Abstract
In sheep, small ruminant lentiviruses cause an incurable, progressive, lymphoproliferative disease that affects millions of animals worldwide. Known as ovine progressive pneumonia virus (OPPV) in the U.S., and Visna/Maedi virus (VMV) elsewhere, these viruses reduce an animal’s health, productivity, and lifespan. Genetic variation in the ovine transmembrane protein 154 gene (TMEM154) has been previously associated with OPPV infection in U.S. sheep. Sheep with the ancestral TMEM154 haplotype encoding glutamate (E) at position 35, and either form of an N70I variant, were highly-susceptible compared to sheep homozygous for the K35 missense mutation. Our current overall aim was to characterize TMEM154 in sheep from around the world to develop an efficient genetic test for reduced susceptibility. The average frequency of TMEM154 E35 among 74 breeds was 0.51 and indicated that highly-susceptible alleles were present in most breeds around the world. Analysis of whole genome sequences from an international panel of 75 sheep revealed more than 1,300 previously unreported polymorphisms in a 62 kb region containing TMEM154 and confirmed that the most susceptible haplotypes were distributed worldwide. Novel missense mutations were discovered in the signal peptide (A13V) and the extracellular domains (E31Q, I74F, and I102T) of TMEM154. A matrix-assisted laser desorption/ionization–time-of flight mass spectrometry (MALDI-TOF MS) assay was developed to detect these and six previously reported missense and two deletion mutations in TMEM154. In blinded trials, the call rate for the eight most common coding polymorphisms was 99.4% for 499 sheep tested and 96.0% of the animals were assigned paired TMEM154 haplotypes (i.e., diplotypes). The widespread distribution of highly-susceptible TMEM154 alleles suggests that genetic testing and selection may improve the health and productivity of infected flocks.
Introduction
Visna/Maedi virus (VMV) and its closely related North American counterpart, ovine progressive pneumonia virus (OPPV), are small ruminant lentiviruses (SRLV) of the retroviridae family that infect sheep around the world (for review see [1]). Infections are life-long and there are no effective treatments or vaccines [2]. The first signs of disease typically appear after age two and often include the loss of body condition and indurative mastitis (hard udder). Disease progression is associated with severe clinical signs that include difficulty breathing, chronic wasting, loss of motor control, and arthritis. Significant transmission occurs horizontally among adult sheep by respiratory routes, and vertically between dam and offspring by ingestion of infected colostrum [3], [4].
The impact of SRLV infection is considerable when the virus is introduced into a naïve flock with susceptible sheep. The mortality in such flocks may reach 30% per year after a few years [5]. Once established in a ewe flock, subclinical infection weakens the resistance to disease, decreases fertility, and reduces lamb production [6]. In the U.S., a quarter of the sheep are infected with OPPV and a third of sheep operations test positive [7]. Infected ewes have difficulty raising lambs and also transmit infection to their offspring and other flockmates. Collectively, complications of SRLV infections lead to animal pain, disability, early culling, and increased labor.
Stepwise strategies for SRLV disease control typically begin with the removal of infected sheep and focus on lowering the infection prevalence [4]. SRLV can also be eradicated in one production cycle by isolating all neonates from their infected dams, raising the lambs on uninfected colostrum and milk, and maintaining them separately from infected animals thereafter. Although these methods have been successful in eradicating SRLV in sheep, they require a significant commitment of time and resources, and flocks remain vulnerable to SRLV outbreaks if exposed to infected animals [8]. Efforts to eradicate SRLV in sheep and maintain an infection-free status would be enhanced by the use of replacement breeding stock that are genetically resistant to lentivirus infections.
The discovery of ovine transmembrane protein gene 154 (TMEM154) as a major OPPV susceptibility gene [9] provides an opportunity to produce sheep that are less susceptible to infection. The function of the TMEM154 protein has not yet been reported for any species and remains unknown. However, the ancestral TMEM154 haplotype in sheep is common and predicted to encode a precursor protein of 191 amino acids that is cleaved to a mature protein with 161 residues. Two other haplotypes encoding polypeptide isoforms are also common in U.S. sheep and, together with the ancestral haplotype, account for more than 97% of those observed [9]. The ancestral TMEM154 haplotype (designated haplotype 3, GenBank accession JX961707) encodes glutamate (E) at position 35 and asparagine (N) at position 70. Haplotype 2 has an isoleucine (I) mutation at position 70 while haplotype 1 has a lysine (K) mutation at position 35. A case-control study with 130 pairs of 4- to 9-year old ewes matched for lifetime exposure showed the odds of being infected were 69 times greater for those with one copy of either haplotype 2 or 3, compared to those homozygous for haplotype 1 (p-value <0.0001, 95% CI 12–2800) [9]. Likewise, a cohort study involving 2705 unmatched U.S. sheep from Nebraska, Idaho, and Iowa showed the relative risk of infection was 2.85 times greater for sheep with one copy of either haplotype 2 or 3, compared to those homozygous for haplotype 1 (p-value <0.0001, 95% CI 2.36–3.43) [9]. Also reported were two naturally occurring TMEM154 deletion mutations, R4A(delta53) and E82Y(delta82), that were predicted to abolish the protein’s function. Although rare, sheep homozygous for R4A(delta53) remained healthy, productive, and uninfected despite a long lifetime of significant exposure. In addition to its association with susceptibility to OPPV infection, TMEM154 was also recently reported to be associated with the abundance of integrated provirus (i.e. viral load) in Rambouillet, Polypay, and Columbia sheep in the U.S. [10]. Taken together, these observations suggest that removing sheep with the most susceptible TMEM154 alleles may help eradicate OPPV and protect flocks from reinfection.
