Abstract
The first occupation-associated cancers to be recognized were the sooty warts (cancers of the scrotum) suffered by chimney sweeps in 18th century England. In the 19th century, high incidences of skin cancers were noted among fuel industry workers. By the early 20th century, malignant skin tumors were produced in laboratory animals by repeatedly painting them with coal tar. The culprit in coal tar that induces cancer was finally isolated in 1933 and determined to be benzo[a]pyrene (BP), a polycyclic aromatic hydrocarbon. A residue of fuel and tobacco combustion and frequently ingested by humans, BP is metabolized in mammals to benzo[a]pyrene diol epoxide (BPDE), which forms covalent DNA adducts and induces tumor growth. In the 70 yr since its isolation, BP has been the most studied carcinogen. Yet, there has been no crystal structure of a BPDE DNA adduct. We report here the crystal structure of a BPDE–adenine adduct base-paired with thymine at a template–primer junction and complexed with the lesion-bypass DNA polymerase Dpo4 and an incoming nucleotide. Two conformations of the BPDE, one intercalated between base pairs and another solvent-exposed in the major groove, are observed. The latter conformation, which can be stabilized by organic solvents that reduce the dielectric constant, seems more favorable for DNA replication by Dpo4. These structures also suggest a mechanism by which mutations are generated during replication of DNA containing BPDE adducts.
Benzo[a]pyrene (BP) in the diet and from combustion of fuel and tobacco is one of the most potent carcinogens to which humans are frequently exposed (1, 2). A metabolic pathway involving cytochrome P450 and epoxide hydrolase converts BP to BP diol epoxides (BPDEs) (Fig. 1A) (3). Cis or trans opening of the epoxide ring of BPDE by the exocyclic amino group N6 of adenine or N2 of guanine results in covalent DNA adducts (4) (Fig. 1 A). Two-dimensional NMR studies (5, 6) suggest that BPDE-dA adducts are intercalated between base pairs whereas BPDE-dG adducts either reside in the minor groove or are intercalated with base displacement depending on the trans or cis stereochemistry of the adducts. These bulky polycyclic aromatic hydrocarbon (PAH) adducts impede DNA replication and induce mutations by perturbing the double-helical structure. When cultured Chinese hamster cells are treated with nonphysiological high doses of BPDE, mutations at dG predominate, but at lower doses likely corresponding to environmental exposure, and the proportion of mutations at dA increases (7). Recent studies suggest that one or more Y-family DNA polymerases may facilitate either accurate or mutagenic bypass of BPDE adducts (8). For example, polymerase (Pol) κ is accurate in bypassing BPDE-dG adducts (9, 10) whereas Pol η is error-prone (11, 12). Pol ι can accurately insert dTMP opposite a BPDE-dA adduct, but then requires Pol κ to extend primers to complete lesion bypass synthesis (13). We report here the crystal structures of a ternary complex composed of an archaeal Y-family DNA polymerase, Dpo4, an incoming dNTP, and a DNA substrate containing a BPDE-dA adduct in the template strand. The cis (10R)-dA adduct (dA*) is derived from (+)-(7R,8S,9S,10R)-BPDE (Fig. 1 A), which is the major and most tumorigenic isomer formed on metabolism of BP in mammals (14).
Fig. 1.
