Abstract
Both BMP2 and BMP4 are involved in tooth development. We examined phenotypes and BMP2 and BMP4 gene variations in two Mexican oligodontia families. Physical and oral examinations and panoramic radiographs were performed on affected and unaffected members in these two families. The affected members lacked six or more teeth. DNA sequencing was performed to detect BMP2 and BMP4 gene variations. Three single nucleotide polymorphisms (SNPs) in BMP2 and BMP4 genes were identified in the two families, including one synonymous and two missense SNPs: BMP2 c261A>G, pS87S, BMP2 c570A>T, pR190S, and BMP4 c455T>C, pV152A. Among the six affected patients, 67% carried “GG” or “AG” genotype in BMP2 c261A>G and four were “TT” or “AT” genotype in BMP2 c570A>T (pR190S). Polymorphism of BMP4 c455T>C resulted in amino acid changes of Val/Ala (pV152A). BMP2 c261A>G and BMP4 c455T>C affect mRNA stability. This was the first time that BMP2 and BMP4 SNPs were observed in Mexican oligodontia families.
Keywords: Bone morphogenetic protein 2, Phenotype, Oligodontia, Bone morphogenetic protein 4, Single nucleotide polymorphisms
INTRODUCTION
Tooth agenesis is the most common craniofacial congenital anomaly in humans. Oligodontia is defined as the congenital absence of six or more teeth, excluding the third molars. This severe form of tooth agenesis is associated with systemic anomalies and multiorgan syndromes and can appear in an isolated sporadic form and as an isolated family characteristic. The prevalence of oligodontia is 0.08–1.1% (Schalk-van der Weide et al., 1992; Stockton et al., 2000; Gabris et al., 2001). Tooth agenesis is a complex phenotype, with variable penetrance and expressivity affecting various numbers of teeth in different regions. Differences in tooth sizes and dentition distribution may also be observed, especially in more severe cases.
Tooth development involves multiple processes that are controlled by many growth and transcription factors (Thesleff, 2003), including the bone morphogenetic protein (BMP), fibroblast growth factor, sonic hedgehog, and WNT signaling pathways. Studies in mice and humans have shown that hundreds of genes control tooth development (Nieminen et al., 1998). Gene function defects can affect tooth formation and lead to congenital missing teeth, although the molecular bases of such defects are not completely understood. Mutations of the msh homeobox 1 (MSX1), paired box gene 9 (PAX9), axis inhibition protein 2 (AXIN2), latent transforming growth factor beta binding protein 3, and ectodysplasin-A (EDA) genes are associated with tooth agenesis (Liu et al., 2000; Stockton et al., 2000; van den Boogaard et al., 2000; Lammi et al., 2003; Kapadia et al., 2007; Han et al., 2008; Noor et al., 2009). However, genetic mutations in MSX1, PAX9, EDA, or AXIN2 do not fully account for all patients with tooth agenesis (Gerits et al., 2006). Tooth development studies in mice have identified many candidate genes responsible for oligodontia.
BMPs are members of the transforming growth factor beta superfamily, and their molecules participate in the cascade of signaling events during early tooth development. BMP2 and BMP4 genes are expressed in the epithelium and mesenchyme in tooth germs (Aberg et al., 1997; Thomadakis et al., 1999; Chen et al., 2008). BMP2 and BMP4 conventional knockout mice die during early gastrulation owing to failure of mesoderm induction (Zhang and Bradley, 1996; Lawson et al., 1999). Recently, studies using conditional knockout mice for BMP2 and BMP4 have revealed critical roles for each BMP in tooth development, particularly in root development (Gluhak-Heinrich et al., 2010; Feng et al., 2011). Furthermore, BMP4 synergizes with MSX1 and PAX9 to control the mesenchymal layer of the developing tooth bud (Bei et al., 2000; Kong et al., 2011). Single nucleotide polymorphisms (SNPs) are the most abundant resource of genetic variation among individuals. Specific allelic variants of SNPs have been reported to be more prevalent in persons with tooth agenesis than in healthy individuals. The purpose of this study was to investigate the involvement of BMP2 and BMP4 in the pathogenesis of tooth agenesis in two Mexican oligodontia families.