While these findings show promise for improving U.S. sheep populations, it is important to identify which other populations around the world may be impacted by highly-susceptible TMEM154 haplotypes. Estimating a population’s relative vulnerability to disease helps establish guidelines for preventative measures before an outbreak occurs. Previous research in U.S. sheep showed [9] the combined frequency of the highly-susceptible TMEM154 haplotypes could be estimated with a DNA marker on the OvineSNP50 BeadChip. This was possible because the “c” nucleotide allele of the single nucleotide polymorphism (SNP) OAR17_5388531 on the OvineSNP50 BeadChip is in strong linkage disequilibrium (LD) with the E35 allele located approximately 10 kb upstream (r 2 = 0.98). The major E35-containing haplotypes include the ancestral form of TMEM154 (haplotype 3) and the I70 variant (haplotype 2). Together, these two alleles accounted for more than 91% of the alleles containing E35 in U.S. sheep [9]. Thus, the frequency of the ‘c’ allele of OAR17_5388531 was a reasonable estimate of genetic predisposition to OPPV infection in U.S. sheep. The overall aim of the present research was to characterize TMEM154 in sheep from around the world to develop an efficient genetic test for reduced susceptibility. The findings indicated that most sheep populations around the world have highly-susceptible forms of TMEM154 and the genetic test described here efficiently detected eight known variant forms and four novel TMEM154 haplotypes discovered during this project. This genetic test, and similar designs adapted to other genotyping platforms, may be useful for preventing or controlling ovine lentivirus infections through selective breeding.
Results
LD between TMEM154 E35K and OAR17_5388531 in an International Panel of 75 Sheep
The availability of next-generation whole genome sequence data from the International Sheep Genomics Consortium (ISGC) provided the opportunity to directly determine TMEM154 genotypes in silico for 75 sheep sampled from 39 breeds and two wild species from around the world (45 groups in total). The data set for each animal represented approximately 10-fold genome coverage and was used to estimate LD between the SNP alleles for TMEM154 E35K and those for OAR17_5388531. In this set of 75 sheep, the r 2 statistic for these two markers was 0.87 and thus indicated relatively strong LD. Only four sheep from three breeds had alleles that were unambiguously in the opposite phase (i.e., the E35 allele was linked to the ‘t’ allele of OAR17_5388531). The high r 2 statistic indicated that SNP OAR17_5388531 could provide a reasonable estimate of highly-susceptible TMEM154 alleles in most sheep populations.
Estimating the Frequency of the Highly-susceptible TMEM154 Alleles in Sheep Breeds from Around the World
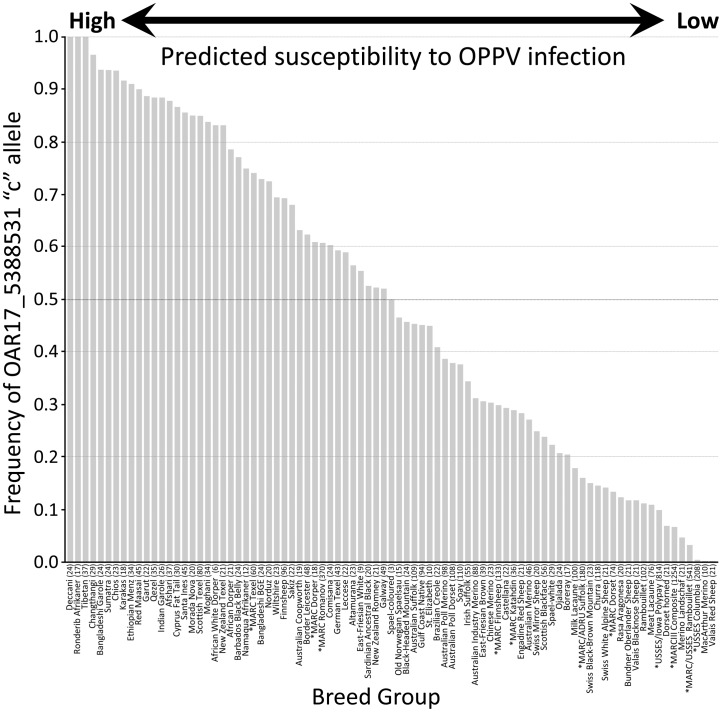
In a collection of 2,759 sheep DNAs from 74 breeds from around the world, the frequency of the “c” nucleotide allele of the c/t SNP OAR17_5388531 ranged from 0.0 to 1.0 with a mean of 0.51 and median of 0.50 among breeds (Figure 1). Breed groups with the highest “c” allele frequencies are predicted to have high frequencies of TMEM154 E35 (haplotypes 2 and 3) and larger proportions of highly-susceptible animals. Conversely, breed groups with low “c” allele frequencies are predicted to have more homozygous K35 animals (haplotype 1) and be less susceptible to OPPV. The results indicated that the highly-susceptible TMEM154 alleles are present in breeds throughout the world.
Figure 1. Estimating the frequency of highly-susceptible TMEM154 alleles in global sheep populations.
The “c” allele of SNP OAR17_5388531 is in linkage disequilibrium with the “g” nucleotide allele in codon 35 (gaa) of TMEM154. Genotypes for OAR17_5388531 were derived from the ISGC ovine SNP50k data set [11]. Numbers in parentheses for each breed group indicate the number of animals genotyped. The 11 breed groups with asterisks were genotyped for TMEM154 E35 by Sanger sequencing [9] and were included for comparison with the 74 ISGC breed groups.
Discovering Novel TMEM154 SNPs and Missense Mutations in an International Panel of 75 Sheep
The fidelity of genetic testing is enhanced by knowing the position and frequency of polymorphisms in the populations to be tested. If not accounted for, nucleotide variation at neighboring sites may cause base-pair mismatching with oligonucleotides used in DNA testing and significantly decrease the genotyping accuracy in some populations. Moreover, characterizing nucleotide variation in many previously untested breeds allows discovery of TMEM154 missense mutations. For these reasons, the same set of 75 whole genome sequence data was also used to identify novel polymorphisms in the TMEM154 gene region. An analysis of nucleotide differences among the 75 animals revealed approximately 1500 variant sites in a 61,663 bp region containing TMEM154, of which, only 128 had been previously reported [9]. The five wild sheep accounted for 13% of the nucleotide differences observed, however, no heterozygous animals were observed in wild sheep and it was assumed these were mostly species-related nucleotide differences. The positions of these 1,500 SNPs and their minor allele frequencies (MAF) are shown in Figure 2A (blue dots). For comparison, frequency data from 96 rams from 10 U.S. sheep breeds are shown for 128 SNPs (red dots). In regions of TMEM154 where data are available from both panels of sheep, there was a trend towards more SNPs and higher MAFs in the international panel of 39 breeds compared to the U.S. sheep from 10 breeds.