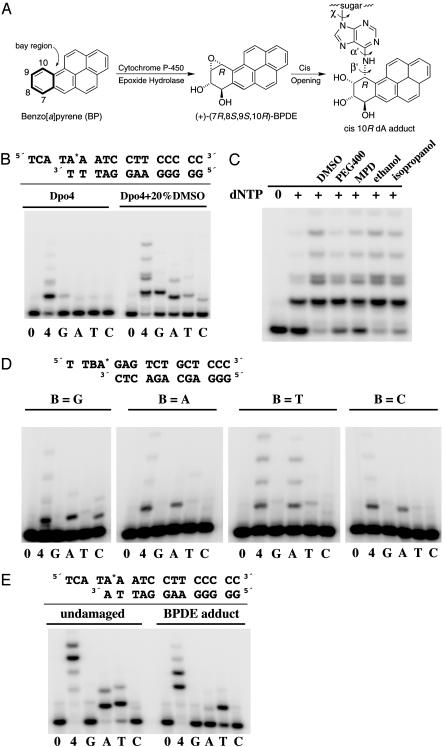
Formation of a benzo[a]pyrene diol epoxide adduct and its effect on DNA synthesis. (A) Structure of BP, its bay-region (+)-(7R,8S,9S,10R) diol epoxide (BPDE) metabolite and the cis (10R) BPDE-dA adduct in DNA resulting from cis opening of the DE by the N6 amino group of adenine. The α′, β′, and χ torsion angles are labeled. (B) The template-primer duplexes used to grow the crystal and primer extension by Dpo4 in solution. The BPDE-dA adduct is indicated as A*. Primer extension by Dpo4 was carried out with 10 nM Dpo4, 10 nM oligonucleotide substrates, and 100 μM dNTP (all four or each kind) at 37°C for 20 min in the absence and presence of 20% DMSO. The primer strand was 32P labeled, and the products were separated on a 20% polyacrylamide gel. (C) Effects of organic solvents on the primer extension by Dpo4. Twenty percent of various organic solvents was added to the reaction mix containing all four dNTPs. (D) Incorporation of each dNTP opposite the BPDE-dA adduct in four sequence contexts by Dpo4. (E) Primer extension of a dA*·dA mismatch in the absence and presence of the BPDE adduct. Reaction conditions are the same as in C.
Materials and Methods
Adducted Oligonucleotide Syntheses. The preparation of suitably protected, diastereomerically pure (10R) and (10S) phosphoramidites corresponding to cis opened BPDE dA adducts is described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. The oligonucleotide used for the present crystal structure, 5′-(TCA TA*A ATC CTT CCC CC)-3′, with the cis (10R) BPDE-dA adduct indicated by the asterisk and underline, was synthesized essentially as described (15) from the pure (10R)-phosphoramidite and purified by reverse-phase HPLC (see Supporting Materials and Methods). Synthesis and characterization of the oligonucleotide 5′-T TTA* GAG TCT GCT CCC-3′ (Fig. 1D) has been reported (16). The other sequences shown in Fig. 1D, in which the T adjacent to A* was replaced by A, G, and C, were synthesized essentially as described (15) by using the mixed (10R)/(10S) phosphoramidite diastereomers. The desired (10R) BPDE-adducted oligonucleotides were purified and separated from their (10S) diastereomers by reverse-phase HPLC (see Supporting Materials and Methods). Their absolute configurations were assigned by measurement of their CD spectra, which exhibit positive bands at 330–350 nm in contrast to the (10S) diastereomers, which exhibit negative bands (16). In all cases, the (10R) diastereomers eluted before the (10S) diastereomers.
Crystallization and Structure Determination. Full-length Dpo4 was overexpressed and purified as described (17). Dpo4 and DNA duplex were mixed at a 1:1.2 molar ratio in 20 mM Hepes (pH 7.0), 0.1 mM EDTA, 1 mM DTT, and 100 mM NaCl. The final protein concentration was ≈8 mg/ml. With the addition of 1 mM dATP, crystals were produced by the hanging drop method at 20°C by using the precipitant solution of 100 mM Hepes (pH 7.0), 100 mM calcium acetate, 12% polyethylene glycol (PEG) 3350, and 2% glycerol. Crystals were transferred to the mother liquor with 25% PEG 3350 and 15% ethylene glycol and flash frozen in liquid propane for data collection. Diffraction data were collected at –178°C by using an R axis IPII detector mounted on an RU 200 generator (Rigaku, Tokyo) and processed by using hkl (18). The structures were determined by molecular replacement by using the type I structure as a search model (17, 19) and refined to 2.70 Å with manual adjustment (19, 20). The final R and Rfree are 0.206 and 0.246, and none of the residues is in the disallowed region in Ramachandran plot (Table 1).
