Abstract
RACK1 is not a G protein but closely resembles the heterotrimeric Gβ-subunit. RACK1 serves as a scaffold, linking protein kinase C to its substrates. We demonstrate that RACK1 physiologically binds inositol 1,4,5-trisphosphate receptors and regulates Ca2+ release by enhancing inositol 1,4,5-trisphosphate receptor binding affinity for inositol 1,4,5-trisphosphate. Overexpression of RACK1 or depletion of RACK1 by interference RNA markedly augments or diminishes Ca2+ release, respectively, without affecting Ca2+ entry. These findings establish RACK1 as a physiologic mediator of agonist-induced Ca2+ release.
RACK1 was originally discovered, by protein-binding assays, to bind phosphorylated, active forms of protein kinase C (PKC) (1). RACK1 acts as a scaffold protein, bringing activated PKC into contact with its various substrates (2–4). RACK1 is not a G protein but resembles the heterotrimeric Gβ-subunit (Gβ) with ≈50% amino acid sequence homology. Moreover, like Gβ, RACK1 possesses seven predicted WD40 repeats. RACK1 has a molecular mass of ≈36 kDa, in contrast to Gβ, whose molecular mass is ≈38 kDa with the decreased size reflecting a truncated N terminus. RACK1 binds to Gβ (5) and occurs in a complex with the Gα- and Gγ-subunits that is promoted and stabilized by the Gα-subunit.
Gβγ binds directly to potassium channels, regulating their response to the activation of G protein-coupled receptors (6). Because of the similarity in structure of RACK1 and Gβ, one might anticipate a role for RACK1 in regulating ion channels. In the present study, we demonstrate that RACK1 binds to inositol 1,4,5 trisphosphate (IP3) receptors (IP3Rs) and is critical for normal IP3-mediated Ca2+ release.
Experimental Procedures
Culture of Cells. Rat PC12 cells (passage numbers 6–15), human embryonic kidney (HEK)293 cells, and rat aortic smooth muscle A7r5 cells (passage numbers 10–25) were cultured as described (7, 8). Expression of RACK1 constructs and small interfering RNA (siRNA) was performed as described (8) in HEK293, PC12, and A7r5 cells by using Lipofectamine 2000 (GIBCO).
Ca2+ Imaging. Ca2+ measurements were as described (8). Fura-2/acetoxymethyl ester loading was for 1 h at 20°C for PC12 cells, 25 min at 20°C for HEK293 cells, and 30 min at 20°C for A7r5 cells. Transfected enhanced yellow fluorescent protein (YFP) served as the transfection marker and was detected at excitation wavelength 485 nm. Resting Ca2+ levels in cell lines were similar, 100–200 nM, and cells with higher basal levels were excluded from data collection because these cells tend to have constitutive Ca2+ entry. All measurements shown are representative of a minimum of three and, in most cases, a larger number of independent experiments.
IP3R and RACK1 Mutagenesis. Δ90–110, Δ580–600 IP3R. Amino acids (aa) 90–110 were deleted from the IP3R by using single overlap extension. This process required PCR of the N terminus of IP3R aa 1–90 and aa 110–421 (an internal KpnI site) separately, then combining the two templates into a single PCR by using a 5′ primer at the beginning of the aa 1–90 KpnI sequence, which has a 6× CAG repeat engineered into the PCR primer and a 3′ primer on the aa 110–421 KpnI sequence. The overlap region was engineered onto the reverse primer (aa 1–90) and forward primer (aa 110–421). The same strategy was used to design the Δ580–600 construct by using the same aa 1–580 KpnI 5′ primer and an aa 600-1384 ApaI 3′ primer, with the overlap region on the reverse primer (aa 1–580) and the forward primer (aa 600–1384), cloned into pCDNA 3.1 by using partial restriction digests.
Δ103–178 RACK1 mutagenesis. aa 103–178 were deleted from GST-RACK1 constructs by using single overlap extension as previously described, with EcoRI and XhoI as restriction sites in p-GEX 4T1.
