by phosphorimaging by using imagequant software. (C) ChIP assays with anti-p53 (FL) (lane 1) and antiacetylated-p53 (lane 2). Western blot with the DO-1 antibody shows the amounts of immunoprecipitated p53 proteins, either by anti-p53 (FL) (lane 4) or by antiacetylated-p53 (lane 5) antibodies, used in the ChIP assays in lanes 1 and 2. Cells were treated with deacetylase inhibitors TSA and nicotinamide before the ChIP assay. (D) The resulting DNA amounts in C were quantitated by phosphorimaging using imagequant software.
To minimize contributions of other factors (e.g., p53 phosphorylation) that may also be involved in binding of p53 to the p21 promoter under DNA damage conditions, we also performed a similar experiment in unstressed cells. Cells were cultured in the presence of deacetylase inhibitors that included TSA (for HDACs) and nicotinamide (for Sir2) to accumulate sufficient levels of acetylated p53. After ChIP with different antibodies, PCRs were performed on samples with normalized amounts of p53 proteins. As indicated in Fig. 4 C and D, acetylated p53 showed high level binding to the p21 promoter, whereas the same amount of total p53 protein revealed only a very weak binding to the promoter in the same cells. Taken together, the above results demonstrate that acetylation of p53 can strongly enhance its binding to the endogenous p21 promoter under physiological conditions.
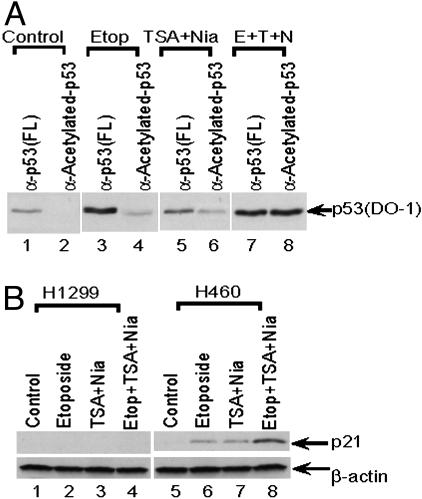
To further investigate the function of p53 acetylation in transcription, we tested whether the levels of p53 acetylation correlate with levels of p53-mediated transcription of the endogenous p21 target gene. Interestingly, in H460 cells treated with etoposide, a comparison of total p53 protein (immunoprecipitated with anti-full-length p53 antibody) and acetylated p53 protein (immunoprecipitated with antiacetylated p53 antibody) indicated that only ≈20% of total p53 is acetylated (Fig. 5A, lane 4 vs. lane 3). These results thus indicate that even under conditions of DNA damage, apparently only a fraction of total p53 is acetylated. Furthermore, the results also indicate that HDAC1- and Sir2α-mediated p53 deacetylation may play critical roles in regulating the levels of p53 acetylation, as treatment with corresponding deacetylase inhibitors (TSA and nicotinamide) significantly increased the levels of acetylated p53 in vivo (Fig. 5A, lane 6 vs. lane 2). More importantly, in the presence of TSA and nicotinamide, p53 was fully acetylated in vivo in response to DNA damage by etoposide (Fig. 5A, lane 8 vs. lane 7). Furthermore, as shown in Fig. 5B, p21 expression levels were significantly enhanced when the cells were treated with DNA damage reagents and/or deacetylase inhibitors. More importantly, the acetylation levels of p53 also correlate well with p21 activation in the H460 human lung carcinoma cells (Fig. 5B, lanes 5–8). There was no obvious p21 activation in p53-null human lung carcinoma H1299 cells under the same conditions (Fig. 5B, lanes 1–4). These data suggest that acetylation of p53, at least in part, contributes to p53-mediated transcriptional activation in vivo.
Fig. 5.
Endogenous p53 is fully acetylated on DNA damage. (A) Western blot with DO-1 antibody showing the amounts of p53 immunoprecipitated with the indicated p53 antibodies from cells with no treatment (lanes 1 and 2), treatment with etoposide (lanes 3 and 4), treatment with TSA and nicotinamide (lanes 5 and 6), or treatment with etoposide, TSA, and nicotinamide (lanes 7 and 8). (B) Western blot showing p21 levels (Upper) in p53-null H1299 cells (lanes 1–4) or wild-type p53-expressing H460 cells (lanes 5–8) after treatment with the indicated drugs. β-Actin (Lower) was used as a loading control.