Table 1. Summary of crystallographic data.
| Space group | P212121 |
| Unit cell (a,b,c), Å | 100.0, 103.6, 106.1 |
| No. of complex in a.u. | 2 |
| Non-hydrogen atoms (water included) | 7143 (397) |
| Resolution range* | 25.6-2.70 Å (2.75-2.70 Å) |
| Rmerge*† | 0.093 (0.485) |
| Unique reflection | 27650 |
| Completeness (%)* | 89.5 (85.7) |
| R value‡ | 0.207 |
| Rfree§ | 0.246 (703 reflections) |
| rms deviation bond length, (Å) | 0.016 |
| rms deviation bond angle, ° | 1.69 |
| Avg B value (Wilson)Å2 | 46.7 (63.4) |
Data completeness in the highest resolution shell is shown in parentheses.
Rmerge = ΣhΣi|Ihi| — 〈Ih〉| /Σ〈Ih〉, where Ihi is the intensity of the ith observation of reflection h, and 〈Ih〉 is the average intensity of redundant measurements of the h reflections.
R value = Σ||Fo| — |Fc|| /Σ|Fo|, where Fo and Fc are the observed and calculated structure-factor amplitudes.
Rfree is monitored with the reflections excluded from refinement in parentheses.
Primer Extension. Primer extension by Dpo4 was carried out with 10 nM Dpo4, 10 nM oligonucleotide substrates with 5′32P-labeled primer, and 100 μM dNTP (all four or each kind) at 37°C for 20 min. The products were separated on a 16% polyacrylamide gel. Effects of organic solvents on the primer extension by Dpo4 were assayed with the addition of 20% (vol/vol) of various organic solvents to the reaction mix containing all four dNTPs.
Results and Discussion
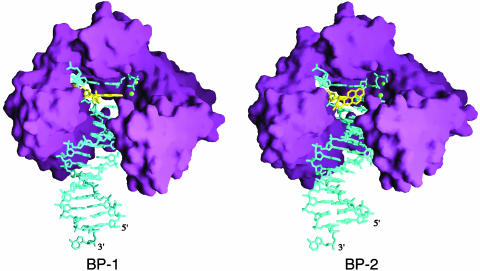
Our attempt to crystallize dA* opposite an incoming nucleotide has not been successful. The crystal structure we obtained is of a primer extension complex, in which the dA* is base paired with dT at the 3′ end of the primer strand, and the incoming dATP pairs with the next undamaged templating base, dT (Fig. 1B). The Dpo4-BPDE adduct crystal structure determined at 2.7-Å resolution (Table 1) not only captures the BPDE adduct adjacent to the active site of Dpo4, but also reveals two strikingly different conformations of the PAH adduct, due to the two Dpo4–DNA ternary complexes (BP-1 and BP-2) in each asymmetric unit (Fig. 2). In the BP-1 complex, the hydrocarbon is intercalated at the 5′ side of the dA* between the adduct-containing (dA*·dT) and the replicating (dT·dATP) base pairs, and, in the BP-2 complex, it is placed in the major groove nearly perpendicular to the DNA base pairs (Fig. 3A).
Fig. 2.
Crystal structures of BP-1 and BP-2. Dpo4 is represented by a purple molecular surface, the DNA and the incoming dATP are shown as blue sticks, and the PAH is highlighted in yellow. The divalent cations (Ca2+) are shown as green spheres. The unpaired dAMP at the 3′ end of the template strand was added by the terminal deoxynucleotide transferase (TdT) activity of Dpo4 that is common to archael D in B-like polymerases (34) (Fig. 4, which is published as supporting information on the PNAS web site). Figs. 2 and 3 were generated by using ribbons and grasp (35, 36).
Fig. 3.
Distortion of DNA by the BPDE adduct. (A) Comparison of the crystal and NMR structures. The dA*·dT and the surrounding base pairs including the replicating base pair are shown as ball-and-stick models. The crystal structures are shown with the Fo – Fc omit electron density maps contoured at 1.0σ in blue. The carbon, oxygen, nitrogen, and phosphorus atoms are colored yellow, red, blue, and purple, respectively. (B) Hydrogen bond formation at dA*·dT and the adjacent replicating base pair dT·dATP. Looking down the DNA helical axis, the two layers of the base pair and the PAH adduct are shown, purple for the replicating base pair, gold for the dA* adduct, and green for its partner dT. The incoming nucleotide in BP-1 is in the syn conformation. In the BP-2 complex, where the PAH is in the major groove, the adenine base of the dA* is shifted to the major groove, disrupting the normal hydrogen bonds with its partner, dT. The location of a normal dA is modeled in gray. (C) Stereo view of the overlay of the DNA structures from BP-1 (blue) and BP-2 (gold) after superimposition of the Dpo4 structures. With the PAH intercalated, the base pair ladder in the BP-1 complex is shifted by one register compared with that in the BP-2 complex.