GST Pull-Down Assays. Cell lysis buffer (150 mM NaCl/50 mM Tris, pH 7.8/1% Triton X-100/1 mM EDTA) was added to 100 μg of HEK or PC12 whole-cell lysates to bring samples to a total volume of 500 μl. Samples were cleared of insoluble debris by centrifugation at 10,000 × g. GST beads bound to purified RACK1 were added and incubated on a rotator for 1 h at 4°C. The GST-Sepharose beads were washed three times with lysis buffer and quenched with 20 μl of SDS sample buffer. Coprecipitates were resolved by SDS/PAGE and analyzed by Western blot analysis.
Calcium Release Measurements. Calcium release through recombinant type I (SII+) IP3R or mutant IP3R was measured as described (9).
IP3 Measurements. IP3 measurements were performed by using [3H]IP3 and rat brain cerebellar membranes as described (10).
Yeast Two-Hybrid. Experiments were performed by using the Matchmaker 3 yeast two-hybrid system (Clontech), with all IP3R fragments cloned into the pGBKT7-binding domain vector by using EcoRI/SalI restriction sites and all RACK1 fragments cloned into the pGADT7 vector by using EcoRI/XhoI restriction sites.
Antibodies and Reagents. Plasmids were obtained from the following sources: Matchmaker 3 yeast two-hybrid kit, enhanced YFP vector, MYC-tagged vector cDNA, and GF-109203X were from Clontech; InsP3R WT type 1 was from Suresh Joseph (Thomas Jefferson University, Philadelphia); pGEX-4T vector was from Amersham Pharmacia; anti-His antibody, carbachol, bradykinin, and vasopressin were from Sigma; Fura-2/ acetoxymethyl ester was from Molecular Probes; polyclonal anti-IP3R was generated in our laboratory (11); and polyclonal RACK1 antibody was from Santa Cruz Biotechnology.
Results
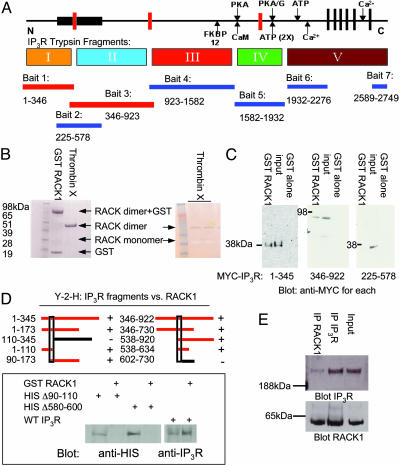
RACK1 Binds IP3R. In yeast two-hybrid analysis, we examined the binding of seven distinct fragments of IP3R as described (11). One of the binding partners observed is RACK1 (Fig. 1A). RACK1 binds to two IP3R bait fragments representing aa 1–346 and aa 346–923, respectively. Surprisingly, an IP3R bait representing the IP3-binding core (aa 225–578) does not bind RACK1. One possible explanation for the failure of RACK1 to bind the aa 225–578 fragment is that it binds to two distinct sites on IP3R contained in the aa 1–346 and aa 346–923 fragments but not fully evident in the aa 225–578 fragment.
Fig. 1.
RACK1 binds to two distinct sites in the N terminus of IP3R. (A) Schematic depiction of the functional domains of the IP3R, including the ligand-binding (bait 2), modulatory and transmembrane regions, and trypsin digest (I–V) domains. The seven regions used as bait in the yeast two-hybrid screen also are indicated below the protein and the corresponding amino acids (rat type 1 sequence). When RACK1 was screened against all IP3R baits, only baits 1 and 3 (N terminus of IP3R containing the ligand-binding core) supported growth on selective media. (B Left) In vitro Coomassie stain of GST-fusion protein binding of purified RACK1 to glutathione Sepharose and free RACK1 that has been cleaved from GST by using biotinylated thrombin followed by clearing with streptavidin beads. (Right) Silver stain of free RACK1 running as a monomer (negative stain) and a dimer (positive stain). (C) GST pull-down assays with HEK293 cells expressing MYC-tagged IP3R fragments, passed over GST-RACK1 columns. Western blot analysis with anti-MYC antibody demonstrates IP3R fragments that can bind to purified RACK1. (D Upper) Yeast two-hybrid analysis (Y-2-H) for RACK1-binding sites. Red bars depict yeast two-hybrid analysis baits capable of interacting with RACK1. (Lower) Full-length HIS-tagged WT and mutant IP3R were expressed in COS7 cells and passed over GST-RACK1 columns. Western blot analysis with anti-HIS antibody demonstrates IP3R mutants are incapable of binding to RACK1. (E) Coimmunoprecipitation experiments from PC12 cell lysates of endogenous RACK1 and type 1 IP3R, visualized by Western blot analysis.