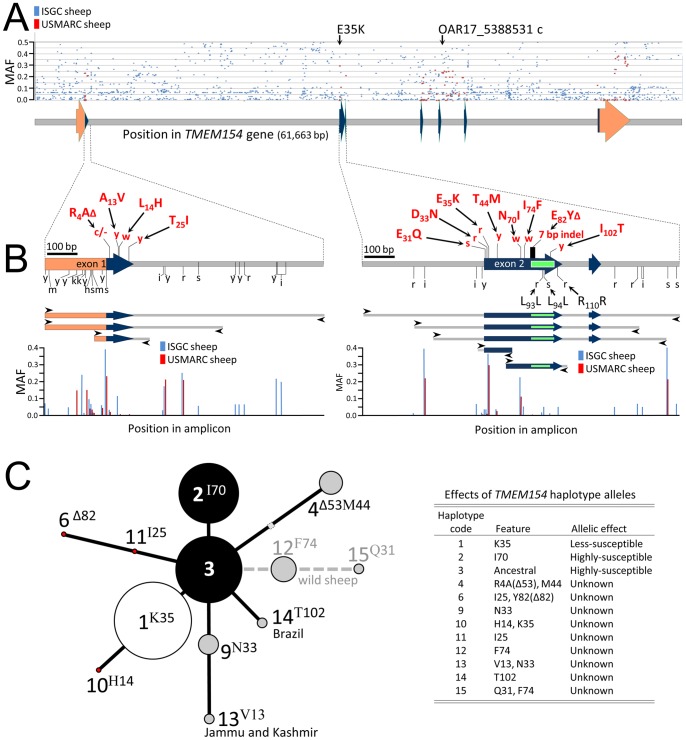
Figure 2. TMEM154 SNP maps and median-joining networks.
Panel A, genomic map of TMEM154: orange arrows, 5′ and 3′untranslated regions of exons; blue arrows, exon coding regions; grey rectangles, introns or intergenic regions. Blue and red tick dots denote position and frequency of SNPs in an international panel of 75 sheep and a panel of 96 U.S. sheep [9], respectively. Panel B, high resolution map of TMEM154 regions targeted for PCR-amplification. PCR amplification primers are indicated with black arrowheads and listed in Table S3. Red lowercase letters above SNP positions are IUPAC/IUBMB ambiguity codes for nucleotides (r = a/g, y = c/t, m = a/c, k = g/t, s = c/g, w = a/t) [17] and indicate 12 sites affected by nonsynonymous substitutions. The red uppercase letters above SNP positions indicate the amino acid polymorphisms encoded at TMEM154 codons 4, 13, 14, 25, 31, 33, 35, 44, 70, 75, 82 and 102. Black lowercase letters below SNPs indicate nucleotide polymorphisms that resulted in synonymous substitutions. Panel C, the areas of circles for haplotypes 1 to 4 are proportional to the frequencies in the international panel of 75 ISGC sheep. The symbols are as follows: black circles, risk factors; white circle, non-risk factor; grey circles, risk factor status unknown; red circles, haplotypes known in U.S. sheep but not observed in the international panel of 75 ISGC sheep (risk factor status unknown); shaded square, TMEM154 haplotype predicted to have occurred but unobserved to date. Dashed grey line, haplotypes observed in wild sheep species but not domestic sheep. Haplotypes 13 and 14 were observed in one animal each and their location of origin is indicated.
Four of the previously unreported SNPs were coding mutations located in the predicted signal peptide (A13V) and extracellular domains (E31Q, I74F, and I102T) of TMEM154. Variants A13V and I102T were discovered in populations of domestic sheep, whereas E31Q and I74F were found in the wild sheep. The inferred haplotypes for these four putative SNPs were placed in the context of the TMEM154 median-joining network to provide a framework for their future validation and evaluation of effects on susceptibility to OPPV (Figure 2C).
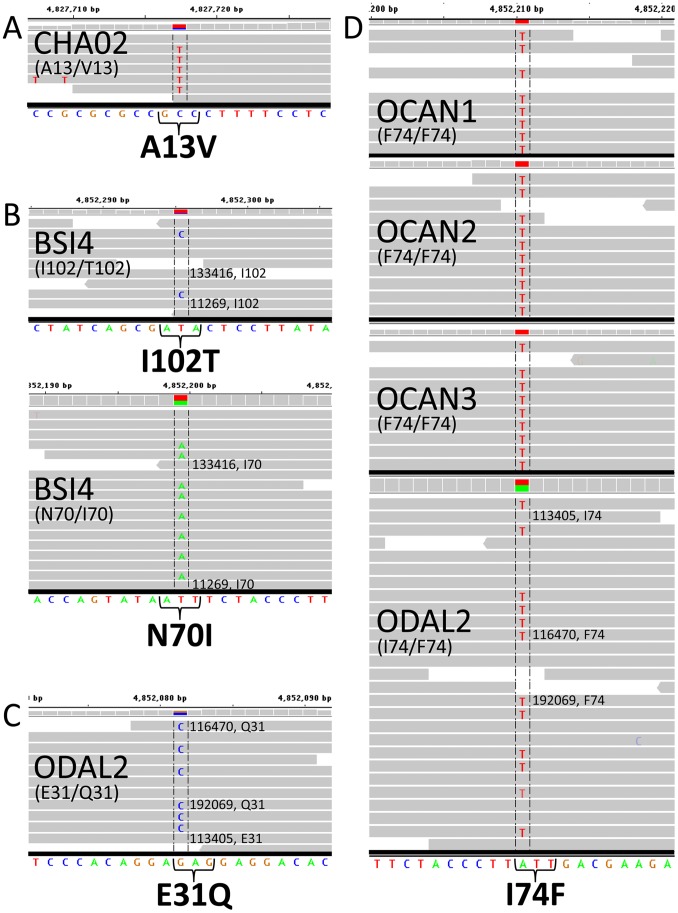
The A13V variant was observed as a heterozygote in a single Changthangi sheep, a local breed in the Changthang area of Leh district of Jammu and Kashmir state (CHA02, Figure 3A). Although the A13V variant was observed in only one animal, five of the seven mapped reads contain the GTC codon for valine. This animal was also homozygous for the TMEM154 haplotype 9, suggesting that this mutation arose on a haplotype that contained the N33 mutation (Figure 2C, haplotype 13).