Discussion
Numerous studies indicate that the C terminus of p53 acts as a critical regulator of p53 function. Deletion of the C terminus, antibodies specific for the C terminus (PAb421), single-stranded DNA, interacting proteins such as HMG-1, and posttranslational modifications of the C terminus have all been reported to induce the transactivation potential of p53 in various assays (34–37). Consistent with this model, C-terminal acetylation of p53 was found to dramatically stimulate its sequence-specific DNA binding activity in vitro, possibly as a result of an acetylation-induced conformational change (19). These results were further confirmed in several similar studies (23, 24). Nevertheless, one recent study (38) reported that the effects of p53 acetylation on its DNA binding vary with the nature of the DNA binding probes. In confirmation of the original idea that acetylation of p53 can enhance its sequence-specific DNA binding, we have shown that a highly purified and fully acetylated (at the CBP/p300 sites) p53 isolated from cultured cells exhibits enhanced site-specific DNA binding to several DNA probes in vitro. Consistent with these findings, ChIP assays have demonstrated that acetylation of p53 augments binding to an endogenous target promoter. Our findings seem to be at odds with the results reported by Espinosa and Emerson (38), who failed to see any effect of in vitro acetylation on p53 binding to long DNA fragments. However, their assays used an in vitro acetylated p53 protein without further purification, and it was not determined whether acetylated p53 comprised a major or a minor fraction in their preparation. In fact, because the PAb421 antibody (directed against the p53 C terminus) does not recognize acetylated p53 but was able to quantitatively supershift the p53/DNA complexes (38), this would argue that the major proportion of p53 in the acetylated p53 protein preparations used by Espinosa and Emerson was indeed unacetylated. As a consequence, these p53 preparations are likely compromised in their ability to show acetylation-dependent binding of p53 to DNA, particularly on the large DNA fragments that show a weak, but significant binding of unacetylated p53. On the other hand, in relation to the observation of a requirement for the p53 C terminus for p53 function in vitro (38), we cannot exclude the possibility that the C terminus of p53 behaves as a positive factor for transcription under certain circumstances (ref. 48; W. An and R.G.R., unpublished results). The reported role of p53 C-terminal acetylation events in coactivator recruitment is consistent with such a function (33).
More than 30 years ago, acetylation was shown to be an important modification of histones that correlated with increased transcriptional activity (20). The significance of histone acetylation for transcriptional regulation seems well accepted, although the precise roles for the diverse acetylation events are still not completely understood. More recently, a large number of nonhistone transcriptional factors have been demonstrated to be bona fide substrates for acetyltransferases, suggesting that acetylation represents another type of general protein modification important for functional regulation of transcriptional factors (45). CBP/p300, a protein possessing histone acetyltransferase activity, acts as a coactivator of p53 and potentiates its transcriptional activity as well as its biological function in vivo (49–51). The significance of CBP/p300, and arguably its interaction with p53, is underscored by the presence of p300 mutations in several types of tumors (52). Additionally, mutations of CBP in human Rubeinstein–Taybi syndrome, as well as CBP-knockout mice, lead to a higher risk of tumorigenesis (52). Although CBP/p300-mediated transcription activation by p53 also involves nucleosomal histone acetylation at the promoter region, our results indicate that acetylation of p53 may contribute, at least in part, to full activation of specific p53 target genes in vivo by increasing its site-specific DNA binding activity.
