Abstract
Herbaspirillum massiliense strain JC206T sp. nov. is the type strain of H. massiliense sp. nov., a new species within the genus Herbaspirillum. This strain, whose genome is described here, was isolated from the fecal flora of a healthy Senegalese patient. H. massiliense is an aerobic rod. Here we describe the features of this organism, together with the complete genome sequence and annotation. The 4,186,486 bp long genome (one chromosome but no plasmid) contains 3,847 protein-coding and 54 RNA genes, including 3 rRNA genes.
Keywords: Herbaspirillum massiliense, genome
Introduction
Herbaspirillum massiliense strain JC206T (= CSUR P159 = DSMZ 25712) is the type strain of H. massiliense sp. nov. This bacterium was isolated from the stool of a healthy Senegalese patient. It is a Gram-negative, aerobic, flagellated, indole-negative bacillus.
The current approach to classification of prokaryotes, generally referred to as polyphasic taxonomy, relies on a combination of phenotypic and genotypic characteristics [1]. However, as more than 3,000 bacterial genomes have been sequenced [2], we recently proposed that genomic information should be integrated in the description of new bacterial species [3,4].
The genus Herbaspirillum (Baldani et al. 1986) was created in 1986 [5,6]. To date, this genus, comprised of nitrogen-fixing, Gram-negative bacilli, contains 13 species and two subspecies, including H. aquaticum (Dobritsa et al. 2010) [7], H. aurantiacum (Carro et al. 2011) [8], H. autotrophicum (Aragno and Schlegel 1978) Ding and Yokota 2004 [9], H. canariense (Carro et al. 2011) [8], H. chlorophenolicum (Im et al. 2004) [10], H. frisingense (Kirchhof et al. 2001) [11], H. hiltneri (Rothballer et al. 2006) [12], H. huttiense subsp. huttiense (Leifson 1962) Ding and Yokota 2004 [9], H. huttiense subsp. putei (Ding and Yokota 2004) Dobritsa et al. 2010 [7], H. lusitanum (Valverde et al. 2003) [13], H. rhizosphaerae (Jung et al. 2007) [14], H. rubrisubalbicans (Christopher and Edgerton 1930) Baldani et al. 1996 [6], H. seropedicae (Baldini et al. 1986) [5], and H. soli (Carro et al. 2011) [8]. Members of the genus Herbaspirillum have mainly been isolated from the environment, in particular from soil, and from plants for which they play the role of growth promoters, but have also occasionally been isolated from humans, either as proven pathogens, causing bacteremia in leukemic patients [15,16], as potential pathogens in aortic aneurysms [17], or in respiratory secretions from cystic fibrosis patients [18,19]. To the best of our knowledge, this is the first to report the isolation of a Herbaspirillum sp. from the normal fecal flora. Here we present a summary classification and a set of features for H. massiliense sp. nov. strain JC206T (= CSUR P159 = DSMZ 25712) together with the description of the complete genomic sequencing and annotation. These characteristics support the circumscription of the species H. massiliense.
Classification and features
A stool sample was collected from a healthy 16-year-old male Senegalese volunteer patient living in Dielmo (a rural village in the Guinean-Sudanian zone in Senegal), who was included in a research protocol. Written assent was obtained from this individual; no written consent was needed from his guardians for this study because he was older than 15 years old (in accordance with the previous project approved by the Ministry of Health of Senegal and the assembled village population and as published elsewhere [20].) Both this study and the assent procedure were approved by the National Ethics Committee of Senegal (CNERS) and the Ethics Committee of the Institut Fédératif de Recherche IFR48, Faculty of Medicine, Marseille, France (agreement numbers 09-022 and 11-017). Several other new bacterial species were isolated from this specimen using various culture conditions [3,4].