MATERIAL AND METHODS
Patients and phenotype analysis
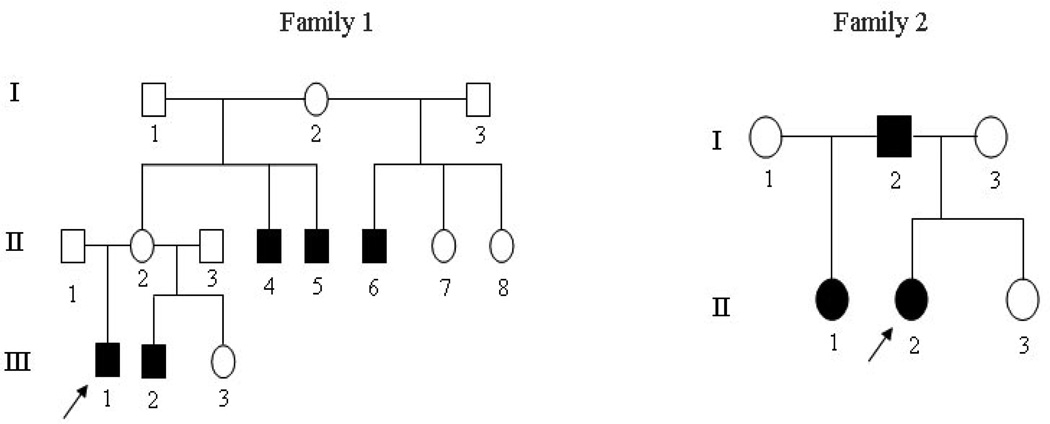
The probands of the two Mexican families were pediatric patients at the University of Texas Health Science Center at San Antonio Dental School. Physical and dental examinations were performed on the members of the families. Pedigree construction was carried out through interviews and clinical examinations (Figure 1). An intra-oral examination was performed to assess the presence of teeth, tooth sizes, tooth morphology, and enamel abnormalities. A panorex radiograph from available members of the families was taken to assess dental development. Extra- and intra-oral pictures of the probands were made. A thorough clinical examination of other tissues of ectodermal origin, including skin, hair, nails, sweat glands, ears, and eyes, was carried out. The institutional review board at the University of Texas Health Science Center at San Antonio Dental School approved this study, and informed consent was obtained from all participants.
Figure 1.
Pedigrees of the two oligodontia families with arrows indicating the proband of each family. Black figures = affected; open figures = unaffected; squares = males; circles = females.
Mutational analysis
Peripheral blood samples were collected from the members of the two families and from healthy Mexicans as negative controls. Genomic DNA was extracted from whole blood using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). Polymerase chain reaction was used to amplify the coding exons and adjacent intronic sequences of the BMP2 and BMP4 genes using BMP2- and BMP4-specific primers (Table 1). DNA sequencing was performed at the University of Texas at Austin Institute for Cellular and Molecular Biology core. Sequencing results and consensus sequences from the National Center for Biotechnology Information human genome database were compared using the SeqManII program in the Lasergene package (DNAstar Inc., Madison, WI, USA).
Table 1.
Primers used for PCR amplification and sequence.
| Gene | Exon | Primer sequence (5'-3') | Amplicon size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| BMP2 | 2F | CTCACGTCGGTCCTGTCC | 392 | 53 |
| 2R | CCCTGCTCCATGCCTCAC | |||
| 3aF | CAAACGTCATTACTTGGCTTACTG | 591 | 51 | |
| 3aR | GCTGTTTGTGTTTGGCTTGA | |||
| 3bF | GCTAGTAACTTTTGGCCATGAT | 600 | 53 | |
| 3bR | TGTTTTCCCAACTTCTTCTTTTCG | |||
| BMP4 | 3F | CCATCTTGCCCCTCCATTTCTA | 570 | 60 |
| 3R | CTTCTTCCCCAGGGCTTTCACT | |||
| 4aF | TGCTTATTTTCCCCCAGTAGGT | 704 | 60 | |
| 4aR | GGCGCCGGCAGTTCTTATTCTT | |||
| 4bF | GGGCCAGCATGTCAGGATTAGC | 575 | 60 | |
| 4bR | TGTGGGTGAGTGGATGGGAACG |
Bioinformatics analysis of BMP2 and BMP4 proteins and messenger RNA (mRNA) structures
Changes in protein structure or function owing to amino acid substitution were predicted using online tools: PolyPhen (http://genetics.bwh.harvard.edu/pph) and SIFT (http://blocks.fhcrc.org/sift/SIFT.html). Computational two-dimensional models of the BMP2 and BMP4 proteins were performed using the bioinformatics tool DNASIS v2.5. Secondary structures of the full-length BMP2 and BMP4 mRNAs were predicted using GeneBee (http://www.genebee.msu.su/genebee.html).