In the BP-1 complex, where the PAH is intercalated, the torsion angles that define the orientation of the hydrocarbon relative to the modified dA are 157° (α′) and 107° (β′) (Fig. 1 A), which perfectly match a predicted favorable conformation of the adduct (21) and are practically identical to those found in NMR structures determined in the absence of a polymerase (Fig. 3A) (6, 22). Because of the intercalation of the PAH, the dA*·dT base pair is distorted by 36° of buckle, 31° of propeller twist, and 10° of base opening (23), and only one hydrogen bond is retained between this base pair (Fig. 3). The close contacts between the BP moiety and the base 3′ to the dA* observed in the NMR structures are alleviated by an 8° unwinding between the two base pairs.
In the BP-2 complex, where the PAH is placed in the major groove, the face of the PAH bearing the 8- and 9-hydroxyl groups is packed perpendicular to the plane of base pairs, and the other face is fully exposed to the solvent. The α′ and β′ torsion angles are –85° and –47°, respectively, which are energetically unfavorable based on molecular simulation (21). The exposed PAH is clearly less stable than the intercalated PAH, as indicated by the incomplete electron densities (Fig. 3A) and higher temperature factors. Structural characterization of BPDE-dA adducts by NMR to date has found the hydrocarbon intercalated only between base pairs and not in the major groove (5, 6, 24). To accommodate the PAH in the major groove without clashing with the neighboring base pairs, the dA* slides toward the major groove by nearly 2 Å (Fig. 3), and the dA*·dT base pair and two adjacent 3′ base pairs are underwound by 6° to 16° (23). In both complexes, the glycosidic torsion angle χ is anti, and the conformation of the tetrahydrobenzo ring of BPDE, which could not be determined by NMR studies, is a distorted half chair (Table 2).
Table 2. Selected dihedral angles (in degrees) for the BP-1 and BP-2 complexes.
| Angle* | BP-1 | BP-2 |
|---|---|---|
| χ | 219.5 | 268.1 |
| α′ | 156.8 | -85.1 |
| β′ | 107.0 | -47.2 |
| N6-C10-C9-O9 | 37.4 | 66.2 |
| O9-C9-C8-O8 | -54.4 | -57.6 |
| O8-C8-C7-O7 | -84.8 | -83.4 |
In both BP-1 and BP-2, the partially saturated ring assumes a distorted half-chair conformation. In BP-1, the C10-N6 bond linking the hydrocarbon to the base makes an angle of ≈50° with the plane of the aromatic rings, corresponding to an orientation for this bond that is about midway between axial and equatorial; whereas, in BP-2, this bond is pseudoequatorial and makes an ≈25° angle with the aromatic system. In both structures, the 7- and 8-hydroxyl groups are close to pseudoequatorial, and the 9-hydroxyl group is pseudoaxial. This orientation of the 9-hydroxyl group closely resembles that proposed in the NMR solution structure of a DNA duplex containing a cis opened (10R) BP tetrahydroepoxide dA adduct (PDB accession code 1N8C, Fig. 3A) that lacked the 7- and 8-hydroxyl groups on the tetrahydro ring (6).
Angles are defined as follows: χ = O4′-C1′-N9-C4; α′ = N1-C6-N6-C10(BP); β′ = C6-N6-C10(BP)-C9(BP).