HDACs have emerged as notable components in regulating transcriptional activation as well. Treatment of cells with the HDAC inhibitor TSA was found to increase the level of acetylated p53 and led to the identification of the adaptor protein PID/MTA2, a component of the HDAC1 complex that can enhance HDAC1-mediated deacetylation of p53 (43, 53). Subsequent work has identified Sir2α (SIRT1), a TSA-resistant, nicotinamide adenine dinucleotide (NAD)-dependent histone deacetylase that can both deacetylate p53 and attenuate its transcriptional activity (54–56). Moreover, SIRT1-deficient cells exhibit p53 hyperacetylation after DNA damage and ionizing radiation-induced thymocyte apoptosis, further supporting a role for SIRT1-mediated p53 deacetylation in p53-dependent stress responses (57). Recent studies indicate that targeted deacetylation and acetylation occur very quickly amidst a global equilibrium of genomic acetylation and deacetylation (58, 59). Although the steady-state level of acetylated p53 is low even in stressed cells, we have found that p53 is fully acetylated (at the p300/CBP sites) in response to DNA damage when p53 deacetylation is inhibited (Fig. 5A, lane 7 vs. 8). Thus, it is possible that acetylation of p53 plays a critical role in initiating the binding of p53 to at least some DNA sites, in addition to the maintenance of this binding and effects on coactivator interactions at the target promoter (33). Because some p53 target promoters such as p21 contain very strong DNA binding sites for p53, it also is possible that acetylation-mediated augmentation of p53 DNA binding contributes less to transcription activation on these promoters relative to other p53 target promoters with weak binding sites. Thus, just as phosphorylation of Ser-46 at the p53 N-terminal domain seems to be important specifically in UV-induced apoptosis through activation of the p53AIP1gene (11), acetylated p53 may preferentially interact with, or function at, specific promoters and induce subsequent cellular responses. In this regard, it was reported that DNA damage-induced acetylation of p73 potentiates its abilities to selectively activate the transcription of proapoptotic genes (60), further supporting this notion. Importantly, acetylation of p53 also regulates its own stability as well as its physical interactions with transcriptional coactivators such as CBP/p300 (33, 41, 45, 46). Taken together, it is very likely that acetylation of p53 acts through multiple mechanisms, including effects on DNA binding, protein stability and protein–protein interactions, to synergistically regulate transcription in vivo.
Acknowledgments
We thank other members of the Gu laboratory and Woojin An of the Roeder laboratory for sharing unpublished data and for critical comments. This work was supported in part by grants from the Leukemia and Lymphoma Society Foundation, the Stewart Trust, the Irma T. Hirschl Trust, and the National Institutes of Health National Cancer Institute (to W.G.), and by The Rockefeller University (to R.G.R.). W.G. is a Leukemia and Lymphoma Society Scholar.
Abbreviations: TSA, trichostatin A; EMSA, electrophoretic mobility-shift assay; ChIP, chromatin immunoprecipitation; HDAC, histone deacetylase complex; CBP, CREB-binding protein.
References
- 1.Levine, A. J. (1997) Cell 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 2.Prives, C. & Hall, P. A. (1999) Pathol. J. 187, 112–126. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 4.Hollstein, M., Rice, K., Greenblatt, M. S., Soussi, T., Fuchs, R., Sorlie, T., Hovig, E., Smith-Sorensen, B., Montesano, R. & Harris, C. C. (1994) Nucleic Acids Res. 22, 3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 5.Lohrum, M. A. & Vousden, K. H. (1999) Cell Growth Differ. 6, 1162–1168. [DOI] [PubMed] [Google Scholar]