Figure 3. Evidence for TMEM154 missense mutations in whole genome sequences data from an international panel of 75 sheep.
Computer screen images of Integrated Genome Viewer software [18] and showing next generation sequencing reads for animals with previously unreported SNPs affecting the TMEM154 coding sequence. Numbers shown on the reads indicated the most distal identification number on the read name when viewed in the IGV software. Direct public links to these data are provided: OCAN1 F74/F74, OCAN2 F74/F74, OCAN3F74/F74, ODAL2 I74/F74, CHA02 A13/V13, BSI4 I102/T102, BSI4N70/I70, ODAL2 E31/Q31.
The I102T variant was observed as a compound heterozygote in a single Santa Inês sheep, a breed of hair sheep found in Brazil (BSI4, Figure 3B). This animal was also heterozygous for N70I (i.e., haplotypes 2 and 3). Two of nine reads provided evidence for the rare T102 allele and both reads had high quality scores (base Phred quality of 38). Haplotype phase was determined by manual inspection of two other paired-end reads and indicated that the rare T102 allele was on haplotype 3 (Figure 2C, haplotype 14).
The most frequent of the new variants, I74F, was identified in wild sheep (Figure 3D; OCAN1, OCAN2, OCAN3, and ODAL2). The F74 variant was also observed in wild bighorn sheep from Wyoming, USA (n = 10, MAF 0.20, data not shown). A single Ovis dali animal from the ISGC set of 75 was also compound heterozygote for I74F and E31Q (ODAL2, Figure 3C). Haplotype phase for these two SNPs was determined by identifying three sets of paired-end reads which each showed that the rare Q31 allele was on the same haplotype as the rare F74 allele (Figure 3C and D). Although F74 and QA31 were only observed in wild sheep, they were placed within the context of the ovine TMEM154 median-joining network because wild sheep are also susceptible to SRLV. The most parsimonious locations of these haplotypes are shown in Figure 2C (haplotype 15). Whether in domestic sheep or wild sheep, the effects of these previously unreported TMEM154 missense mutations on susceptibility to OPPV infection are unknown.
Assigning TMEM154 Haplotype Pairs (Diplotypes) to Animals
TMEM154 polymorphisms were scored in all 75 animals at 12 sites: R4A(delta53), A13V, L14H, T25I, E31Q, D33N, E35K, T44M, N70I, I74F, E82Y(delta82), and I102T. Haplotype phase for these 12 sites was unambiguous in 68% of the animals because they had less than two heterozygous sites. Paired haplotypes (i.e., diplotypes) were unambiguously assigned for 100% of the animals by comparing each animal’s genotype to the 78 possible paired combinations of 12 haplotypes (Tables S1 and S2). However, the 20 bp GC–rich region of R4A(delta53) was underrepresented by sequencing reads in 24 of 75 animals. In those animals, the occurrence of M44 was used to infer haplotype 4 by assuming complete linkage disequilibrium with A4(delta53). This assumption is based on the observation that an A4(delta53) allele was always present with an T44 allele, and vice versa, in more than 8,000 U.S. sheep previously genotyped by Sanger sequencing at USMARC [9] (and unpublished data). The most common highly-susceptible haplotypes (2 and 3) were present in 35 of 45 groups in spite of the small sample sizes (Table 1). The average combined frequency of TMEM154 haplotypes 2 and 3 in the 75 sheep was 0.51±0.08 (Table 2). These results were consistent with those for SNP OAR17_5388531 and confirmed that the highly-susceptible TMEM154 haplotypes 2 and 3 were widely distributed among the world’s sheep.
Table 1. TMEM154 haplotypes and mutations identified in whole genome sequence data from an international panel of 75 sheep.
| Breed or species group | Region of prominence | Animals diplotyped | TMEM154haplotypesobserved | Novel missense variants |
| African White Dorper | South Africa | 2 | 1, 2, 3 | –1 |
| Afrikaner, Namaqua | South Africa | 1 | 1, 3 | – |
| Afrikaner, Ronderib | South Africa | 2 | 2, 3 | – |
| Afshari | NW Iran | 2 | 1, 2, 3, 4 | – |
| Awassi | Middle east | 1 | 2, 3 | – |
| Awassi, Turkish | Turkey | 2 | 3 | – |
| Bangladeshi | Bangladesh | 2 | 2, 3 | – |
| Brazilian Creole | Brazil | 2 | 1, 3 | – |
| Castellana | Spain | 2 | 1 | – |
| Changthangi | Jammu and Kashmir | 2 | 3, 9 | A13V |
| Cheviot | England-Scotland | 2 | 1, 2 | – |
| Churra | Spain | 2 | 1 | – |
| Cine Capari | Turkey | 1 | 3, 9 | – |
| Dorset, Poll | USA | 1 | 1 | – |
| Ethiopian Menz | Ethiopia | 1 | 3 | – |
| Finnsheep | Finland | 2 | 1, 2, 3 | – |
| Garole, Banglegdeshi | Bangledesh | 1 | 2 | – |
| Garole, Indian | India | 1 | 2, 3 | – |
| Garut | Indonesia | 2 | 3 | – |
| Gulf Coast native | USA Gulf Coast | 2 | 1, 2 | – |
| Karakas | Turkey | 2 | 1, 2, 3 | – |
| Karya | Turkey | 1 | 2, 4 | – |
| Lacaune, Meat | France | 1 | 1 | – |
| Lacaune, Milk | France | 1 | 1, 4 | – |
| Merino | Spain | 3 | 1, 3 | – |
| Morada Nova | Brazil | 2 | 1, 3 | – |
| Norduz | Turkey | 2 | 1, 3, 4 | – |
| Ojalada | Spain | 2 | 1, 4 | – |
| Ovis canadensis | Canada-USA | 3 | 3 | I74F |
| Ovis dalli | Canada-USA | 2 | 3 | E31Q, I74F |
| Romney | England | 1 | 1, 2 | – |
| Sakiz | Turkey | 2 | 1, 2, 9 | – |
| Salz | Spain | 3 | 1, 2, 3 | – |
| Santa Inês | Brazil | 2 | 1, 2, 3 | I102T |
| Scottish Blackface | United Kingdom | 1 | 1, 2 | – |
| Sumatra | Sumatra | 2 | 1, 2 | – |
| Swiss Mirror | Switzerland | 1 | 1 | – |
| Swiss White Alpine | Switzerland | 4 | 1, 2, 3 | – |
| Texel | Netherlands | 1 | 2, 3 | – |
| Tibetan, Eastern | Tibet | 1 | 2, 9 | – |
| Tibetan, Northern | Tibet | 1 | 3 | – |
| Valais Blacknose | Switzerland | 1 | 1 | – |
| Welsh Hardy Speckled Face | Wales | 1 | 1 | – |
| Welsh Mountain, Dolgellau | Wales | 1 | 1 | – |
| Welsh Mountain, Tregaron | Wales | 1 | 2 | – |
Not detected.