The fecal specimen was preserved at -80°C after collection and sent to Marseille. Strain JC206T (Table 1) was isolated in June 2011 after passive filtration of the stool sample to select motile species using companion plate, cell culture inserts with 0.4 μm-pore membranes (Becton Dickinson, Heildeberg, Germany) and Leptospira broth (BioMerieux, Marcy l’Etoile, France). Subsequently, we cultivated strain JC206T on 5% sheep blood agar in an aerobic atmosphere at 37°C. This strain exhibited a 96.7% 16S rDNA nucleotide sequence similarity with H. aurantiacum (Carro et al. 2012), the phylogenetically closest validly published Herbaspirillum species (Figure 1), that was cultivated from volcanic soil in Canary Islands. This value was lower than the 98.7% 16S rRNA gene sequence threshold recommended by Stackebrandt and Ebers to delineate a new species without carrying out DNA-DNA hybridization [29].
Table 1. Classification and general features of Herbaspirillum massiliense strain JC206T according to the MIGS recommendations [21].
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain: Bacteria | TAS [22] | |
| Phylum Proteobacteria | TAS [23] | ||
| Class Betaproteobacteria | TAS [24,25] | ||
| Order Burkholderiales | TAS [24,26] | ||
| Family Oxalobacteriaceae | TAS [24,27] | ||
| Genus Herbaspirillum | TAS [5,6] | ||
| Species Herbaspirillum massiliense | IDA | ||
| Type strain JC206T | IDA | ||
| Gram stain | Negative | IDA | |
| Cell shape | Rod | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Nonsporulating | IDA | |
| Temperature range | Mesophile | IDA | |
| Optimum temperature | 37°C | IDA | |
| MIGS-6.3 | Salinity | Growth in BHI medium + 5% NaCl | IDA |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| Carbon source | Unknown | ||
| Energy source | Unknown | ||
| MIGS-6 | Habitat | Human gut | IDA |
| MIGS-15 | Biotic relationship | Free living | IDA |
| MIGS-14 | Pathogenicity Biosafety level Isolation |
Unknown 2 Human feces |
|
| MIGS-4 | Geographic location | Senegal | IDA |
| MIGS-5 | Sample collection time | September 2010 | IDA |
| MIGS-4.1 | Latitude | 13.7167 | IDA |
| MIGS-4.1 | Longitude | – 16.4167 | IDA |
| MIGS-4.3 | Depth | Surface | IDA |
| MIGS-4.4 | Altitude | 51 m above sea level | IDA |
Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [28]. If the evidence is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
Figure 1.
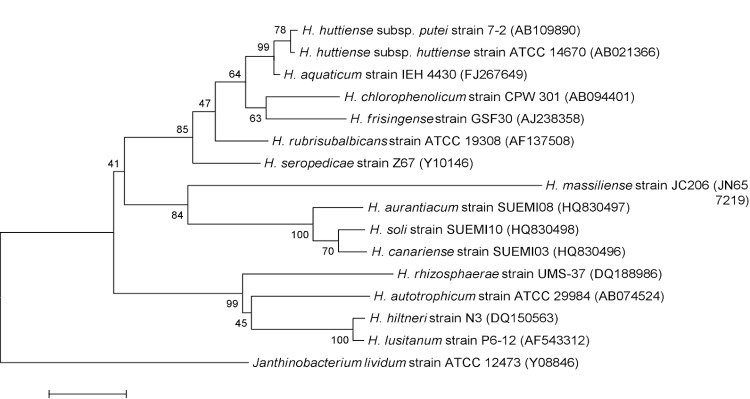
Phylogenetic tree highlighting the position of Herbaspirillum massiliense strain JC206T relative to other type strains within the Herbaspirillum genus. GenBank accession numbers are indicated in parentheses. Sequences were aligned using CLUSTALW, and phylogenetic inferences obtained using the maximum-likelihood method within the MEGA software. Numbers at the nodes are bootstrap values obtained by repeating 500 times the analysis to generate a majority consensus tree. Janthinobacterium lividum was used as an outgroup. The scale bar represents a 0.5% nucleotide sequence divergence.