To monitor direct binding of IP3R and RACK1, we prepared a GST fusion form of RACK1 (Fig. 1B). GST-RACK1 and free RACK1, from which GST has been cleaved, migrate both as dimers and monomers in SDS/PAGE gels, even under denaturing conditions. We conducted binding experiments by using cell lysates from HEK293 cells expressing MYC-IP3R fragments passed over GST-RACK1 columns (Fig. 1C). RACK1 binds to IP3R fragments aa 1–345 and aa 346–922 but not to aa 225–578, confirming the results of the yeast two-hybrid analysis experiments. Thus, RACK1 appears to bind to two distinct sites in the N-terminal area of the IP3R.
We conducted additional yeast two-hybrid analysis to narrow down sites on IP3R that bind RACK1 (Fig. 1D). Areas required for binding to RACK1 are in aa 90–110 and aa 580–600 of IP3R. To confirm the importance of these areas, we deleted these regions and examined direct binding of the mutated IP3R to RACK1 (Fig. 1D). Deletion of aa 90–110 or aa 580–600 in full-length His-tagged IP3R abolished binding to RACK1. Thus, although RACK1 interacts with two sites on IP3R, disruption of either of these sites abolishes binding.
To ascertain whether IP3R and RACK1 interact in vivo, we conducted immunoprecipitation of the native proteins in PC12 cells (Fig. 1E). IP3R robustly immunoprecipitates RACK1, with the reciprocal immunoprecipitation having similar efficacy.
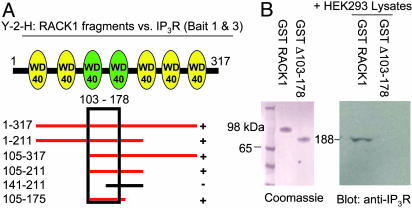
We next examined portions of RACK1 that are critical for binding to IP3R. Yeast two-hybrid analysis reveals that the third and forth WD40 repeats of RACK1 are critical for binding (Fig. 2A). Smaller fragments coding for the third repeat only were unable to bind to IP3R in yeast two-hybrid analysis (data not shown). We then examined the direct binding of the IP3R to full-length RACK1 or a mutant RACK1 containing deletions in the third and fourth WD40 repeats (Δ103–178) (Fig. 2B). Whereas full-length RACK1 binds to IP3R, binding is abolished by deletion of the two WD40 repeats.
Fig. 2.
RACK1 binds to IP3R within WD40 repeats 3 and 4. (A) Cartoon of RACK1 depicting the seven WD40 repeats within the protein and the yeast two-hybrid analysis (Y-2-H) mapping of RACK1 to baits 1–345 and 346–922 of the IP3R. Red bars indicate positive binding to both IP3R fragments. (B Left) Coomassie stain of GST-purified RACK1 and mutant RACK1 Δ103–178. (Right) HEK293 cell lysates were passed over WT and Δ103–178 GST-RACK1 columns. Western analysis with anti-IP3R antibody demonstrates IP3R binding only to full-length RACK1.