Table 2. Occurrence of TMEM154 haplotypes in an international panel of 75 sheep.
| Haplotype code | Key feature of haplotype | Haplotypes observeda | Groups with haplotypeb |
| 1 | K35 | 56 | 26 |
| 2 | I70 | 34 | 22 |
| 3c | E35 | 42 | 25 |
| 4 | A4(delta53) | 5 | 5 |
| 6 | Y82(delta82) | 0 | 0 |
| 9 | N33 | 4 | 4 |
| 10 | H14 | 0 | 0 |
| 11 | I25 | 0 | 0 |
| 12 | F74 | 6 | 2 |
| 13 | V13 | 1 | 1 |
| 14 | T102 | 1 | 1 |
| 15 | Q31 | 1 | 1 |
In 75 sheep.
Of 45 total groups.
Ancestral haplotype.
Reference DNAs for TMEM154 Testing
The development of genotyping assays for routine high-throughput testing required a set of reference DNAs. Homozygous DNAs were useful as PCR and genotyping controls because they provided uncomplicated results. A minimal set of DNAs was available from animals with the four most common homozygous diplotypes (1,1; 2,2; 3,3; and 4,4) and three rare diplotypes (1,6; 1,9; and 1,10). Together, these seven DNAs provided examples of the eight most common polymorphisms: R4A(delta53), L14H, T25I, D33N, E35K, T44M, N70I, and E82Y(delta82). DNA is not readily available from sheep with the four newly-discovered rare alleles (A13V, E31Q, I74F, and I102T) and thus representative DNA sequences were synthesized and used as controls (Materials and Methods). An empirically determined mixture of synthetic control DNA and reference DNA with TMEM154 diplotype 1,1 was used to produce a heterozygous MALDI-TOF MS genotype and demonstrated that animals with these rare alleles were detectable in the assay. The combination of the seven reference DNAs, together with four synthetic alleles, provided a minimal set of controls for 12 coding polymorphisms for TMEM154 for test development and routine assay quality control.
Essential Regions of TMEM154 for Genetic Testing
Based on the locations of missense mutations in TMEM154, two key regions were targeted for testing: the complete 30-amino acid signal peptide region encoded by exon 1, and residues 31 to 102 of the extracellular domain encoded by exon 2. Three amplicons were designed to encompass the 12 coding polymorphisms on these two exons. One amplicon corresponded to the region of exon 1 containing the R4A(delta53), A13V, L14H, and T25I (Figure 2B). Two other amplicons overlapped each other and corresponded to a region of exon 2 containing E31Q, D33N, E35K, T44M, N70I, I74F, E82Y(delta82), and I102T (Figure 2B).
MALDI-TOF MS Assay Design and Validation
A three-phase iterative strategy was used to validate the assay development and check concordance of diplotypes derived from MALDI-TOF MS with those derived from Sanger sequencing. In each phase, the samples were blinded, scored, and decoded. Adjustments in assay conditions were made between phases of development. In the first phase, a U.S. panel of 96 rams was genotyped and showed 100% concordance between MALDI-TOF MS and Sanger genotyping. In the second phase, a U.S. panel of 95 tetrad families was used to detect genotyping errors as revealed by non-Mendelian inheritance patterns. One error was detected where the R4 allele was not evident in some animals known to be heterozygous for R4 and A4(delta53) alleles. This apparent allele “dropout” phenomenon was also occasionally observed with R4 alleles scored by Sanger sequencing (data not shown). Although the cause(s) of the R4 allele dropout was unknown, this region of TMEM154 contains significant secondary structures with melting temperatures near 99 °C and a c/g polymorphism located 10 bp upstream of the R4A(delta53) site. Neither the addition of dimethyl sulfoxide in the PCR cocktail, nor the use of alternative primers in the MALDI-TOF MS extension reaction completely alleviated the occasional R4 allele dropout. Thus, homozygous A4(delta53) genotypes were scored only when they occurred in animals that were also homozygous for M44 in exon 2.
In the last phase of validation, results were obtained for 499 sheep in a single blinded genotyping trial to measure efficiency and accuracy of the genotype assay. Genotyping statistics were calculated for the eight most common polymorphic sites because the remaining four SNPs were monomorphic in these sheep. The call rate for the eight sites was 99.4% for the 499 sheep tested and 96.0% of the animals received a TMEM154 diplotype assignment (Table 3). Comparing diplotypes from MALDI-TOF MS and Sanger sequencing for 479 animals showed seven discordances (1.46%). Close inspection of raw tracefiles revealed the discordances were due to problems in the Sanger data, including: unamplified alleles, poor quality reads, and errors in manual scoring. These results indicate this MALDI-TOF MS-based test for TMEM154 provides an accurate alternative to Sanger-based genotyping.