The placement of substrates in the BP-1 complex indicates that the nucleotidyl transfer reaction could not occur when the BPDE is intercalated. Although the overall Dpo4 structure is similar to that complexed with undamaged DNA (17) (rms deviation of 341 Cα atoms under 0.8 Å) (Movie 1, which is published as supporting information on the PNAS web site), the 3′ OH of the primer strand and the α phosphate of the dATP are >10 Å apart due to the PAH intercalated between them. The incoming dATP is in the syn conformation, which enables the adenine to maintain a Hoogsteen base pair with the templating dT and maximize the aromatic ring stacking with the PAH (Fig. 3B). The placement of the triphosphate moiety is permutated from the normal; the α phosphate takes the β position, β takes the γ position, and γ takes the α position (Fig. 3A). If a chemical reaction occurs, it would be hydrolysis of dATP to dADP rather than covalent linkage between the primer strand and incoming nucleotide. In fact, hydrolysis of the incoming ddATP to ddADP was previously observed in the type I Dpo4 ternary complex (17), in which the primer strand contained a dideoxynucleotide at its 3′ end. Presumably, when the 3′ hydroxyl groups of the primer strand and the incoming nucleotide are absent, the triphosphate moiety of an incoming nucleotide can be rearranged, resulting in its hydrolysis rather than nucleotidyl transfer.
Placement of the PAH in the major groove in the BP-2 complex, although energetically unfavorable without Dpo4, allows the modified and the replicating base pairs to stack similarly as in undamaged DNA (Fig. 3). The catalytic core of Dpo4 encompassing the palm, finger, and thumb domains is entirely superimposable between the BP-1 and BP-2 complexes; only the little finger domain shifts by ≈0.5 Å (Movie 1). The Dpo4 active site of BP-2 contains both metal ions essential for the catalysis (25), and the 3′ hydroxyl group of the primer strand and the α phosphate of the incoming dATP are <5 Å apart (Fig. 3A). Dpo4, however, extends DNA beyond the BPDE adduct rather poorly (Fig. 1B), perhaps because this reactive but energetically “unfavorable” conformation of the adduct exists rather infrequently in solution. To stabilize the PAH in the major groove and so to enhance the activity of Dpo4, we reduced the dielectric constant of the reaction buffer by addition of organic solvents. Most alcohols, PEG 400, and DMSO enhance Dpo4's ability to extend DNA beyond the adducts (Fig. 1C). In the presence of 20% DMSO, which potentially influences the protein structure in addition to stabilizing the solvent-exposed PAH, Dpo4 also exhibits altered base preference for extension beyond the adduct (Fig. 1B). We suspect that, for any of the Y-family polymerases to insert a nucleotide opposite the BPDE adduct, the PAH moiety probably has to be flipped out into the major or minor groove. If the PAH is intercalated on either the 5′ or 3′ side, it would clash with the finger domain of Dpo4 or separate the 3′ end of a primer strand from the incoming nucleotide. The unusually small finger and thumb domains of the Y-family polymerases leave the major and minor groove wide open (26–29) and thus may facilitate such adduct placement.
The crystal structure, although representing primer extension beyond the dA* lesion, offers insights into how base substitution and frameshift mutations are induced by BPDE adducts at dA and dG (30–32). When the PAH is placed in the major groove in the BP-2 complex, the adenine base of the dA* shifts ≈2 Å toward the major groove, juxtaposing two hydrogen bond acceptors, the N1 of dA* and the O4 of its base pair partner dT (Fig. 3B). dC and dA, each possessing an exocyclic amino group in the place of the O4 of dT, would be better suited than dT to base pair with the dislocated dA*. Indeed, both Pol κ (9, 10) and Dpo4 favor incorporation of dAMP opposite dA* (Fig. 1D). By analogy, the PAH moiety of a BPDE-dG adduct (dG*) is likely placed in the minor groove rather than intercalated (5) to allow replication to occur. The guanine base may have to shift toward the minor groove (33), thus inducing mispairing with dT or dA. In support of this hypothesis, it has been reported that, depending on the sequence context, BPDE-dG adducts induce either G → A or G → T mutations (30, 31).