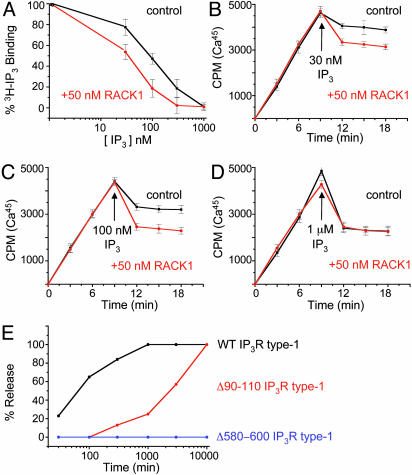
RACK1 Regulates Receptor Binding of IP3 and Ca2+ Release. To explore an influence of RACK1 on IP3R function, we first evaluated the influences of 50 nM RACK1 on [3H]IP3 binding to IP3R in rat brain membranes (Fig. 3A). RACK1 increases the potency of unlabeled IP3 in competing for [3H]IP3 binding ≈2-fold. We next evaluated the influence of 50 nM RACK1 on IP3-induced Ca2+ release from microsomal membrane preparations of COS7 cells (11) (Fig. 3 B–D). RACK1 approximately doubles Ca2+ release by 30 and 100 nM IP3. At 1 μM, IP3 produces a maximal release of Ca2+, and RACK1 does not provide any further augmentation. We do not observe activation of IP3R by RACK1 alone, even with high concentration of RACK1 (10 μM) (data not shown). These observations suggest that RACK1 acts by enhancing the affinity of IP3 for IP3R rather than by altering the channel itself, which would lead to a greater maximal release.
Fig. 3.
RACK1 modulates IP3R binding to IP3R. (A) Influence of unlabeled IP3 on binding of [3H]IP3 to rat cerebellar membranes in the absence or presence of 50 nM free RACK1. (B–D) Ca45 uptake into IP3R-containing microsomes in the presence of oxalate. IP3 at varying concentrations (arrow) was added in the presence (red lines) or absence (black lines) of 50 nM RACK1, and Ca2+ release was monitored. (E) IP3 concentration response in microsomes containing WT IP3R (black line), Δ90–110 IP3R (red line), or Δ580–600 (blue line). Ca2+ concentration was held at 250 nM.
If RACK1 is critical for IP3-induced Ca2+ release, then deletions within IP3R that bind RACK1 would be expected to alter release. Accordingly, we transfected COS7 cells with WT IP3R or with truncation mutant IP3R, missing either aa 90–110 (Δ90–110) or aa 580–600 (Δ580–600). These mutants represent the domains that are critical for RACK1 binding (see Fig. 1D). Compared with WT IP3R (Fig. 3E, black line), IP3R Δ90–110 displays a 100-fold lesser sensitivity to IP3-induced Ca2+ release (Fig. 3E, red line) and IP3R Δ580–600 fails to release Ca2+ in response to any IP3 concentration (Fig. 3E, blue line). Although these deletions may influence other functions of IP3R besides RACK1 binding, they are consistent with an important role for RACK1 in regulating IP3R Ca2+ release.
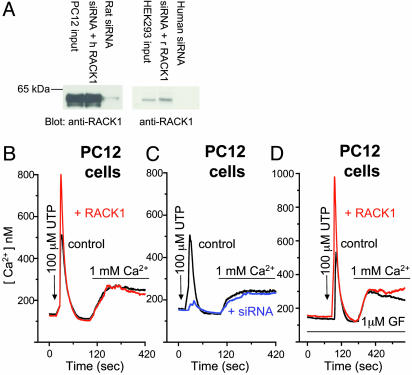
IP3R Ca2+ Release in Intact Cells Depends on RACK1. We used overexpression constructs and siRNA duplexes specific to RACK1 to ascertain whether RACK1 is required for IP3R-mediated Ca2+ release in intact cells. Because siRNA procedures can have nonspecific effects (12), rescue experiments using the same protein from a different species that would not be affected by siRNA are important to demonstrate direct effects of protein depletion. siRNA treatment depletes RACK1 from rat PC12 cells, which is rescued by human RACK1 expression (Fig. 4A Left). siRNA also depletes RACK1 from human HEK293 cells, and protein expression is restored with rat RACK1 (Fig. 4A Right).
Fig. 4.