Table 3. Call rates and concordance of TMEM154 genetic testing.
| Animal group | Yearsampled | Sheep | Missing SNPgenotypes | SNP callratea (%) | Missingdiplotypes | Diplotypecall rate | Discordant diplotypes | MALDI-TOFMS errors | Sanger errors |
| 4- to 9-year-old ewesb | 2003 | 260 | 7 | 99.7 | 7 | 0.973 | 3 | 0 | 3 |
| Composite lambsc | 2011 | 239 | 16 | 99.2 | 13 | 0.946 | 4 | 0 | 4 |
| Total | nad | 499 | 23 | 99.4 | 20 | 0.960 | 7 | 0 | 7 |
Genoptypic data were collected in a single pass for R4A(delta53), H14L, T25I, D33N, E35K, T44M, N70I, and E82Y(delta82).
This animal group was composed of 160 OPP case-control pairs as previously described [9].
Breed composition: 1/2 Romanov, 1/4 Rambouillet, 1/8 White Dorper, and 1/8 Katahdin.
Not applicable.
Discussion
This report characterizes TMEM154 in sheep from around the world by estimating the distribution of highly-susceptible haplotypes and identifying novel nucleotide variants. Genotype results from SNP OAR17_5388531 of the ovine SNP50 BeadChip indicated that the two most susceptible alleles (haplotypes 2 and 3) were common and distributed widely. Analysis of next generation sequence for the complete TMEM154 gene in an international panel of 75 sheep confirmed that the three most common haplotypes described in U.S. sheep populations (haplotypes 1, 2, and 3) are also abundant in groups of sheep sampled from other countries. The most common TMEM154 haplotype in the international panel of 75 sheep was the less susceptible haplotype 1 encoding lysine at position 35 with a frequency of 0.37. Thus, the opportunity exists in many populations to reduce susceptibility by selectively breeding animals.
The distribution of rare mutant haplotype alleles in populations may provide insight to their history. For example, TMEM154 haplotypes 6, 10, and 11 (i.e., those with Y82(delta82), H14, and I25 mutations, respectively) were not detected in the international panel of 75 sheep. To date, haplotype 6 has been observed only in Suffolk, and haplotypes 10 and 11 have only been observed in animals with Rambouillet germplasm. In contrast, haplotypes 4 and 9 (i.e., those with A4(delta53)/M44 and N33 mutations, respectively) were observed in breed groups from Turkey, Iran, Spain, France, Tibet, Jammu and Kashmir, suggesting that these haplotypes existed before sheep were brought to the U.S. The occurrence of 12 coding mutations in the predicted signal peptide and extracellular domains of TMEM154 is remarkable considering that none have been detected in the putative cytosolic domain of TMEM154 in 309 total sheep from 55 breed groups. However, the biological function of the TMEM154 protein and the impact of these mutations on its function, remain unknown.
The diversity of protein isoforms encoded by the TMEM154 gene in sheep presents a number of challenges for genetic testing and selective breeding. One challenge is to define a framework for describing the alleles and understanding their action. A frequency-based haplotype numbering system was combined with a rooted median-joining network to put the haplotype-encoded peptide isoforms in a simplified context. Another challenge is to test the haplotypes for their effects on susceptibility to infection while accounting for potential confounders like virus strain, animal exposure, breed type, location, climate, geography, and animal husbandry practices. These factors may potentially overcome the effects of TMEM154 haplotypes in a given setting. A third challenge is to develop and implement effective TMEM154 allele management strategies. This will require deploying genetic tests, tracking alleles, and measuring the incidence of infection in a wide variety of settings relevant to commercial sheep production.
The genetic information and assay designs described here were developed so they could be adapted to other platforms and technologies around the world to measure the effects of TMEM154 haplotypes in local populations. This report also provides the first commercially-available high-throughput genetic test for ovine TMEM154 haplotypes. The availability of a commercial test is essential for producers to make immediate genetic progress in their respective sheep-improvement programs. As more information becomes available about the effects of TMEM154 alleles in specific populations and environments, the most appropriate haplotypes can be selected for reducing OPPV susceptibility.
Materials and Methods
Ethics Statement
Prior to their implementation, all animal procedures were reviewed and approved by the Care and Use Committee at the United States Department of Agriculture (USDA), Agricultural Research Service, Meat Animal Research Center (USMARC) in Clay Center, Nebraska.
Animal Samples and Genotypes
The ISGC collected and genotyped 2,819 sheep from 74 breeds as part of a large study into genetic diversity and the impact of selection after domestication [11]. Samples were collected from multiple flocks to minimize relationships within breed. Breeds were collected from the Americas, Africa, Asia, Europe, and the domestication center in present day Iran and Turkey. The geographic origin, breed identity, and number of animals per breed has been previously described [11]. DNA samples were genotyped with the Illumina (San Diego, California USA) ovine SNP50 Beadchip. Genotypes for SNP OAR17_5388531 were available for 2,759 sheep and used for analysis.
The ISGC selected 75 animals for whole genome sequencing to extend its investigation of genetic diversity and selection in the world’s sheep breeds [10]. The majority of the animals (61%) were drawn from the previous study [10] to capture the diversity present across O. aries. Additional animals were recruited that either: 1) had previously been used in the construction of genomic resources for the sheep genome [12], 2) carried disease genes, or 3) were wild sheep sampled from the bighorn (O. canadensis) and thinhorn (O. dalli) populations of North America. Each genome was sequenced and mapped to a read-depth coverage of approximately10-fold with Illumina GAII (unpublished). Prepublication access to the raw sequence data (.bam files) was provided under the Toronto guidelines for data users [13]. These 75 sheep from 39 breed groups and two species groups were used to derive genotypes for the 62 kb genomic region containing TMEM154.
The USMARC Sheep Diversity Panel, version 2.4 consists of 96 rams from nine breeds, a composite population, and one Navajo-Churro ram with a rare prion haplotype allele (ARK) as previously described [14]. These rams were part of a set of 96 tetrad families consisting of a ram, a ewe, and twin offspring and used to confirm the haplotype phase of TMEM154 alleles and to further evaluate the accuracy of genotype scoring. Since the first report of this panel in 2010 [14], family number 47 has been removed because the genotypes from multiple disperse loci for this ram (USMARC Finn no. 200117718) indicate it is not the sire of these offspring. The remaining 95 families were used for testing the accuracy, reproducibility, and segregation of MALDI-TOF MS assays for 12 TMEM154 polymorphisms affecting 12 codons.