RESULTS
Family and phenotype analysis
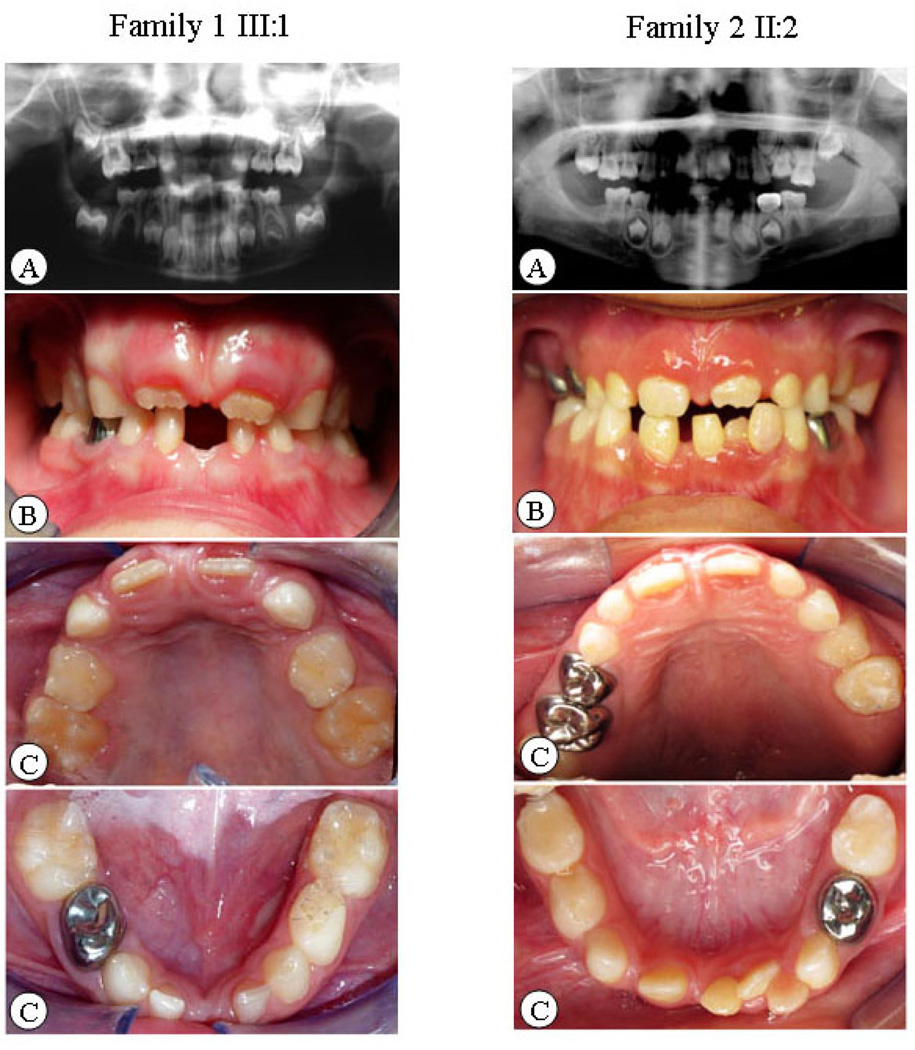
The two Mexican families in the study resided in Texas, USA. The pedigree analysis is shown in Figure 1. Eleven individuals in the families underwent clinical and oral examinations. No abnormalities in other ectodermal tissues were found in these individuals. The phenotypes of the families showed isolated non-syndromic oligodontia. The extra-oral examination indicated gross craniofacial asymmetry but no obvious maxillofacial deformities. The intra-oral examination displayed normal crown sizes, no oral cleft, and no enamel hypoplasia, except in subject II: 2 of family 2, who had a peg-shaped upper later incisor. The diagnosis of congenitally missing teeth depended not only on the panoramic radiographs of dentition but also on the inter-oral examination and dental history. Figure 2 shows the panoramic radiographs as well as intra-oral and extra-oral photographs of the probands from the families. Six affected members in the families had multiple missing teeth. Excluding for third molars, the average number of missing teeth in each person was six in the affected individuals of family 1. In family 2, the number of missing teeth was 14, which was more than twice the number in family 1 (Table 2).
Figure 2.
Clinical and radiographic oral-facial manifestation of oligodontia in proband of each family. A. Panoramic radiograph of dentition. B. Intra-oral bite photos. C. Intra-oral dentition photos.
Table 2.
Phenotypes of affected family members.
| Right |
Left |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 7 | 6 | 5 | 4 | 3 | 2 | 1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| F1 | ||||||||||||||||
| III:1 | ||||||||||||||||
| Maxilla | ? | * | * | * | * | ? | ||||||||||
| Mandibles | ? | * | * | * | * | * | ? | |||||||||
| III:2 | ||||||||||||||||
| Maxilla | ? | * | * | * | * | ? | ||||||||||
| Mandibles | ? | * | * | * | ? | |||||||||||
| II:4 | ||||||||||||||||
| Maxilla | * | * | * | * | ||||||||||||
| Mandibles | * | * | ||||||||||||||
| F2 | ||||||||||||||||
| II:1 | ||||||||||||||||
| Maxilla | * | * | * | * | * | * | * | * | ||||||||
| Mandibles | * | * | * | * | * | * | * | * | ||||||||
| II:2 | ||||||||||||||||
| Maxilla | ? | * | * | * | * | * | * | * | * | ? | ||||||
| Mandibles | ? | * | * | * | * | * | * | * | * | ? | ||||||
| I:1 | ||||||||||||||||
| Maxilla | * | * | * | * | * | * | * | * | ||||||||
| Mandibles | * | * | * | * | * | * | * | * | * | |||||||
III:1, III:2, II:4 in family 1 and II:1, II:2, I:1 in family 2.