Regarding frameshift mutations, the intercalated PAH (dA*) in the BP-1 complex retains only one hydrogen bond with its partner dT and shifts the DNA base pair by one register compared with the DNA in the BP-2 complex (Fig. 3C). Misalignment by one nucleotide may occur after a wrong nucleotide has been inserted opposite the adduct and then displaced by intercalation of the PAH 5′ to it. When the adduct (dA*) is opposite a mismatched dA at the template-primer junction (Fig. 1E), the dA may actually pair with the dT 5′ to the dA* by –1 misalignment. Dpo4 clearly favors incorporation of dTMP that most likely pairs with dA two bases 5′ to the dA*, and the major product of the primer extension after the dA*·dA mismatch is 1 to 2 nucleotides shorter than the template strand (Fig. 1E). In contrast, human Pol κ is capable of faithfully incorporating dC opposite dG* and extending primers after Pol ι inserts dT opposite dA* (9, 13). We suspect that Pol κ may possess an active site specially configured to stabilize a nonintercalated PAH yet at the same time maintain Watson–Crick base pairs surrounding the adduct.
Supplementary Material
Acknowledgments
We thank Drs. M. Gellert and K. Mizuuchi for suggesting the experiments of reducing dielectric constants and Drs. R. Craigie and D. Leahy for critical reading of the manuscript.
Abbreviations: PAH, polycyclic aromatic hydrocarbon; BP, benzo[a]pyrene; BPDE, benzo-[a]pyrene diol epoxide; Pol, polymerase.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1S0M).
References
- 1.Phillips, D. H. (1983) Nature 303, 468–472. [DOI] [PubMed] [Google Scholar]
- 2.Phillips, D. H. (1999) Mutat. Res. 443, 139–147. [DOI] [PubMed] [Google Scholar]
- 3.Sims, P., Grover, P. L., Swaisland, A., Pal, K. & Hewer, A. (1974) Nature 252, 326–328. [DOI] [PubMed] [Google Scholar]
- 4.Jerina, D. M., Chadha, A., Cheh, A. M., Schurdak, M. E., Wood, A. W. & Sayer, J. M. (1991) Adv. Exp. Med. Biol. 283, 533–553. [DOI] [PubMed] [Google Scholar]
- 5.Geacintov, N. E., Cosman, M., Hingerty, B. E., Amin, S., Broyde, S. & Patel, D. J. (1997) Chem. Res. Toxicol. 10, 111–146. [DOI] [PubMed] [Google Scholar]
- 6.Volk, D. E., Thiviyanathan, V., Rice, J. S., Luxon, B. A., Shah, J. H., Yagi, H., Sayer, J. M., Yeh, H. J., Jerina, D. M. & Gorenstein, D. G. (2003) Biochemistry 42, 1410–1420. [DOI] [PubMed] [Google Scholar]
- 7.Wei, S. J., Chang, R. L., Wong, C. Q., Bhachech, N., Cui, X. X., Hennig, E., Yagi, H., Sayer, J. M., Jerina, D. M., Preston, B. D., et al. (1991) Proc. Natl. Acad. Sci. USA 88, 11227–11230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedberg, E. C., Wagner, R. & Radman, M. (2002) Science 296, 1627–1630. [DOI] [PubMed] [Google Scholar]
- 9.Ogi, T., Shinkai, Y., Tanaka, K. & Ohmori, H. (2002) Proc. Natl. Acad. Sci. USA 99, 15548–15553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki, N., Ohashi, E., Kolbanovskiy, A., Geacintov, N. E., Grollman, A. P., Ohmori, H. & Shibutani, S. (2002) Biochemistry 41, 6100–6106. [DOI] [PubMed] [Google Scholar]
- 11.Rechkoblit, O., Zhang, Y., Guo, D., Wang, Z., Amin, S., Krzeminsky, J., Louneva, N. & Geacintov, N. E. (2002) J. Biol. Chem. 277, 30488–30494. [DOI] [PubMed] [Google Scholar]