- 6.Vousden, K. H. (2000) Cell 103, 691–694. [DOI] [PubMed] [Google Scholar]
- 7.El-Deiry, W. S., Tokino, T., Velculescu, V. E., Levy, D. B., Parsons, R., Trent, J. M., Lin, D., Mercer, W. E., Kinzler, K. W. & Vogelstein, B. (1993) Cell 75, 817–825. [DOI] [PubMed] [Google Scholar]
- 8.Nakano, K. & Vousden, K. (2001) Mol. Cell 7, 683–694. [DOI] [PubMed] [Google Scholar]
- 9.Yu, J., Zhang, L., Hwang, P., Kinzler, K. & Vogelstein, B. (2001) Mol. Cell 7, 673–682. [DOI] [PubMed] [Google Scholar]
- 10.Oda, E., Ohki, R., Murasawa, H., Nemoto, J., Shibue, T., Yamashita, T., Tokino, T., Taniguchi, T. & Tanaka, N. (2000) Science 288, 1053–1058. [DOI] [PubMed] [Google Scholar]
- 11.Oda, K., Arakawa, H., Tanaka, T., Matsuda, K., Tanikawa, C., Mori, T., Nishimori, H., Tamai, K., Tokino, T., Nakamura, Y. & Taya, Y. (2000) Cell 102, 849–862. [DOI] [PubMed] [Google Scholar]
- 12.Wu, X., Bayle, J. H., Olson, D. & Levine, A. J. (1993) Genes Dev. 7, 1126–1132. [DOI] [PubMed] [Google Scholar]
- 13.Barak, Y., Juven, T., Haffner, R. & Oren, M. (1993) EMBO J. 12, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kastan, M. B., Zhan, Q., el-Deiry, W. S., Carrier, F., Jacks, T., Walsh, W. V., Plunkett, B. S., Vogelstein, B. & Fornace, A. J., Jr. (1992) Cell 71, 587–597. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto, K. & Beach, D. (1994) EMBO J. 13, 4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freedman, D. A., Wu, L. & Levine, A. J. (1999) Cell Mol. Life Sci. 55, 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Appella, E. & Anderson, C. W. (2000) Pathol. Biol. 48, 227–245. [PubMed] [Google Scholar]
- 18.Giaccia, A. J. & Kastan, M. B. (1998) Genes Dev. 12, 2973–2983. [DOI] [PubMed] [Google Scholar]
- 19.Gu, W. & Roeder, R. G. (1997) Cell 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 20.Kouzarides, T. (2000) EMBO J. 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sterner, D. E. & Berger, S. L. (2000) Microbiol. Mol. Biol. 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muth, V., Nadaud, S., Grummt, I. & Voit, R. (2001) EMBO J. 20, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakaguchi, K., Herrera, J. E., Saito, S., Miki, T., Bustin, M., Vassilev, A., Anderson, C. W. & Appella, E. (1998) Genes Dev. 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, L., Scolnick, D. M., Trievel, R. C., Zhang, H. B., Marmorstein, R., Halazonetis, T. D. & Berger, S. L. (1999) Mol. Cell. Biol. 19, 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prives, C. & Manley, J. L. (2001) Cell 107, 815–818. [DOI] [PubMed] [Google Scholar]
- 26.Liu, Y, Colosimo, A. L., Yang, X. J. & Liao, D. (2000) Mol. Cell. Biol. 20, 5540–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson, M., Carbone, R., Sebastiani, C., Cioce, M., Fagioli, M., Saito, S., Higashimoto, Y., Appella, E., Minucci, S., Pandolfi, P. P. & Pelicci, P. G. (2000) Nature 406, 207–210. [DOI] [PubMed] [Google Scholar]
- 28.Ferbeyre, G., de Stanchina, E., Querido, E., Baptiste, N., Prives, C. & Lowe, S. W. (2000) Genes Dev. 14, 2015–2027. [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez, M. S., Desterro, J. M., Lain, S., Lane, D. P. & Hay, R. T. (2000) Mol. Cell. Biol. 20, 8458–8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura, S., Roth, J. A. & Mukhopadhyay, T. (2000) Mol. Cell. Biol. 20, 9391–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu, J., Nie, L., Wiederschain, D. & Yuan, Z. M. (2001) Mol. Cell. Biol. 21, 8533–8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohrum, M. A., Woods, D. B., Ludwig, R. L., Balint, E. & Vousden, K. H. (2001) Mol. Cell. Biol. 21, 8521–8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barlev, N. A., Liu, L., Chehab, N. H., Mansfield, K., Harris, K. G., Halazonetis, T. D. & Berger, S. L. (2001) Mol. Cell 8, 1243–1254. [DOI] [PubMed] [Google Scholar]