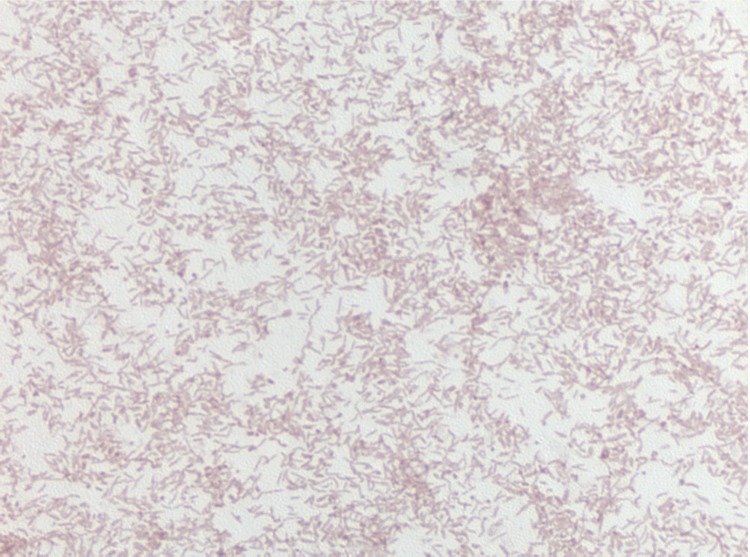
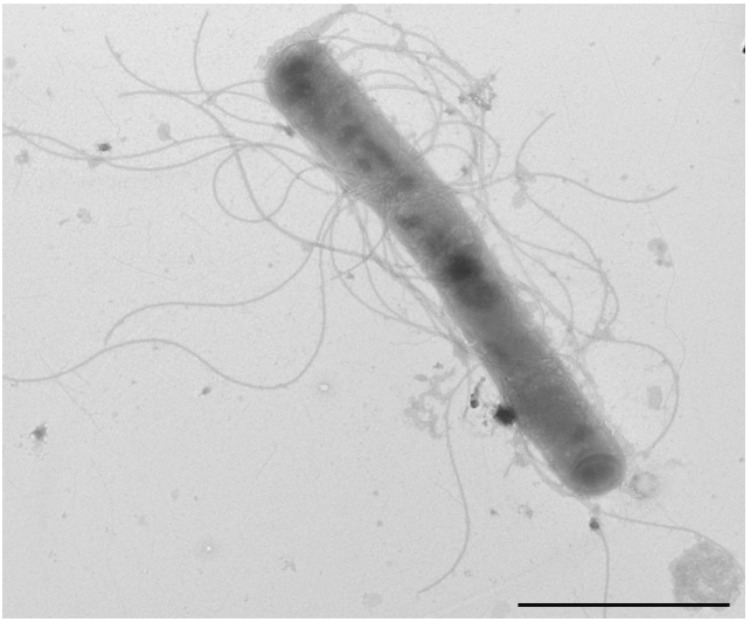
Different growth temperatures (25, 30, 37, 45°C) were tested. No growth occurred at either 25°C or 45°C, growth occurred at either 30 or 37°C. Optimal growth was observed at 37°C. Colonies were light brown, opaque and 0.5 mm in diameter on blood-enriched Columbia agar and Brain Heart Infusion (BHI) agar. Growth of the strain was tested under anaerobic and microaerophilic conditions using GENbag anaer and GENbag microaer systems, respectively (BioMérieux), and in the presence of air, of 5% CO2 and in anaerobic conditions. Optimal growth was obtained aerobically, with weak growth being observed under microaerophilic condition and with 5% CO2. No growth occurred under anaerobic conditions. Gram staining showed Gram negative curved rods (Figure 2). A motility test was positive. Cells grown on agar have a mean diameter of 0.44 µm by electron microscopy and have several polar flagella (Figure 3).
Figure 2.
Gram staining of H. massiliense strain JC206T.
Figure 3.
Transmission electron microscopy of H. massiliense strain JC206T, using a Morgani 268D (Philips) at an operating voltage of 60kV.The scale bar represents 900 nm.
Strain 206T exhibited catalase and oxidase activities. Using an API 20 NE strip (BioMerieux), nitrate reduction, indole formation, glucose fermentation and urease were negative. Arginine dihydrolase and esculin hydrolysis were positive. H. massiliense is susceptible to ticarcillin, imipenem, trimethoprim/sulfamethoxazole, gentamicin, amikacin, and colimycin but resistant to fosfomycin and nitrofurantoin.