RACK1 modifies purinergic influences on IP3R function. (A) Western blots of rat PC12 cell lysates and human HEK cell lysates, respectively, transfected for 56 h with YFP alone (input), YFP plus siRNA RACK1 duplex (specific to cell species), or YFP plus siRNA duplex plus RACK1 (opposite to siRNA species). Gels were transferred and blotted with a polyclonal antibody specific for RACK1. Free Ca2+ measurements were made in rat PC12 cells transfected for 60 h with either YFP alone (black trace), YFP plus RACK1 (red trace), or YFP plus siRNA (blue trace). (B and C) Ca2+ pools were released in cells by UTP (100μM; arrow) in nominally Ca2+-free medium followed by replacement with UTP and normal 1 mM Ca2+ medium (bar). (D) As in B and C except with pretreatment with the PKC inhibitor GF-109203X (GF, 1 μM; first bar). Traces in B–D are averaged from multiple (30–50) cells.
We examined IP3R function by Ca2+ imaging in PC12 cells (Fig. 4B). Compared with YFP-expressing control cells (Fig. 4B, black trace) overexpression of RACK1 (Fig. 4B, red trace) almost doubles Ca2+ release in response to purinergic receptor activation by UTP, although exerting no effect on subsequent agonist-induced Ca2+ entry. These findings fit with previous evidence that IP3R-mediated Ca2+ release and Ca2+ entry are distinct processes (13). Depletion of RACK1 by siRNA profoundly reduces IP3R Ca2+ release in response to UTP with no effect on subsequent Ca2+ entry (Fig. 4C, blue trace). Ca2+-release kinetics are unaltered by RACK1, with the time to activation and deactivation being essentially the same with or without RACK1.
RACK1 is best known for its interactions with PKC (1, 4). Accordingly, we examined the influence of the pan-specific PKC inhibitor GF-109203X on UTP-induced Ca2+ responses (Fig. 4D). GF-109203X treatment fails to alter Ca2+ release or entry in control (Fig. 4D, black trace) or RACK1-expressing (Fig. 4D, red trace) cells, indicating a PKC-independent effect of RACK1. GF-109203X decreases basal and RACK1-stimulated IP3RCa2+ release in response to bradykinin but increases responses to muscarinic cholinergic and vasopressin activation (data not shown).
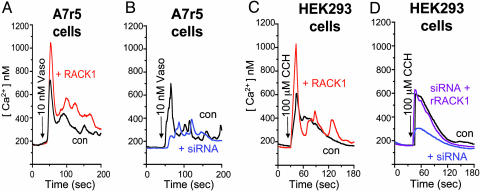
To ascertain the generality of our findings with purinergic receptors in PC12 cells, we examined vasopressin and muscarinic cholinergic responses in A7r5 cells, a rat line of aortic smooth muscle cells, and HEK293 cells, respectively. Compared with YFP-expressing control A7r5 cells (Fig. 5 A and B, black traces), RACK1 overexpression (Fig. 5A, red trace) leads to a doubling of vasopressin-elicited Ca2+ release, which is profoundly reduced after siRNA-induced deletion of RACK1 (Fig. 5B, blue trace). Similarly, muscarinic cholinergic stimulation of Ca2+ release is doubled by overexpression of RACK1 (Fig. 5C, red trace) in HEK293 cells, whereas siRNA treatment greatly reduces the effect of cholinergic activation (Fig. 5D, blue trace). Transfection of rat RACK1 into HEK293 cells restores Ca2+ release (Fig. 5D, purple trace). In all of these experiments, we observe no effects of RACK1 overexpression or its deletion on Ca2+ entry (data not shown). This independent regulation of Ca2+ entry and release accords with other studies (8, 13).
Fig. 5.
RACK1 modifies IP3R function in response to muscarinic cholinergic and vasopressin stimulation. Free Ca2+ measurements were made in rat A7r5 and human A7r5 cells transfected for 60 h with YFP alone (black traces), YFP plus RACK1 (red traces), YFP plus siRNA (specific to cell species; blue traces), or YFP plus human siRNA plus rat RACK1 (purple traces). Ca2+ pools were released by vasopressin (Vaso, 10 nM; arrow) (A and B) in nominally Ca2+-free medium and carbachol (CCH, 100 μM; arrow) (C and D) in nominally Ca2+-free medium. All traces are averaged from multiple cells.