- 34.Hupp, T. R., Sparks, A. & Lane, D. P. (1995) Cell 83, 237–245. [DOI] [PubMed] [Google Scholar]
- 35.Selivanova, G., Iotsova, V., Okan, I., Fritsche, M., Strom, M., Groner, B., Grafstrom, R. C. & Wiman, K. G. (1997) Nat. Med. 6, 632–638. [DOI] [PubMed] [Google Scholar]
- 36.Hupp, T. R., Meek, D. W., Midgley, C. A. & Lane, D. P. (1992) Cell 71, 875–886. [DOI] [PubMed] [Google Scholar]
- 37.Jayaraman, J. & Prives, C. (1995) Cell 81, 1021–1029. [DOI] [PubMed] [Google Scholar]
- 38.Espinosa, J. M. & Emerson, B. M. (2001) Mol. Cell 8, 57–69. [DOI] [PubMed] [Google Scholar]
- 39.Jayaraman, L., Moorthy, N. C., Murthy, K. G., Manley, J. L., Bustin, M. & Prives, C. (1998) Genes Dev. 12, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crook, T., Marston, N. J., Sara, E. A. & Vousden, K. H. (1994) Cell 79, 817–827. [DOI] [PubMed] [Google Scholar]
- 41.Li, M., Luo, J., Brooks, C. & Gu, W. (2002) J. Biol. Chem. 277, 50607–50611. [DOI] [PubMed] [Google Scholar]
- 42.Weinmann, A. S., Bartley, S. M., Zhang, T., Zhang, M. Q. & Farnham, P. J. (2001) Mol. Cell. Biol. 21, 6820–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo, J., Su, F., Chen, D., Shiloh, A. & Gu, W. (2000) Nature 408, 377–381. [DOI] [PubMed] [Google Scholar]
- 44.Grönroos, E., Hellman, U., Heldin, C.-H. & Ericsson, J. (2002) Mol. Cell 10, 483–493. [DOI] [PubMed] [Google Scholar]
- 45.Brooks, C. L. & Gu, W. (2003) Curr. Opin. Cell Biol. 15, 164–171. [DOI] [PubMed] [Google Scholar]
- 46.Ito, A., Lai, C. H., Zhao, X., Saito, S., Hamilton, M. H., Appella, E. & Yao, T. P. (2001) EMBO J. 20, 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hecht, A., Laroche, T., Strahl-Bolsinger, S., Gasser, S. M. & Grunstein, M. (1995) Cell 80, 583–592. [DOI] [PubMed] [Google Scholar]
- 48.McKinney, K. & Prives, C. (2002) Mol. Cell. Biol. 22, 6797–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu, W., Shi, X. L. & Roeder, R. G. (1997) Nature 387, 819–823. [DOI] [PubMed] [Google Scholar]
- 50.Lill, N. L., Grossman, S. R., Ginsberg, D., DeCaprio, J. & Livingston, D. M. (1997) Nature 387, 823–827. [DOI] [PubMed] [Google Scholar]
- 51.Avantaggiati, M. L., Ogryzko, V., Gardner, K., Giordano, A., Levine, A. S. & Kelly, K. (1997) Cell 89, 1175–1184. [DOI] [PubMed] [Google Scholar]
- 52.Goodman, R. H. & Smolik, S. (2000) Genes Dev. 14, 1553–1577. [PubMed] [Google Scholar]
- 53.Juan, L. J., Shia, W. J., Chen, M. H., Yang, W. M., Seto, E., Lin, Y. S. & Wu, C. W. (2000) J. Biol. Chem. 275, 20436–20443. [DOI] [PubMed] [Google Scholar]
- 54.Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L. & Gu, W. (2001) Cell 107, 137–148. [DOI] [PubMed] [Google Scholar]
- 55.Vaziri, H., Dessain, S. K., Eaton, N. E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L. & Weinberg, R. A. (2001) Cell 107, 149–159. [DOI] [PubMed] [Google Scholar]
- 56.Langley, E., Pearson, M., Faretta, M., Bauer, U. M., Frye, R. A., Minucci, S., Pelicci, P. G. & Kouzarides, T. (2002) EMBO J. 21, 2383–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng, H., Mostoslavsky, R., Saito, S., Manis, J. P., Gu, Y., Patel, P., Bronson, R., Appella, E., Alt, F. W. & Chua, K. F. (2003) Proc. Natl. Acad. Sci. USA 100, 10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katan-Khaykovich, Y. & Struhl, K. (2002) Genes Dev. 16, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robyr, D. & Grunstein, M. (2003) Methods 31, 83–89. [DOI] [PubMed] [Google Scholar]
- 60.Costanzo, A., Merlo, P., Pediconi, N., Fulco, M., Sartorelli, V., Cole, P. A., Fontemaggi, G., Fanciulli, M., Schiltz, L., Blandino, G., et al. (2002) Mol. Cell 9, 175–196. [DOI] [PubMed] [Google Scholar]