Congenitally missing teeth;
? = diagnosis impossible for young age patient.
F1 and F2 = family 1 and family 2.
Genotype analysis
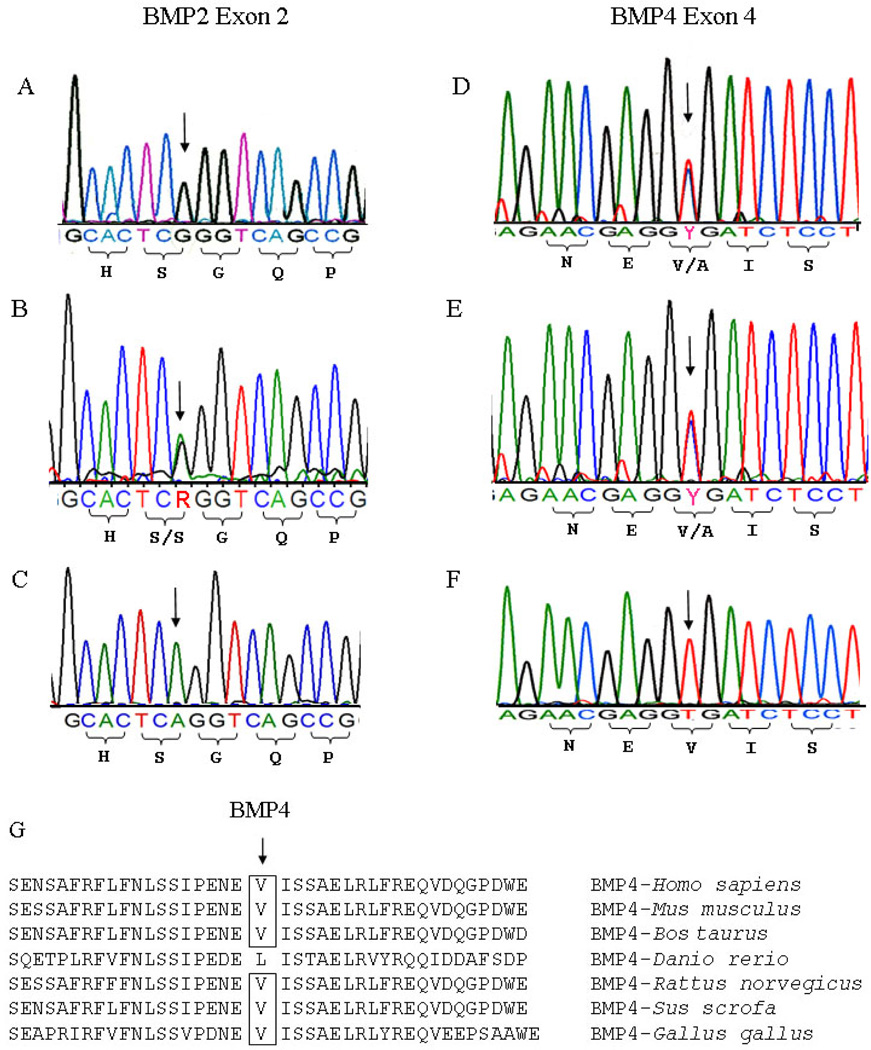
The sequence analysis of exon and exon-intron boundaries of BMP2 and BMP4 was performed on the members of the two Mexican oligodontia families and revealed three SNPs: BMP2 c261A>G, pS87S; c570A>T, pR190S, and BMP4 c455 T>C, pV152A (Table 3). Among the six affected patients in the two families, 67% carried either the GG or AG genotypes of BMP2 c261A>G. The TT and AT genotypes of BMP2 c570 alleles accounted for 66% of the BMP2 c570 SNPs. In the BMP4 gene, valine at position 152 had a high degree of homology across species with the exception of Danio rerio (Figure 3A). A sequence chromatogram of BMP2 c261A>G and BMP4 c455 T>C in the oligodontia patients of each family compared with the sequences of people without missing teeth is shown in Figure 3B–G.
Table 3.
Sequence variations found in BMP2 and BMP4 genes.
| Gene | Exon | Sequence variation | Amino acid change | Family 1 |
Family 2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| III:1 | III:2 | II:2 | II:4 | I:2 | I:1 | I:2 | I:3 | II:1 | II:2 | ||||
| BMP2 | 2 | c.261A>G | S87S | G | AG | G | AG | AG | AG | A | G | A | AG |
| 3 | c.570A>T | R190S | T | T | T | AT | T | AT | A | T | A | AT | |
| BMP4 | 4 | c.455T>C | V152A | T | T | T | TC | T | T | TC | T | TC | T |
Figure 3.