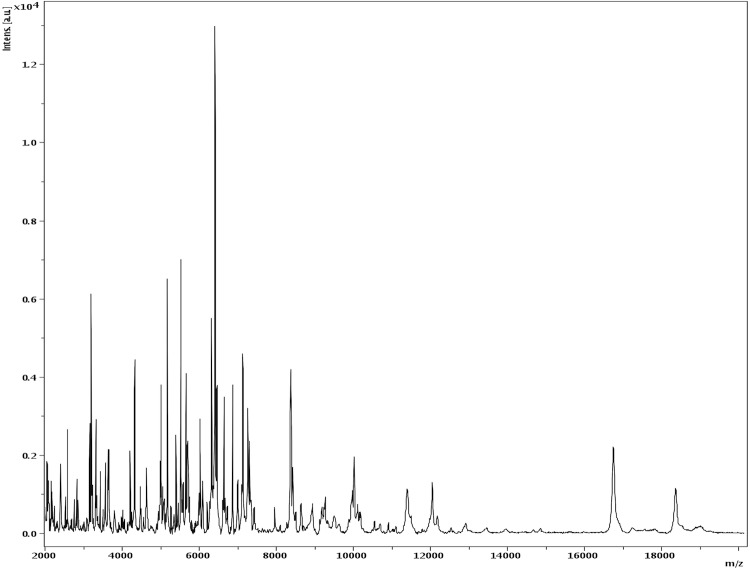
Matrix-assisted laser-desorption/ionization time-of-flight (MALDI-TOF) MS protein analysis was carried out as previously described [30] using a Microflex spectrometer (Bruker Daltonics, Germany). Spectra were compared with the Bruker database that contained no spectrum from Herbaspirillum species. No significant score was obtained with any other taxon. We incremented our database with the spectrum from strain JC206 T (Figure 4).
Figure 4.
Reference mass spectrum from H. massiliense strain JC206T. Spectra from 12 individual colonies were compared and a reference spectrum was generated.
Genome sequencing information
Genome project history
The organism was selected for sequencing on the basis of its phylogenetic position and 16S rRNA similarity to other members of the genus Herbaspirillum, and is part of a “culturomics” study of the human digestive flora aiming at isolating all bacterial species within human feces. It was the second genome of a Herbaspirillum species and the first genome of H. massiliense sp. nov. A summary of the project information is shown in Table 2. The Genbank accession number of the genome is CAHF00000000 and consists of 27 contigs. Table 2 shows the project information and its association with MIGS version 2.0 compliance.
Table 2. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | Shot Gun, Paired-end 3 Kb library |
| MIGS-29 | Sequencing platforms | 454 GS FLX Titanium |
| MIGS-31.2 | Fold coverage | 29× |
| MIGS-30 | Assemblers | Newbler version 2.5.3 |
| MIGS-32 | Gene calling method | Prodigal |
| Genbank ID | CAHF00000000 | |
| Genbank Date of Release | June 1, 2012 | |
| Project relevance | Study of the human gut microbiome |
Growth conditions and DNA isolation
H. massiliense sp. nov. strain JC206T (= CSUR P159, = DSM 25712), was grown aerobically on 5% sheep blood-enriched Columbia agar at 37°C. Cell growth from eight petri dishes (“spread plates”) was resuspended in 4×100µl of G2 buffer (EZ1 DNA Tissue kit, Qiagen). A first mechanical lysis was performed by glass powder on the Fastprep-24 device (MP Biomedicals, USA) during 2×20 seconds. DNA was then incubated with lysozyme for 30 minutes at 37°C and extracted using the EZ 1 Advanced XL BioRobot (Qiagen). DNA was concentrated and purified using the QiAmp kit (Qiagen). The yield and concentration were measured using the Quant-it Picogreen kit (Invitrogen) and the Genios_Tecan fluorometer at 52.5 ng/µl.
