Abstract
For the first time, we have investigated the bacterial toxicity or compatibility properties of Pt nanoparticles (NPs) with different sizes (P1, P2, P3, P4 and P5). The bacterio-toxic or compatible properties of these five different sized Pt NPs with the clinical pathogen, Pseudomonas aeruginosa were explored by many analytical methods such as the conventional plate count method, matrix assisted laser desorption/ionization mass spectrometry (MALDI-MS), fluorescence microscopy and fluorescence sensoring techniques. The results revealed that the 1–3 nm sized (P1 and P2) Pt NPs showed bacterio-toxic properties while the 4–21 nm (P3, P4 and P5) Pt NPs exhibited bacterio-compatible properties. This is the first study which reports the bacterial toxicity of Pt NPs. The information released from this study is significantly important to future clinical, medical, biological and biomedical applications of Pt NPs.
Metal nanoparticles (NPs) have been widely studied due to their large surface-to-volume ratios and quantum effects comparable to atoms and bulk materials. These features make them attractive in various applications where they are used as catalysts, owing to the large number of active atoms on their surfaces, corners and edges1,2,3, as well as their potential applications in photography, nonlinear optics, electronics, photonics, information storage, chemical and biological sensing, surface-enhanced Raman scattering (SERS), nanoscale communications, clinical diagnosis and cancer therapy4,5,6,7,8,9,10,11,12,13,14,15,16.
The size and shape effects of the NPs exhibit a direct relationship between the catalytic activity and particle morphologies. Among various metals, noble metal NPs are particularly interesting due to their close lying conduction and valence bands in which electrons move freely. The free electrons in metals can generate surface plasmon resonance (SPR) bands which change with particle size, shape and the corresponding medium. The fascinating color of noble metal NPs also depends on their size and shapes as well as the refractive index of the surrounding medium. The synthesis of metal NPs with well-defined sizes and shapes has been investigated but still remains a challenging task. Among the different noble metals studied, platinum NPs have attracted significant attention due to their distinctive ability in catalyzing partial oxidation17, hydrogenation18 and dehydrogenation19 of a variety of important molecules that are essential for many industrial applications. Therefore, the synthesis of the bare and stable platinum NPs are particularly important in different catalytic reactions involving platinum elements.
Recently, a number of chemical methods have been developed for the synthesis of platinum (Pt) NPs. The first study on shape controlled Pt NPs using a linear polymer as stabilizing agent was reported by Ahmadi et al. in 199620. They selectively synthesized cubic and tetrahedral Pt NPs by changing the molar concentration of the stabilizing agent of the Pt precursors. Recently, various other methods have been developed for the synthesis of size- and shape-controlled Pt NPs rods, wires, chains, quasi-sphericals, tetrahedrals, cubics and polypods21,22,23,24,25, etc. El-Sayed's group reported earlier that the shape effect of Pt NPs can change the reaction pathway to a great extent26,27. Colloidal Pt nanowires were synthesized by Fenske et al. using dodecylamine as ligands28. Previously, Jana et al. synthesized quasi-spherical Pt NPs at higher yields24. Mafune et al. developed a laser irradiation method for the synthesis of Pt NPs in an aqueous solution of sodium dodecyl sulfate (SDS)29. Kinge et al. synthesized Pt NPS using templates30. Herricks et al. studied the morphological change of Pt NPs with the addition of NaNO3 in a polyol process at 160°C31. Chen et al. modified the polyol process by introduction of trace amounts of iron species to the reaction mixture and generated branched multipods32. Recently, Lim et al. further modified the polyol process by adding iron species to produce multioctahedral Pt nanostructures33. Tsuji et al. studied the role of poly(vinylpyrrolidone) in the formation of branched Pt nanostructures in polyol process34. A few other methods have also been reported in literature for the synthesis of Pt NPs35,36,37. Most of the above conventional heating methods required either harsh reducing conditions, addition of separate seed particles, long reaction time, produce only one particle shape, or sometimes generate a mixture of multiple shaped particles with lower yields. There are also reports on the synthesis of polyhedral Pt NPs using poly (vinylpyrrolidone) (PVP) as capping agent in the presence of Pt seed particles38. Song et al. synthesized Pt nanowire networks using a soft template in the presence of sodium borohydride39. Kundu and Liang reported the photoinduced formation of shape selective Pt NPs40.
The synthesis of Pt NPs has been intensively reported. Among their studies, several reports focused on probing the interactions of Pt NPs with living cells. Asharani et al.41 investigated the uptake and bioactivity (e.g., cytotoxicity, genotoxicity and protein expression) of Pt NPs (5–8 nm in size) on human cells. They synthesized Pt NPs capped with polyvinyl alcohol and incubated them with human cells. Their uptake and biological properties were evaluated through metabolic activity, genome integrity, cell cycle and protein expression. They reported that the Pt NPs entered the cells through diffusion and localized inside the cytoplasm. Exposure to the Pt NP also increased DNA damage, accumulation of cells at the S-phase of the cell cycle and apoptosis. Protein-expression levels uncovered upregulation of p53, phosphorylated p53, p21 and downregulation of proliferating cell nuclear antigen following Pt NP treatment. Pro-caspase 3 and poly-ADP ribose polymerase and cyclin B levels were not altered in both the cell types after Pt NP exposure. This study recommended the development of Pt NP based anticancer agents by appropriate surface modifications to augment its innate anticancer activity.