DNA sequencing of BMP2 and BMP4 genes in the two Mexican oligodontia families. BMP2 c261A>G variation. Sequence analysis demonstrated c261A>G. A. c261 “GG” genotype in proband III:1 patient in family 1. B. c261 “AG” heterozygous genotype in proband II:2 patient in family 2. C. c261 “AA” genotype in an unaffected person BMP4 c455T>C. D. c455 “TC” heterozygous genotype in II:4 patient in family 1. E. c455 “TC” heterozygous genotype in II:1 patient in family 2. F. c455 “TT” homozygous genotype in an unaffected person. G. Protein alignment of human BMP4 (residues between 133 and 172) with other BMP4 species from Mus musculus, Bos taurus, Danio rerio, Rattus norvegicus, Sus scrofa, and Gallus gallus. Valine (V) at position 152 in human BMP4 has a high degree of homology across species except for D. rerio.
Predicted functional SNPs BMP2 c261A>G and BMP4 c455T>C
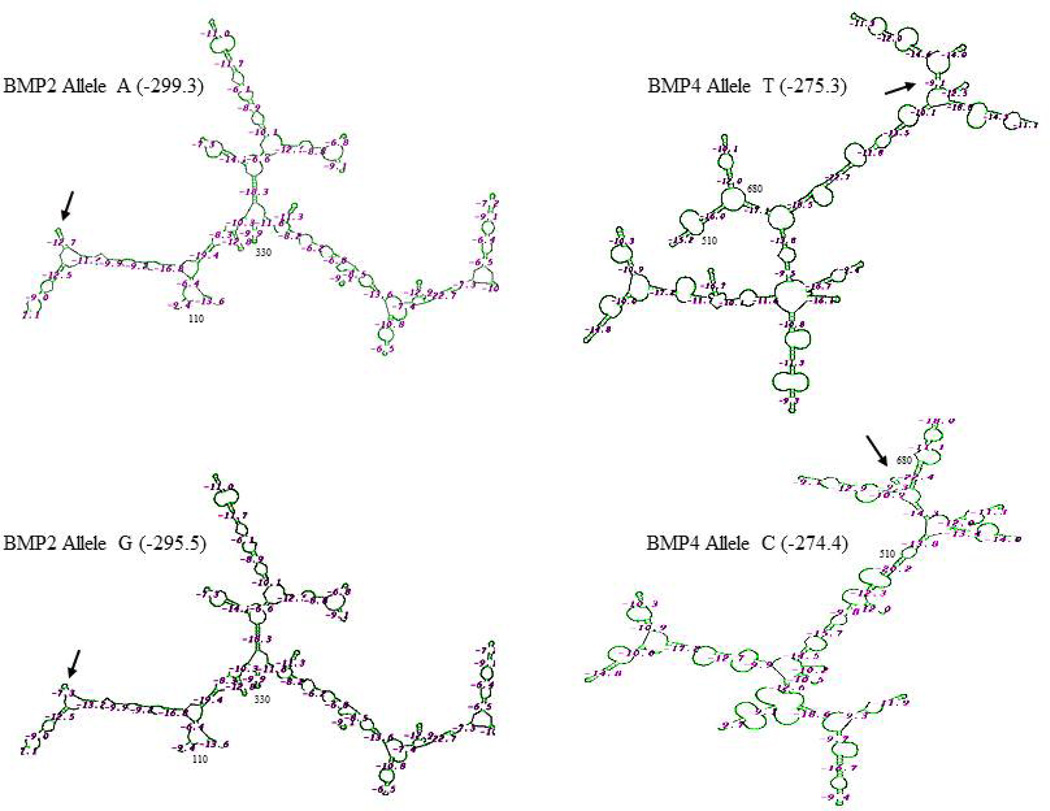
Using GeneBee, we predicted the RNA secondary structures of the SNPs of the BMP2 and BMP4 genes. The results showed that BMP2 c261A>G and BMP4 c455T>C substitutions led to different mRNA structures between c261A and c261G alleles and between c455T and c455C alleles, respectively (Figure 4). The free energy of BMP2 mRNAs was affected by A/G substitution (−299.3 kcal/mol for the A allele and −295.5 kcal/mol for the G allele), and the free energy of BMP4 was −275.3 for the T allele and −274.4 for the C allele, although these protein secondary structures are unmodified (data not shown).Capasso et al. (2009) have observed that BMP4 c455T>C changes the BMP4 mRNA secondary structures between c455T and c455C and that BMP4 mRNA and protein expression levels were higher in the T allele in a southern Italy population. The above data imply that the BMP2 G allele and BMP4 C allele require higher energy for mRNA stability.
Figure 4.
Predicted mRNA secondary structures of BMP2 and BMP4 coding region alleles. SNPs of BMP2 c261A>G and BMP4 c455T>C cause changes of the mRNA secondary structures and free-energy parameters. Higher free energy is need for the secondary formation of BMP2 G-allele and BMP4 C-allele. The differences of the secondary structures of BMP2 and BMP4 SNPs are indicated by arrows.