- 12.Chiapperino, D., Kroth, H., Kramarczuk, I. H., Sayer, J. M., Masutani, C., Hanaoka, F., Jerina, D. M. & Cheh, A. M. (2002) J. Biol. Chem. 277, 11765–11771. [DOI] [PubMed] [Google Scholar]
- 13.Frank, E. G., Sayer, J. M., Kroth, H., Ohashi, E., Ohmori, H., Jerina, D. M. & Woodgate, R. (2002) Nucleic Acids Res. 30, 5284–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buening, M. K., Wislocki, P. G., Levin, W., Yagi, H., Thakker, D. R., Akagi, H., Koreeda, M., Jerina, D. M. & Conney, A. H. (1978) Proc. Natl. Acad. Sci. USA 75, 5358–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroth, H., Yagi, H., Sayer, J. M., Kumar, S. & Jerina, D. M. (2001) Chem. Res. Toxicol. 14, 708–719. [DOI] [PubMed] [Google Scholar]
- 16.Page, J. E., Pilcher, A. S., Yugi, H., Sayer, J. M., Jerima, D. M. & Dipple, A. (1999) Chem. Res. Toxicol. 12, 258–263. [DOI] [PubMed] [Google Scholar]
- 17.Ling, H., Boudsocq, F., Woodgate, R. & Yang, W. (2001) Cell 107, 91–102. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 19.Brünger, A. T., Adams, P. D., Clore, G. M., Delane, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 20.Jones, T. A., Zou, J.-Y. & Cowan, S. W. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 21.Tan, J., Geacintov, N. E. & Broyde, S. (2000) Chem. Res. Toxicol. 13, 811–822. [DOI] [PubMed] [Google Scholar]
- 22.Mao, B., Gu, Z., Gorin, A., Chen, J., Hingerty, B. E., Amin, S., Broyde, S., Geacintov, N. E. & Patel, D. J. (1999) Biochemistry 38, 10831–10842. [DOI] [PubMed] [Google Scholar]
- 23.Lavery, R. & Sklenar, H. (1989) J. Biomol. Struct. Dyn. 6, 655–667. [DOI] [PubMed] [Google Scholar]
- 24.Pradhan, P., Tirumala, S., Liu, X., Sayer, J. M., Jerina, D. M. & Yeh, H. J. (2001) Biochemistry 40, 5870–5881. [DOI] [PubMed] [Google Scholar]
- 25.Steitz, T. A. & Steitz, J. A. (1993) Proc. Natl. Acad. Sci. USA 90, 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou, B. L., Pata, J. D. & Steitz, T. A. (2001) Mol. Cell 8, 427–437. [DOI] [PubMed] [Google Scholar]
- 27.Trincao, J., Johnson, R. E., Escalante, C. R., Prakash, S., Prakash, L. & Aggarwal, A. K. (2001) Mol. Cell 8, 417–426. [DOI] [PubMed] [Google Scholar]
- 28.Silvian, L. F., Toth, E. A., Pham, P., Goodman, M. F. & Ellenberger, T. (2001) Nat. Struct. Biol 8, 984–989. [DOI] [PubMed] [Google Scholar]
- 29.Yang, W. (2003) Curr. Opin. Struct. Biol. 13, 23–30. [DOI] [PubMed] [Google Scholar]
- 30.Page, J. E., Zajc, B., Oh-hara, T., Lakshman, M. K., Sayer, J. M., Jerina, D. M. & Dipple, A. (1998) Biochemistry 37, 9127–9137. [DOI] [PubMed] [Google Scholar]
- 31.Fernandes, A., Liu, T., Amin, S., Geacintov, N. E., Grollman, A. P. & Moriya, M. (1998) Biochemistry 37, 10164–10172. [DOI] [PubMed] [Google Scholar]
- 32.Lenne-Samuel, N., Janel-Bintz, R., Kolbanovskiy, A., Geacintov, N. E. & Fuchs, R. P. (2000) Mol. Microbiol. 38, 299–307. [DOI] [PubMed] [Google Scholar]
- 33.Feng, B., Gorin, A., Kolbanovskiy, A., Hingerty, B. E., Geacintov, N. E., Broyde, S. & Patel, D. J. (1997) Biochemistry 36, 13780–13790. [DOI] [PubMed] [Google Scholar]
- 34.Potapova, O., Grindley, N. D. & Joyce, C. M. (2002) J. Biol. Chem. 277, 28157–28166. [DOI] [PubMed] [Google Scholar]
- 35.Carson, M. (1987) J. Mol. Graphics 5, 103–106. [Google Scholar]
- 36.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins Struct. Funct. Genet. 11, 281–296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.