Genome sequencing and assembly
Both a shotgun and a 3-kb paired end sequencing were performed on a 454 GS FLX pyrosequencer. Both projects were loaded on a ¼ and a 1/8 regions of a PTP Picotiterplate. The shotgun library was constructed with 500 ng DNA as recommended by the manufacturer (Roche). For paired end sequencing, five µg of DNA were mechanically fragmented using the Hydroshear device (Digilab, Holliston, MA) with an enrichment size at 3-4kb. The DNA fragmentation was visualized using the BioAnalyzer 2100 on a DNA labchip 7500 (Agilent) with an optimal size of 3.944 kb. The library was constructed according to the 454 GS FLX Titanium paired end protocol. Circularization and nebulization were performed and generated a pattern with an optimal at 418 bp. After PCR amplification through 15 cycles followed by double size selection, the single stranded paired end library was then quantified on the Quant-it Ribogreen kit (Invitrogen) on the Genios Tecan fluorometer at 128 pg/µL. The library concentration equivalence was calculated as 5.62 × 108 molecules/µL. The library was stored at -20°C until further use.
The library was clonally amplified with 2 cpb and 3 cpb, respectively, in 2 × 8 emPCR reactions with the GS Titanium SV emPCR Kit (Lib-L) v2 (Roche). The yields of the emPCR were 13.75 and 2.65% for the shotgun and paired end strategies, respectively.
Approximately 790,000 beads were loaded on the GS Titanium PicoTiterPlate PTP Kit 70×75 and sequenced with the GS FLX Titanium Sequencing Kit XLR70 (Roche). The run was performed overnight and then analyzed on the cluster through the gsRunBrowser and Newbler assembler (Roche). A total of 504,311 passed filter wells were obtained and generated 4.69 Mb with a length average of 312 bp. The passed filter sequences were assembled Using Newbler with 90% identity and 40 bp as overlap. The final assembly identified 5 scaffolds and 27 contigs (>100 bp).
Genome annotation
Open Reading Frames (ORFs) were predicted using Prodigal [31] with default parameters but the predicted ORFs were excluded if they were spanning a sequencing GAP region. The predicted bacterial protein sequences were searched against the National Center for Biotechnology Information (NCBI) nonredundant (NR) and the Clusters of Orthologous Groups (COG) databases using BLASTP. The tRNAScanSE tool [32] was used to find tRNA genes, whereas ribosomal RNAs were found by using RNAmmer [33] and BLASTn against the NR database. ORFans were identified if their BLASTP E-value were lower than 1e-03 for alignment length greater than 80 amino acids. If alignment lengths were smaller than 80 amino acids, we used an E-value of 1e-05. Such parameter thresholds have already been used in previous works to define ORFans.
Genome properties
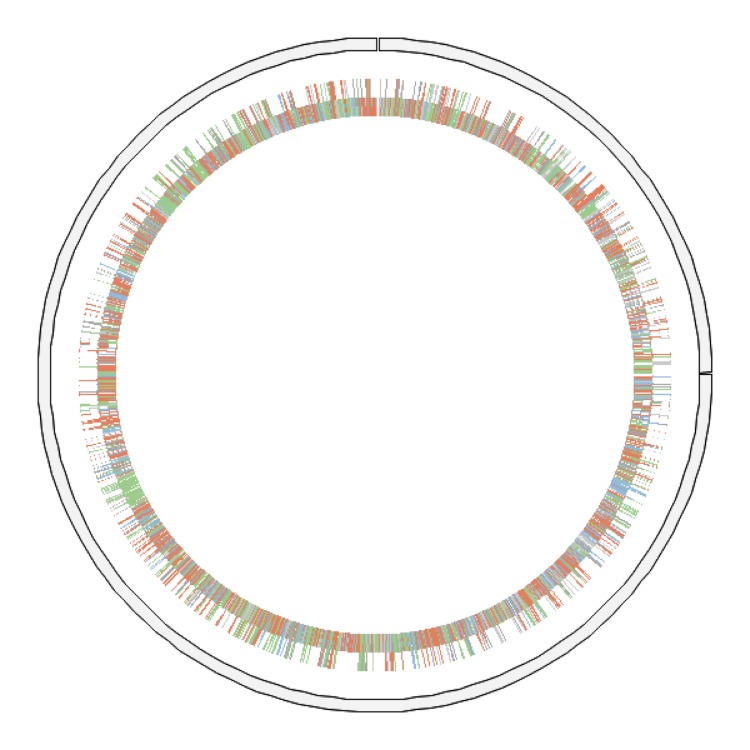
The genome is 4,186,486 bp long (one chromosome but no plasmid) with a 59.73% GC content (Table 3 and Figure 5). Of the 3,901 predicted genes, 3,847 were protein-coding genes, and 54 were RNAs. A total of 2,924 genes (74.95%) were assigned a putative function. ORFans accounted for 312 (8.0%) of the genes. The remaining genes were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4. The properties and the statistics of the genome are summarized in Tables 3 and 4.