DISCUSSION
The phenotype of dental agenesis is complicated and displays distinct patterns in the permanent dentition. Familial tooth agenesis can occur either as an isolated anomaly (non-syndromic hypodontia) involving one, several, or many teeth or as part of a systemic condition or syndrome (syndromic hypodontia). Aside from tooth agenesis, anomalies such as reduced tooth dimension and morphologic abnormalities can also occur. In this study, neither family displayed abnormalities in other organs. Therefore, their phenotypes were classified as non-syndromic oligodontia.
The number and positions of missing teeth differed between the families. In family 1, the missing teeth of the affected subjects were identified mainly as incisors and premolars, whereas affected members of family 2 exhibited missing molars as well as missing incisors and premolars (see Figure 2 and Table 2). Additionally, subject II:2 of family 2 had a peg-shaped upper lateral incisor, and the contralateral incisor was absent. According to the literature, the peg shape of the upper lateral incisor is usually accompanied by absence of the contralateral incisor. Therefore, we concluded that a peg-shaped lateral incisor and the absence of lateral incisors are different patterns of the same genetic trait (Nieminen, 2009).
Analysis of the phenotypic characteristics of the affected members revealed several traits in the congenitally missing teeth: 1) The maxillary lateral incisors were the most frequently teeth involved in oligodontia or hypodontia, followed by the maxillary second premolar and mandibular central incisor; on the contrary, the canine was the least frequently absent. 2) The number of missing teeth on the left and right arches was similar. 3) No significant difference occurred between the number of missing teeth in the upper and lower dentition. These findings are not completely in accordance with observations from other reports, especially concerning the positions of the most frequently missing teeth, which may be caused by different gene defects (Arte et al., 2001).
At present, tooth agenesis is known to be associated with several gene variations, including MSX1, PAX9, AXIN2, and EDA (Liu et al., 2000; Stockton et al., 2000; van den Boogaard et al., 2000; Lammi et al., 2003; Kapadia et al., 2007; Han et al., 2008). However, many oligodontia families reported in the literature could not be identified as having any mutations in these genes (Scarel et al., 2000; Frazier-Bowers et al., 2002; Gerits et al., 2006). This finding suggests that oligodontia may be linked to other gene defects. BMP2 or BMP4 mutant mice display abnormal tooth formation and root development (Gluhak-Heinrich et al., 2010; Feng et al., 2011). In this study, the BMP2 and BMP4 coding regions and surrounding introns were analyzed in two Mexican oligodontia families.
Three alterations of the BMP2 and BMP4 genes were identified among the family members: BMP2 c261A>G, pS87S; c570A>T, pR190S, and BMP4 c455T>C, pV152A. BMP2 pS87S is a synonymous polymorphism, whereas BMP2 pR190S and BMP4 pV152A are missense polymorphisms. Among the six oligodontia patients examined, 67% carried either the GG or AG genotype of BMP2 c261A>G. Using bioinformatics analysis, we found that the BMP2 c261A>G polymorphism exhibited different BMP2 mRNA structures, and the G allele required more energy for mRNA secondary structure stabilization than the A allele did. Recently,Wang et al. (2008) have compared patients with ossification of the posterior longitudinal ligament (OPLL) and control subjects in a Chinese population and found that the BMP2 c261A>G polymorphism is associated with more extensive OPLL (Wang et al., 2008). The BMP2 c261A>G SNP has been implicated in the maintenance of bone mineral density and is associated with an increased risk of knee osteoarthritis in the Caucasian population (Valdes et al., 2006). However, extended analyses with a large sample size should be carried out with oligodontia patients and normal subjects for further verification of this association between BMP2 c261A>G and oligodontia.
A BMP2 c570A>T SNP with an amino acid-altering pR190S was also found, and the amino acid character was changed. Arginine is a basic amino acid, whereas serine is a hydrophilic amino acid. Although we are uncertain whether the change caused by the polymorphism is relevant to oligodontia, we found that the BMP2 c570A>T polymorphism accounted for 66% of the affected members (AT or TT allele) in the two Mexican oligodontia families. Valdes et al. (2004) have reported that the BMP2 c570A>T polymorphism is a risk factor of osteoarthritis in Caucasian women. However, further study has shown that BMP2 c570A>T SNP is not associated with osteoarthritis (Valdes et al., 2006).