Discussion
In this study, we demonstrate that RACK1-binding to the IP3R is associated with enhanced IP3-binding affinity and augmented Ca2+ release. RACK1 binds to two distinct sites in the N terminus of IP3R, adjacent to the IP3-binding core. Interestingly, even under denaturing conditions we observe RACK1 as both a dimer and as a monomer. This finding suggests that two molecules of a RACK1 dimer bind to the two separate sites. Because IP3R is a tetramer, the two portions of the RACK1 dimer might bind to separate IP3R monomers.
The affinity of IP3R for IP3 is enhanced ≈2-fold by 50 nM RACK1, well within physiological ranges. Most striking are the dramatic in vivo influences of RACK1 on IP3R-mediated Ca2+ release. Overexpression of RACK1 markedly increases Ca2+ release. Depletion of RACK1 by siRNA profoundly reduces and in some instances virtually abolishes Ca2+ release. Thus, RACK1 is required for normal agonist-induced Ca2+ release. Regulation of IP3 R-mediated Ca2+ release by RACK1 is a general phenomenon evident in cells from diverse species and organs, as well as with multiple receptor systems: purinergic, muscarinic cholinergic, and vasopressin receptors. Interestingly, RACK1 overexpression does not alter bradykinin-induced Ca2+ release in PC12 cells (data not shown), suggesting that a distinct protein related to RACK1 mediates effects of bradykinin and that RACK1 does not act by elevating IP3 levels.
RACK1 closely resembles Gβ in amino acid sequence and its possession of seven WD40 repeats (1). Muallem and colleagues (14) observed coprecipitation of IP3R with Gβ and enhancement of IP3R-induced Ca2+ release after Gβ administration. We do not observe gating of IP3R by RACK1 alone. Because of the similarities between RACK1 and Gβ, we would expect that the binding sites within Gβ for IP3R (and vice versa) will resemble such domains for RACK1 and the IP3R. IP3R-mediated Ca2+ release is greatly reduced after siRNA-induced deletion of RACK1. Conceivably, a Gβ–IP3R interaction mediates the remaining Ca2+ release. We hypothesize that Gβ may be responsible for bradykinin-elicited Ca2+ release, which is not affected by RACK1.
RACK1 was initially identified as a scaffold for PKC, linking the enzyme to many of its substrates. IP3R is a substrate for PKC (15, 16). We failed to find any effect of the PKC inhibitor GF-109203X on basal or RACK1-stimulated Ca2+ release in response to purinergic receptor activation. However, GF-109203X did decrease bradykinin activation and augmented muscarinic cholinergic and vasopressin responses. Because these effects of GF-109203X were similar in basal and RACK1-activated conditions, they may reflect IP3R phosphorylation by PKC. However, it is difficult to interpret the marked variations in GF-109203X effects with various agonists.
RACK1 has been implicated in neuronal physiology and neurodegenerative diseases. Decreased levels of RACK1 are associated with deficient PKC signaling in Alzheimer's disease (17, 18). Conversely, increased association of PKC with RACK1 occurs in bipolar affective disorder (19). RACK1 has been linked to synaptic plasticity because it binds to N-methyl-d-aspartate receptors, inhibiting their activation (20, 21). Fear-induced conflict behavior also is influenced by RACK1–acetylcholinesterase interactions (2). All of the aforementioned processes involve, in part, changes in intracellular Ca2+ levels. Therefore, by regulating IP3R, RACK1 may control Ca2+ disposition in seemingly disparate processes.
Acknowledgments
We thank Dr. Robert E. Rothe for useful discussion. Research was supported by U.S. Public Health Service Grants MH-18501 and DA-000266 and Research Scientist Award DA-00074 (to S.H.S.) and National Research Service Award NH65090 (to R.L.P.).
Abbreviations: IP3, inositol 1,4,5-trisphosphate; IP3R, IP3 receptor; Gβ, Gβ-subunit; siRNA, small interfering RNA; YFP, yellow fluorescent protein; aa, amino acids; HEK, human embryonic kidney.