Table 3. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 4,186,486 | |
| DNA coding region (bp) | 3,655,584 | 87.32 |
| DNA G+C content (bp) | 2,500,588 | 59.73 |
| Total genes | 3,901 | 100 |
| RNA genes | 54 | 1.4 |
| Protein-coding genes | 3,847 | 98,61 |
| Genes with function prediction | 2,924 | 74.95 |
| Genes assigned to COGs | 3,135 | 80.36 |
| Genes with peptide signals | 378 | 9.68 |
| Genes with transmembrane helices | 955 | 24.48 |
aThe total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
Figure 5.
Graphical circular map of the chromosome. Genes are colored according to their COG categories as follows: information storage and processing (blue), cellular processing and signaling (green), metabolism (red) and poorly characterized (grey).
Table 4. Number of genes associated with the 25 general COG functional categories.
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 183 | 4.69 | Translation |
| A | 2 | 0.05 | RNA processing and modification |
| K | 199 | 5.10 | Transcription |
| L | 183 | 4.69 | Replication, recombination and repair |
| B | 2 | 0.05 | Chromatin structure and dynamics |
| D | 45 | 1.15 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 44 | 1.13 | Defense mechanisms |
| T | 234 | 6.00 | Signal transduction mechanisms |
| M | 272 | 6.97 | Cell wall/membrane biogenesis |
| N | 142 | 3.64 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 141 | 3.61 | Intracellular trafficking and secretion |
| O | 168 | 4.31 | Posttranslational modification, protein turnover, chaperones |
| C | 230 | 5.90 | Energy production and conversion |
| G | 192 | 4.92 | Carbohydrate transport and metabolism |
| E | 263 | 6.74 | Amino acid transport and metabolism |
| F | 67 | 1.72 | Nucleotide transport and metabolism |
| H | 141 | 3.61 | Coenzyme transport and metabolism |
| I | 176 | 4.51 | Lipid transport and metabolism |
| P | 147 | 3.77 | Inorganic ion transport and metabolism |
| Q | 93 | 2.38 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 444 | 11.38 | General function prediction only |
| S | 384 | 9.84 | Function unknown |
| - | 95 | 2.44 | Not in COGs |
The total is based on the total number of protein coding genes in the annotated genome.
Comparison with Herbaspirillum seropedicae
To date, the genome from H. seropedicae strain SmR1 is the only genome from Herbaspirillum species that has been sequenced [34]. By comparison with H. seropedicae, H. massiliense exhibited a smaller genome (4,186,486 bp vs 5,513,887 bp, respectively), a lower G+C content (59.73% vs 63.4%, respectively) and a smaller number of genes (3,901 vs 4,804). In contrast, H. massiliense had higher ratios of genes per Mb (0.93 vs 0.87) and genes with assigned functions (74.9% vs 64.7%).
Conclusion
On the basis of phenotypic, phylogenetic and genomic analyses, we formally propose the creation of Herbaspirillum massiliense sp. nov. that contains the strain JC206T. This bacterium has been found in Senegal.
Description of Herbaspirillum massiliense sp. nov.
Herbaspirillum massiliense (mas.il.ien’se. L. gen. neutr. n. massiliense, of Massilia, the Latin name of Marseille where strain JC206T was cultivated).
Colonies are 0.5 mm in diameter on blood-enriched Columbia agar and Brain Heart Infusion (BHI) agar. Cells are rod-shaped with a mean diameter of 0.44 µm. Motile with tufts of polar flagellae optimal growth occurs under aerobic conditions. Weak growth is observed under microaerophilic conditions and with 5% CO2. No growth is observed under anaerobic conditions. Growth occurs between 30-37°C, with optimal growth observed at 37°C.