In this study, we identified only one polymorphism at exon 4 of the BMP4 gene in the two Mexican oligodontia families. The results showed that at c455T>C, pV152A, the amino acid valine was substituted by alanine. The replacement did not change the secondary structures of the protein, but different mRNA secondary structures occurred in patients with the SNP. The C allele of c455T>C required more energy for mRNA secondary formation than the T allele. This result agrees with previous observations (Capasso et al., 2009). In a case-control study in a Southern Italian population,Capasso et al. (2009) compared melanoma patients with cancer-free controls and found that in patients with BMP4 SNPs, c455T>C is significantly associated with cutaneous melanoma. c455T>C (pV152A) does not change the three-dimensional structure of the BMP4 protein but instead affects mRNA stability and expression. BMP4 mRNA expression in C allele carriers was higher than that in T allele carriers. Recently, several reports have described an association between c455T>C and other disorders, including hip bone density in postmenopausal Caucasian women (Ramesh et al., 2005), bone mineral density in young Korean men and women (Choi et al., 2006), non-syndromic cleft lip with or without cleft palate in Chinese children (Lin et al., 2008), otosclerosis in Belgian-Dutch and French origin populations (Schrauwen et al., 2008), and OPLL in Chinese Han (Meng et al., 2010). In future research, a large sample size should be collected for statistical analysis of the association between the BMP4 c455T>C variant and oligodontia. We cannot exclude mutations in the intronic and regulatory regions of BMP2 and BMP4 that may be implicated in oligodontia in these families.
In conclusion, three polymorphisms of the BMP2 and BMP4 genes were observed in two isolated Mexican families with non-syndromic oligodontia. The association of these SNPs with oligodontia requires further study in a large population. Differences in the genotype/phenotype correction of the BMP2 and BMP4 genes associated with oligodontia among various races are open questions for further research.
ACKNOWLEDGMENTS
We thank Dr. Howard Dang for his critical reading of the manuscript. Research supported by grants from the National Institute of Dental and Craniofacial Research (#RO1 DE019892).
REFERENCES
- Aberg T, Wozney J, Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev. Dyn. 1997;210:383–396. doi: 10.1002/(SICI)1097-0177(199712)210:4<383::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Arte S, Nieminen P, Apajalahti S, Haavikko K, et al. Characteristics of incisor-premolar hypodontia in families. J. Dent. Res. 2001;80:1445–1450. doi: 10.1177/00220345010800051201. [DOI] [PubMed] [Google Scholar]
- Bei M, Kratochwil K, Maas RL. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development. 2000;127:4711–4718. doi: 10.1242/dev.127.21.4711. [DOI] [PubMed] [Google Scholar]
- Capasso M, Ayala F, Russo R, Avvisati RA, et al. A predicted functional single-nucleotide polymorphism of bone morphogenetic protein-4 gene affects mRNA expression and shows a significant association with cutaneous melanoma in Southern Italian population. J. Cancer Res. Clin. Oncol. 2009;135:1799–1807. doi: 10.1007/s00432-009-0628-y. [DOI] [PubMed] [Google Scholar]
- Chen S, Gluhak-Heinrich J, Martinez M, Li T, et al. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J. Biol. Chem. 2008;283:19359–19370. doi: 10.1074/jbc.M709492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Shin CS, Hong YC, Kang D. Single-nucleotide polymorphisms and haplotypes of bone morphogenetic protein genes and peripheral bone mineral density in young Korean men and women. Calcif. Tissue Int. 2006;78:203–211. doi: 10.1007/s00223-005-0139-z. [DOI] [PubMed] [Google Scholar]
- Feng J, Yang G, Yuan G, Gluhak-Heinrich J, et al. Abnormalities in the enamel in bmp2-deficient mice. Cells Tissues Organs. 2011;194:216–221. doi: 10.1159/000324644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier-Bowers SA, Scott MR, Cavender A, Mensah J, et al. Mutational analysis of families affected with molar oligodontia. Connect. Tissue Res. 2002;43:296–300. doi: 10.1080/03008200290000961. [DOI] [PubMed] [Google Scholar]
- Gabris K, Tarjan I, Csiki P, Konrad F, et al. Prevalence of congenital hypodontia in the permanent dentition and its treatment. Fogorv. Sz. 2001;94:137–140. [PubMed] [Google Scholar]
- Gerits A, Nieminen P, De MS, Carels C. Exclusion of coding region mutations in MSX1, PAX9 and AXIN2 in eight patients with severe oligodontia phenotype. Orthod. Craniofac. Res. 2006;9:129–136. doi: 10.1111/j.1601-6343.2006.00367.x. [DOI] [PubMed] [Google Scholar]