References
- 1.Ron, D., Chen, C. H., Caldwell, J., Jamieson, L., Orr, E. & Mochly-Rosen, D. (1994) Proc. Natl. Acad. Sci. USA 91, 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birikh, K. R., Sklan, E. H., Shoham, S. & Soreq, H. (2003) Proc. Natl. Acad. Sci. USA 100, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, B. Y., Harte, R. A. & Cartwright, C. A. (2002) Oncogene 21, 7619–7629. [DOI] [PubMed] [Google Scholar]
- 4.Ceci, M., Gaviraghi, C., Gorrini, C., Sala, L. A., Offenhauser, N., Marchisio, P. C. & Biffo, S. (2003) Nature 426, 579–584. [DOI] [PubMed] [Google Scholar]
- 5.Dell, E. J., Connor, J., Chen, S., Stebbins, E. G., Skiba, N. P., Mochly-Rosen, D. & Hamm, H. E. (2002) J. Biol. Chem. 277, 49888–49895. [DOI] [PubMed] [Google Scholar]
- 6.Wickman, K. D., Iniguez-Lluhl, J. A., Davenport, P. A., Taussig, R., Krapivinsky, G. B., Linder, M. E., Gilman, A. G. & Clapham, D. E. (1994) Nature 368, 255–257. [DOI] [PubMed] [Google Scholar]
- 7.Patterson, R. L., van Rossum, D. B. & Gill, D. L. (1999) Cell 98, 487–499. [DOI] [PubMed] [Google Scholar]
- 8.Patterson, R. L., van Rossum, D. B., Ford, D. L., Hurt, K. J., Bae, S. S., Suh, P. G., Kurosaki, T., Snyder, S. H. & Gill, D. L. (2002) Cell 111, 529–541. [DOI] [PubMed] [Google Scholar]
- 9.Boehning, D. & Joseph, S. K. (2000) J. Biol. Chem. 275, 21492–21499. [DOI] [PubMed] [Google Scholar]
- 10.Bredt, D. S., Mourey, R. J. & Snyder, S. H. (1989) Biochem. Biophys. Res. Commun. 159, 976–982. [DOI] [PubMed] [Google Scholar]
- 11.Boehning, D., Patterson, R. L., Sedaghat, L., Glebova, N. O., Kurosaki, T. & Snyder, S. H. (2003) Nat. Cell Biol. 5, 1051–1061. [DOI] [PubMed] [Google Scholar]
- 12.Check, E. (2003) Nature 425, 10–12. [DOI] [PubMed] [Google Scholar]
- 13.van Rossum, D. B., Patterson, R. L., Kiselyov, K., Boehning, D., Barrow, R. K., Gill, D. L. & Snyder, S. H. (2004) Proc. Natl. Acad. Sci. USA 101, 2323–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng, W., Mak, D. O., Li, Q., Shin, D. M., Foskett, J. K. & Muallem, S. (2003) Curr. Biol. 13, 872–876. [DOI] [PubMed] [Google Scholar]
- 15.Ferris, C. D., Huganir, R. L., Bredt, D. S., Cameron, A. M. & Snyder, S. H. (1991) Proc. Natl. Acad. Sci. USA 88, 2232–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matter, N., Ritz, M. F., Freyermuth, S., Rogue, P. & Malviya, A. N. (1993) J. Biol. Chem. 268, 732–736. [PubMed] [Google Scholar]
- 17.Pascale, A., Fortino, I., Govoni, S., Trabucchi, M., Wetsel, W. C. & Battaini, F. (1996) J. Neurochem. 67, 2471–2477. [DOI] [PubMed] [Google Scholar]
- 18.Battaini, F., Pascale, A., Paoletti, R. & Govoni, S. (1997) Trends Neurosci. 20, 410–415. [DOI] [PubMed] [Google Scholar]
- 19.Wang, H. & Friedman, E. (2001) Biol. Psychiatry 50, 364–370. [DOI] [PubMed] [Google Scholar]
- 20.Yaka, R., Thornton, C., Vagts, A. J., Phamluong, K., Bonci, A. & Ron, D. (2002) Proc. Natl. Acad. Sci. USA 99, 5710–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaka, R., He, D. Y., Phamluong, K. & Ron, D. (2003) J. Biol. Chem. 278, 9630–9638. [DOI] [PubMed] [Google Scholar]