Cells stain Gram-negative. Catalase, oxidase and arginine dihydrolase activities, as well as esculin hydrolysis are present. Nitrate reduction and indole production are absent. Cells are susceptible to ticarcillin, imipenem, trimethoprim/sulfamethoxazole, gentamicin, amikacin, and colimycin. The G+C content of the genome is 59.73%. The 16S rRNA and genome sequences are deposited in Genbank under accession numbers JN657219 and CAHF00000000, respectively. The type strain JC206T (= CSUR P159 = DSMZ 25712) was isolated from the fecal flora of a healthy patient in Senegal.
References
- 1.Tindall BJ, Rossello-Mora R, Busse HJ, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 2010; 60:249-266 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- 2.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier JC, El Karkouri K, Nguyen TT, Armougom F, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci 2012; 6:116-125 10.4056/sigs.2415480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Clostridium senegalense sp. nov. Stand Genomic Sci 2012; 6:386-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldani JI, Baldini VL, Seldin L, Döbereiner J. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root-associated nitrogen-fixing bacterium. Int J Syst Bacteriol 1986; 36:86-93 10.1099/00207713-36-1-86 [DOI] [Google Scholar]
- 6.Baldani JI, Pot B, Kirchhof G, Falsen E, Baldani VL, Olivares FL, Hoste B, Kersters K, Hartmann A, Gillis M, Dobereiner J. Emended description of Herbaspirillum; inclusion of [ Pseudomonas] rubrisubalbicans, a milk plant pathogen, as Herbaspirillum rubrisubalbicans comb. nov.; and classification of a group of clinical isolates (EF group 1) as Herbaspirillum species 3. Int J Syst Bacteriol 1996; 46:802-810 10.1099/00207713-46-3-802 [DOI] [PubMed] [Google Scholar]
- 7.Dobritsa AP, Reddy MC, Samadpour M. Reclassification of Herbaspirillum putei as a later heterotypic synonym of Herbaspirillum huttiense, with the description of H. huttiense subsp. huttiense subsp. nov. and H. huttiense subsp. putei subsp. nov., comb. nov., and description of Herbaspirillum aquaticum sp. nov. Int J Syst Evol Microbiol 2010; 60:1418-1426 10.1099/ijs.0.009381-0 [DOI] [PubMed] [Google Scholar]
- 8.Carro L, Rivas R, Leon-Barrios M, Gonzalez-Tirante M, Velazquez E, Valverde A. 2011. Herbaspirillum canariense sp. nov., Herbaspirillum aurantiacum sp. nov. and Herbaspirillum soli sp. nov., three new species isolated in Tenerife (Canary Islands). Int J Syst Evol Microbiol 2012; 62:1300-1306 10.1099/ijs.0.031336-0 [DOI] [PubMed] [Google Scholar]
- 9.Ding L, Yokota A. Proposals of Curvibacter gracilis gen. nov., sp. nov. and Herbaspirillum putei sp. nov. for bacterial strains isolated from well water and reclassification of [ Pseudomonas] huttiensis, [ Pseudomonas] lanceolata, [ Aquaspirillum] delicatum and [ Aquaspirillum] autotrophicum as Herbaspirillum huttiense comb. nov., Curvibacter lanceolatus comb. nov., Curvibacter delicatus comb. nov. and Herbaspirillum autotrophicum comb. nov. Int J Syst Evol Microbiol 2004; 54:2223-2230 10.1099/ijs.0.02975-0 [DOI] [PubMed] [Google Scholar]
- 10.Im WT, Bae HS, Yokota A, Lee ST. Herbaspirillum chlorophenolicum sp. nov., a 4-chlorophenol-degrading bacterium. Int J Syst Evol Microbiol 2004; 54:851-855 10.1099/ijs.0.02812-0 [DOI] [PubMed] [Google Scholar]
- 11.Kirchhof G, Eckert B, Stoffels M, Baldani JI, Reis VM, Hartmann A. Herbaspirillum frisingense sp. nov., a new nitrogen-fixing bacterial species that occurs in C4-fibre plants. Int J Syst Evol Microbiol 2001; 51:157-168 [DOI] [PubMed] [Google Scholar]