The size dependant biotoxic/biocompatible properties of Pt NPs with respect to bacterial cells, till date remains unexplored. We venture to investigate the probability of the existence of bacterial toxicity or bacterial compatibility for Pt NPs. We have extended an existing method for synthesizing Pt NPs of varying sizes (P1, P2, P3, P4, P5) and investigating their bacterial toxicity/compatibility properties. The bacterio-toxic/compatible properties of the five different sized Pt NPs were carefully investigated using the plate count method, MALDI-MS, fluorescence microscopy and fluorescence sensoring techniques.
Results
Characterization of the Pt NPs
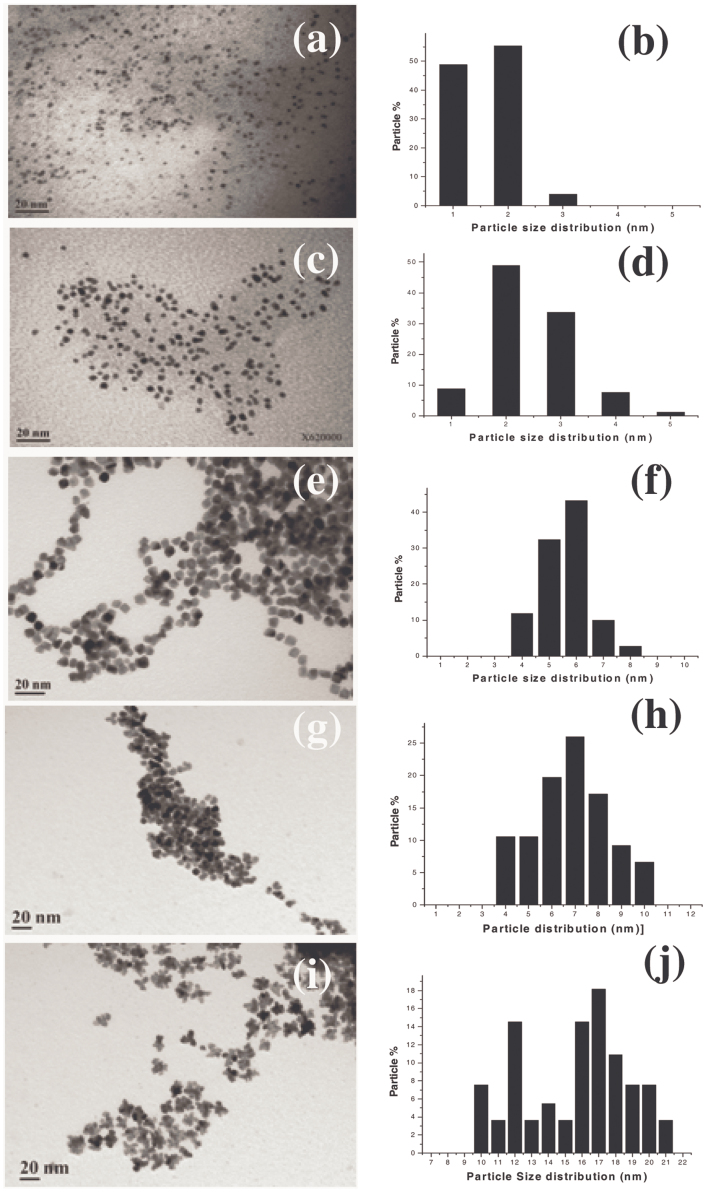
Five different sized and shaped Pt NPs were synthesized using an extension of a existing protocol reported by Koebel et al42. Supporting figure S1 gives the schematic representation of the methodology followed in this study. We named these Pt NPs as P1, P2, P3, P4 and P5 with respect to the order of increasing concentrations of precursor added to the reaction mixture during synthesis. Fig. 1 displays the TEM micrographs and the particle size distribution histograms of the Pt NPs. As observed from Fig. 1(a) the P1 were spherical shaped and were extremely small equivalent to nanodot sizes with particle sizes predominating in the range of 1–2 nm (Fig. 1(b)). P2 were also spherical morphologies, where mostly 2–3 nm particle sizes predominated (Fig. 1(c–d)). P3 were cuboidal shaped (Fig. 1(e)). In this case, as observed from Fig. 1(f), the 5–6 nm particles predominated. Fig. 1(g) presents the TEM micrographs of P4 showing particle shapes which appeared more oval to spherical than cuboidal. In P4 (Fig. 1(h)), 6–8 nm sizes dominated. P5 showed a distinctly different morphology having a floral shape, these nanoflowers (Fig. 1(i)) were observed to have 16–18 nm sizes dominating (Fig. 1(j)). Fig. S2A and S2B display the TEM micrograph and particle size distribution of P6 which were also attempted using 600 mg precursor concentration, but as we can observe from the TEM image, since the morphology remained similar to the P5 nanoflowers and also the sizes had decreased (13 nm predominating), we did not take up P6 for further experiments.