Sheep used for blinded MALDI-TOF MS genotyping trials included 260 USMARC OPP case-control sheep composed of 130 pairs of 4- to 9-year-old ewes [9], and 239 lambs from a 2011 cohort that were 1/2 Rambouillet,1/4 Romanov, 1/8 White Dorper, and 1/8 Katahdin (499 total).
The Wyoming, USA bighorn sheep samples consisted of DNA from 10 wild animals taken from different wildlife management areas across Wyoming prior to 2001.
Determining TMEM154 Diplotypes from Whole Genome Sequences
DNA sequence reads from sheep representing 39 O. aries breeds plus O. canadensis and O. dalli were previously mapped to an ovine reference genome to an average sequencing depth of ten-fold. The UnifiedGenotyper analysis tool from the Genome Analysis Toolkit [15] was used to identify variants, and to genotype the samples at those variant sites. SAMTools viewer [16] was used to extract the subset of the.bam files from each animal’s whole genome sequence files for the locus TMEM154. The genomic coordinates used were chromosome 17 positions 4,822,280 to 4,884,987 on the Oar v2.0 map (http://www.livestockgenomics.csiro.au/sheep/). These locus-specific.bam files, their corresponding index files, and genotypes for the variants identified, were loaded into the Intrepid Bioinformatics data management system. Cross reference hyperlinks (/db_xref) were also added to the GenBank file accession HM355886. Diplotypes for the TMEM154 polypeptide isoforms were constructed by concatenating the genotypes at the 12 polymorphic sites predicted to alter the coding sequence and then matching the concatenated set of genotypes to those listed in Table S1. Because loop structures were not observed in the median-joining network depicted in Figure 2C, the diplotype could be arrived at by only one combination of haplotypes. In rare cases where animals with novel missense SNPs were also heterozygous for nearby missense SNPs, haplotype phase was determined with information from paired-end reads that was accessible in the SAMTools viewer.
MALDI-TOF MS Genotyping and Synthetic DNA Controls for Rare Alleles
Genotyping was performed at GeneSeek with the Sequenom MassARRAY platform and iPLEX GOLD chemistry according to the manufacturer’s instructions (Sequenom, San Diego, CA USA). Briefly, multiplex assays were designed with commercial software and adjusted manually (see Table S3 for list of oligonucleotides). TMEM154 exons 1 and 2 were amplified from genomic DNA by PCR and residual oligonucleotides were subsequently dephosphorylated prior to thermocycling in the single-base extension reaction. The oligonucleotide extension products were desalted with size-exclusion resin and transferred to a 384-well chip for MALDI-TOF MS. High-fidelity, long oligonucleotides (Integrated DNA technologies, Coralville, IA USA) with DNA sequences of the 12 minor alleles were synthesized and used as positive control templates in PCR amplification. The lyophilized oligonucleotides were dissolved in 10 mM TrisCl, 1 mM EDTA (pH 8.0) to 25 µM and then empirically diluted to between 1×103 to 1×106 copies per µl before mixing 1∶1 vol/vol with genomic DNA. These samples were subjected to PCR and genotyping as described above. Sequences used for oligonucleotide synthesis included those for TMEM154 exon 1 (174 bp): tttcagcgggactgacaccgcg[t]gcagcagcatcgc[g]atgccgggg[deleted ‘c’,A4]gcgcgccccccgaggctccgcgcgccg[t,V13]cc[a,H14]tttcctcgccgcggtcctcgcgtcccttccca[t,I25]ccgcccggcgcagggtaagcacccctcggctttccactcccggcgaggaggatgaggaaggctt; TMEM154exon 2a (103 bp): gtctcaattttgtatgtgttcccacagga[c,Q31]aggag[a,N33]acaca[g,E35]aactgtcaggagacgtgcccccaggca [t,M44]ggaaggcctggatgaagagtcagaggccctaag; TMEM154exon 2b (114 bp): cacacttgcttcagtgaccacagaaccttacatcaccagtataa[t,I70]ttctaccctt[t,F74]ttgacgaagacacagaccagtta [deleted ‘gagttta’,Y82]tattaatggtgttgatcccagtgatttt; and TMEM154exon2c (98 bp): actctctctcctgcttctatcagcga[c,T102]actccttataatataccataaaag[g]aaaaggaataaacaaggtaaatattttgcctgttctcatttctaga.
Supporting Information
List of the 78 TMEM154 diplotypes possible from 12 known haplotypes.
(XLSX)
Haplotypes encoding polypeptide isoforms of ovine TMEM154.
(XLSX)
Oligonucleotide information for TMEM154 genetic testing.
(XLSX)
Acknowledgments
We thank J. Carnahan for outstanding technical assistance, J. Watts for secretarial support, the USMARC sheep crew for production and management of sheep, and D.A Hawk of the Wyoming Game and Fish Department for providing bighorn sheep samples. This work was conducted in part using the resources of the University of Louisville’s research computing group and the Cardinal Research Cluster.