- 12.Rothballer M, Schmid M, Klein I, Gattinger A, Grundmann S, Hartmann A. Herbaspirillum hiltneri sp. nov., isolated from surface-sterilized wheat roots. Int J Syst Evol Microbiol 2006; 56:1341-1348 10.1099/ijs.0.64031-0 [DOI] [PubMed] [Google Scholar]
- 13.Valverde A, Velazquez E, Gutierrez C, Cervantes E, Ventosa A, Igual JM. Herbaspirillum lusitanum sp. nov., a novel nitrogen-fixing bacterium associated with root nodules of Phaseolus vulgaris. Int J Syst Evol Microbiol 2003; 53:1979-1983 10.1099/ijs.0.02677-0 [DOI] [PubMed] [Google Scholar]
- 14.Jung SY, Lee MH, Oh TK, Yoon JH. Herbaspirillum rhizosphaerae sp. nov., isolated from rhizosphere soil of Allium victorialis var. platyphyllum. Int J Syst Evol Microbiol 2007; 57:2284-2288 10.1099/ijs.0.64666-0 [DOI] [PubMed] [Google Scholar]
- 15.Ziga ED, Druley T, Burnham CA. Herbaspirillum species bacteremia in a pediatric oncology patient. J Clin Microbiol 2010; 48:4320-4321 10.1128/JCM.01479-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Su Z, Liu Y, Sandoghchian S, Zheng D, Wang S, Xu H. Herbaspirillum species: a potential pathogenic bacteria isolated from acute lymphoblastic leukemia patient. Curr Microbiol 2011; 62:331-333 10.1007/s00284-010-9703-5 [DOI] [PubMed] [Google Scholar]
- 17.Marques da Silva R, Caugant DA, Eribe ER, Aas JA, Lingaas PS, Geiran O, Tronstad L, Olsen I. Bacterial diversity in aortic aneurysms determined by 16S ribosomal RNA gene analysis. J Vasc Surg 2006; 44:1055-1060 10.1016/j.jvs.2006.07.021 [DOI] [PubMed] [Google Scholar]
- 18.Spilker T, Uluer AZ, Marty FM, Yeh WW, Levison JH, Vandamme P, Lipuma JJ. Recovery of Herbaspirillum species from persons with cystic fibrosis. J Clin Microbiol 2008; 46:2774-2777 10.1128/JCM.00460-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coenye T, Goris J, Spilker T, Vandamme P, Lipuma JJ. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J Clin Microbiol 2002; 40:2062-2069 10.1128/JCM.40.6.2062-2069.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trape JF, Tall A, Diagne N, Ndiath O, Ly AB, Faye J, Dieye-Ba F, Roucher C, Bouganali C, Badiane A, et al. Malaria morbidity and pyrethroid resistance after the introduction of insecticide-treated bednets and artemisinin-based combination therapies: a longitudinal study. Lancet Infect Dis 2011; 11:925-932 10.1016/S1473-3099(11)70194-3 [DOI] [PubMed] [Google Scholar]
- 21.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archae, Bacteria, and Eukarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 24.Validation List No 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1-6 10.1099/ijs.0.64188-0 [DOI] [PubMed] [Google Scholar]
- 25.Garrity GM, Bell JA, Lilburn T. Class II. Betaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 575. [Google Scholar]
- 26.Garrity GM, Bell JA, Lilburn T. Order I. Burkholderiales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 575. [Google Scholar]
- 27.Garrity GM, Bell JA, Lilburn T. Family II. Oxalobacteraceae fam. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 623. [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Drolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 2006; 33:152-155 [Google Scholar]
- 30.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 2009; 49:543-551 10.1086/600885 [DOI] [PubMed] [Google Scholar]
- 31.Prodigal. http://prodigal.ornl.gov
- 32.Lowe TM, Eddy SR. t-RNAscan-SE: a program for imroved detection of transfer RNA gene in genomic sequence. Nucleic Acids Res 1997; 25:955-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pedrosa FO, Monteiro RA, Wassem R, Cruz LM, Ayub RA, Colauto NB, Fernandez MA, Fungaro MH, Grisard EC, Hungria M. Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet 2011; 7:e1002064 10.1371/journal.pgen.1002064 [DOI] [PMC free article] [PubMed] [Google Scholar]