- Gluhak-Heinrich J, Guo D, Yang W, Harris MA, et al. New roles and mechanism of action of BMP4 in postnatal tooth cytodifferentiation. Bone. 2010;46:1533–1545. doi: 10.1016/j.bone.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Gong Y, Wu H, Zhang X, et al. Novel EDA mutation resulting in X-linked non-syndromic hypodontia and the pattern of EDA-associated isolated tooth agenesis. Eur. J. Med. Genet. 2008;51:536–546. doi: 10.1016/j.ejmg.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Kapadia H, Mues G, D’Souza R. Genes affecting tooth morphogenesis. Orthod. Craniofac. Res. 2007;10:237–244. doi: 10.1111/j.1601-6343.2007.00407.x. [DOI] [PubMed] [Google Scholar]
- Kong H, Wang Y, Patel M, Mues G, et al. Regulation of bmp4 expression in odontogenic mesenchyme: from simple to complex. Cells Tissues Organs. 2011;194:156–160. doi: 10.1159/000324747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammi L, Halonen K, Pirinen S, Thesleff I, et al. A missense mutation in PAX9 in a family with distinct phenotype of oligodontia. Eur. J. Hum. Genet. 2003;11:866–871. doi: 10.1038/sj.ejhg.5201060. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Chen YJ, Huang YL, Tang GP, et al. Association of bone morphogenetic protein 4 gene polymorphisms with nonsyndromic cleft lip with or without cleft palate in Chinese children. DNA Cell Biol. 2008;27:601–605. doi: 10.1089/dna.2008.0777. [DOI] [PubMed] [Google Scholar]
- Liu W, Dong X, Mai M, Seelan RS, et al. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat. Genet. 2000;26:146–147. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- Meng XL, Wang H, Yang H, Hai Y, et al. T allele at site 6007 of bone morphogenetic protein-4 gene increases genetic susceptibility to ossification of the posterior longitudinal ligament in male Chinese Han population. Chin. Med. J. 2010;123:2537–2542. [PubMed] [Google Scholar]
- Nieminen P. Genetic basis of tooth agenesis. J. Exp. Zool. B Mol. Dev. Evol. 2009;312B:320–342. doi: 10.1002/jez.b.21277. [DOI] [PubMed] [Google Scholar]
- Nieminen P, Pekkanen M, Aberg T, Thesleff I. A graphical www-database on gene expression in tooth. Eur. J. Oral Sci. 1998;106:7–11. doi: 10.1111/j.1600-0722.1998.tb02146.x. [DOI] [PubMed] [Google Scholar]
- Noor A, Windpassinger C, Vitcu I, Orlic M, et al. Oligodontia is caused by mutation in LTBP3, the gene encoding latent TGF-beta binding protein 3. Am. J. Hum. Genet. 2009;84:519–523. doi: 10.1016/j.ajhg.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh BL, Wilson SG, Dick IM, Islam FM, et al. Bone mass effects of a BMP4 gene polymorphism in postmenopausal women. Bone. 2005;36:555–561. doi: 10.1016/j.bone.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Scarel RM, Trevilatto PC, Di Hipólito O, Jr, Camargo LE, et al. Absence of mutations in the homeodomain of the MSX1 gene in patients with hypodontia. Am. J. Med. Genet. 2000;92:346–349. doi: 10.1002/1096-8628(20000619)92:5<346::aid-ajmg10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Schalk-van der Weide Y, Steen WH, Bosman F. Distribution of missing teeth and tooth morphology in patients with oligodontia. ASDC J. Dent. Child. 1992;59:133–140. [PubMed] [Google Scholar]
- Schrauwen I, Thys M, Vanderstraeten K, Fransen E, et al. Association of bone morphogenetic proteins with otosclerosis. J. Bone Miner. Res. 2008;23:507–516. doi: 10.1359/JBMR.071112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockton DW, Das P, Goldenberg M, D’Souza RN, et al. Mutation of PAX9 is associated with oligodontia. Nat. Genet. 2000;24:18–19. doi: 10.1038/71634. [DOI] [PubMed] [Google Scholar]
- Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J. Cell Sci. 2003;116:1647–1648. doi: 10.1242/jcs.00410. [DOI] [PubMed] [Google Scholar]
- Thomadakis G, Ramoshebi LN, Crooks J, Rueger DC, et al. Immunolocalization of bone morphogenetic protein-2 and -3 and osteogenic protein-1 during murine tooth root morphogenesis and in other craniofacial structures. Eur. J. Oral Sci. 1999;107:368–377. doi: 10.1046/j.0909-8836.1999.eos107508.x. [DOI] [PubMed] [Google Scholar]
- Valdes AM, Hart DJ, Jones KA, Surdulescu G, et al. Association study of candidate genes for the prevalence and progression of knee osteoarthritis. Arthritis Rheum. 2004;50:2497–2507. doi: 10.1002/art.20443. [DOI] [PubMed] [Google Scholar]
- Valdes AM, Van Oene M, Hart DJ, Surdulescu GL, et al. Reproducible genetic associations between candidate genes and clinical knee osteoarthritis in men and women. Arthritis Rheum. 2006;54:533–539. doi: 10.1002/art.21621. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu D, Yang Z, Tian B, et al. Association of bone morphogenetic protein-2 gene polymorphisms with susceptibility to ossification of the posterior longitudinal ligament of the spine and its severity in Chinese patients. Eur. Spine J. 2008;17:956–964. doi: 10.1007/s00586-008-0651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 1996;122:2977–2986. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]