The members of the International Sheep Genomics Consortium who contributed samples and expertise towards the design and execution of ovine SNP50k genotyping and next generation sequencing, and/or coauthors of [11] include: Juan Jose Arranz, Universidad de Leon; Georgios Banos, Aristotle University of Thessaloniki; William Barendse, CSIRO Livestock Industries; Ahmedn El Beltagy, Animal Production Research Institute; Jorn Benenwitz, University of Hohenheim; Steven Bishop, The Roslin Institute; Simon Boitard, INRA; Lutz Bunger, Scottish Agricultural College; Jorge Calvo, CITA; Antonello Carta, Agris Sardegna; Ibrahim Cemal, Adnan Menderes University; Elena Ciani, University of Bari; Noelle Cockett, University of Utah; Dave Coltman, University of Alberta; Brian Dalrymple, CSIRO Livestock Industries; Mariasilvia D’Andrea, Università degli Studi del Molise; Ottmar Distl, University of Veterinary Medicine Hannover; Cord Drogemuller, Institute of Genetics, University of Berne; Georg Erhardt, Institut für Tierzucht und Haustiergenetik Justus-Liebig-Universität Gießen; Emma Eythorsdottir, Agricultural University of Iceland; Kimberly Gietzen, Illumina Inc.; Clare Gill, Texas A&M University; Elisha Gootwine, The Volcani Center; Vidya Gupta, National Chemical Laboratory; Olivier Hanotte, University of Nottingham; Ben Hayes, Department of Primary Industries Victoria; Michael Heaton, USDA MARC; Stefan Hiendleder, University of Adelaide; Han Jialin, ILRI and CAAS; Juha Kantanen, MTT Agrifood Research; Matthew Kent, CiGene; James Kijas, CSIRO Livestock Industries; Denis Larkin, University of Aberystwyth; Johannes A. Lenstra, Utrecht University; Kui Li, Lhasa People Hospital, Tibet; Terry Longhurst, Meat and Livestock Australia; Runlin Ma, Chinese Academy of Science; Russell McCulloch, CSIRO Livestock Industries; David MacHugh, University College Dublin; Sean McWilliam, CSIRO Livestock Industries; John McEwan, AgResearch; Jillian Maddox, University of Melbourne; Massoud Malek, IAEA; Faruque Mdomar, Bangladesh Agriculture University; Despoina Miltiadou, Cyprus University of Technology; Luis V. Monteagudo Ibáñez, Universidad de Zaragoza; Carole Moreno, INRA; Frank Nicholas, University of Sydney; Kristen Nowak, University of Western Australia; V. Hutton Oddy, University of New England; Samuel Paiva, Embrapa; Varsha Pardeshi, National Chemical Laboratory; Josephine Pemberton, University of Edinburgh; Fabio Pilla, Università degli Studi del Molise; Laercio R. Porto Neto, CSIRO Livestock Industries; Herman Raadsma, University of Sydney; Cyril Roberts, Caribbean Agricultural Research and Development Institute; Magali San Cristobal, INRA; Tiziana Sechi, Agris Sardegna; Paul Scheet, University of Texas M. D. Anderson Cancer Center; Bertrand Servin, INRA; Mohammad Shariflou, University of Sydney; Pradeepa Silva, University of Peradeniya; Henner Simianer, University of Goettingen; Jon Slate, University of Sheffield; Mikka Tapio, MTT; and Selina Vattathil, University of Texas M. D. Anderson Cancer Center; Vicki Whan, CSIRO Livestock Industries.
Funding Statement
Funding for this research was provided by the United States Department of Agriculture, Agricultural Research Service. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. There were no current external funding sources for this study.
References
- 1. Thormar H (2005) Maedi-visna virus and its relationship to human immunodeficiency virus. AIDS Rev 7: 233–245. [PubMed] [Google Scholar]
- 2. Patel JR, Heldens JG, Bakonyi T, Rusvai M (2012) Important mammalian veterinary viral immunodiseases and their control. Vaccine 30: 1767–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blacklaws BA, Berriatua E, Torsteinsdottir S, Watt NJ, de Andres D, et al. (2004) Transmission of small ruminant lentiviruses. Vet Microbiol 101: 199–208. [DOI] [PubMed] [Google Scholar]
- 4. Peterhans E, Greenland T, Badiola J, Harkiss G, Bertoni G, et al. (2004) Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet Res 35: 257–274. [DOI] [PubMed] [Google Scholar]
- 5. Sigurdsson B, Grimsson H, Palsson PA (1952) Maedi, a chronic, progressive infection of sheep’s lungs. J Infect Dis 90: 233–241. [DOI] [PubMed] [Google Scholar]
- 6. Keen JE, Hungerford LL, Littledike ET, Wittum TE, Kwang J (1997) Effect of ewe ovine lentivirus infection on ewe and lamb productivity. Prev Vet Med 30: 155–169. [DOI] [PubMed] [Google Scholar]
- 7.USDA (2003) Ovine Progressive Pneumonia: Awareness, Management, and Seroprevalence. In: Animal and Plant Health Inspection Service VS, Centers for Epidemiology and Animal Health, editor. Fort Collins.
- 8. Gjerset B, Rimstad E, Teige J, Soetaert K, Jonassen CM (2009) Impact of natural sheep-goat transmission on detection and control of small ruminant lentivirus group C infections. Vet Microbiol 135: 231–238. [DOI] [PubMed] [Google Scholar]
- 9. Heaton MP, Clawson ML, Chitko-McKown CG, Leymaster KA, Smith TP, et al. (2012) Reduced Lentivirus Susceptibility in Sheep with TMEM154 Mutations. PLoS Genet 8: e1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. White SN, Mousel MR, Herrmann-Hoesing LM, Reynolds JO, Leymaster KA, et al. (2012) Genome-wide association identifies multiple genomic regions associated with susceptibility to and control of ovine lentivirus. PloS one 7: e47829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kijas JW, Lenstra JA, Hayes B, Boitard S, Porto Neto LR, et al. (2012) Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biol 10: e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Archibald AL, Cockett NE, Dalrymple BP, Faraut T, Kijas JW, et al. (2010) The sheep genome reference sequence: a work in progress. Animal genetics 41: 449–453. [DOI] [PubMed] [Google Scholar]
- 13. Birney E, Hudson TJ, Green ED, Gunter C, Eddy S, et al. (2009) Prepublication data sharing. Nature 461: 168–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heaton MP, Leymaster KA, Kalbfleisch TS, Freking BA, Smith TP, et al. (2010) Ovine reference materials and assays for prion genetic testing. BMC Vet Res 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nomenclature for incompletely specified bases in nucleic acid sequences. Recommendations 1984. Nomenclature Committee of the International Union of Biochemistry (NC-IUB). Proc Natl Acad Sci U S A 83: 4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, et al. (2011) Integrative genomics viewer. Nature biotechnology 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the 78 TMEM154 diplotypes possible from 12 known haplotypes.
(XLSX)
Haplotypes encoding polypeptide isoforms of ovine TMEM154.
(XLSX)
Oligonucleotide information for TMEM154 genetic testing.
(XLSX)