Figure 1. TEM micrographs showing morphology and size distribution histograms of (a–b) P1, (c–d) P2, (e–f) P3, (g–h) P4 and (i–j) P5 Pt NPs respectively.
Functionalization of PVP over the Pt NPs irrespective of the size/shape was confirmed using FTIR (Fig.S3). The peak region-1 (3650– 3050 cm−1) in the spectra confirms the presence of –OH/ = NH vibration stretching on all the Pt nanoparticles (Fig. S3 (P1–P6)) which confirms the presence of PVP on nanoparticles. Bare Pt nanoparticles also show a small broad band (3600–3400 cm−1) stretching in this region. This broad band is anticipated to be due to –OH stretching and indicates the presence of hydroxyl (-OH) functionality on the surface of bare Pt nanoparticles. But, the major difference lies in the vibration stretching at 2958 cm−1 observed in all the PVP stabilized Pt NPs (absent in bare Pt NPs) resulting from -CH2 groups (Fig. S3, region-2) from PVP, which again confirms the presence of PVP on all the Pt NP surfaces used in the present study. The presence of a heteroatom and a carbonyl group (=C=O) in the pyrrolidone ring sufficiently decreases the symmetry on the surface of Pt NPs. Vibrational modes with new bends at 1765 cm−1 and 1658 cm−1 were present in all the Pt NPs except bare Pt nanoparticles (Fig. S3B region 3). FTIR spectrum of PVP alone exhibits a broad bend at 1750 -1600 cm−1 which splits in two separate distinct bend with functionality on Pt NPs as shown in region-3. As we observe, the FTIR pattern is similar in the case of PVP and the PVP capped Pt NPs with distinct changes in the carbonyl group and heteroatom symmetry. However, these bends were absent in the case of bare Pt NPs (without PVP) indicating the successful capping of PVP on the P1, P2, P3, P4 and P5 Pt NPs used in this study.
Evaluating the bacteriotoxic/compatible property of the Pt NPs
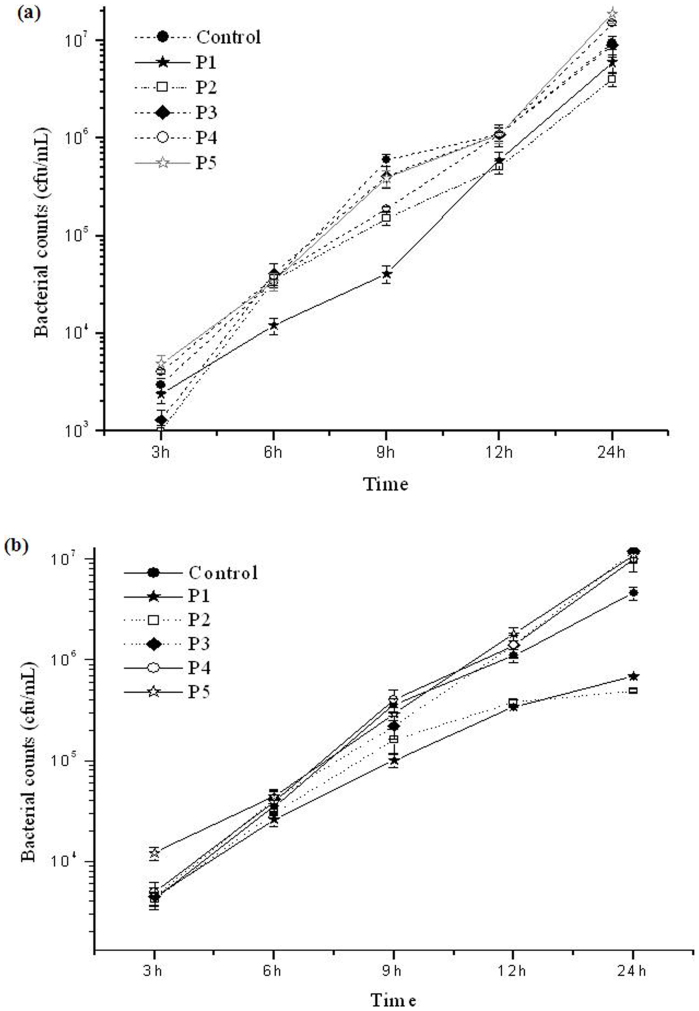
Cells belonging to the clinical pathogen, P. aeruginosa were incubated with different sized Pt NPs P1, P2, P3, P4 and P5 for 3 h, 6 h, 9 h, 24 h and the bacterial counts following incubation with Pt NPs was determined by the plate count method. Fig. 2a and Fig. 2b display the growth curve of the bacterium in the presence of 8.5 mg/L (Fig. 2a) and 15 mg/L (Fig. 2b) concentrations of the various Pt NPs sizes. As can be observed from these figures, P1 and P2 showed reduction in the bacterial counts, especially after 6 h incubation, while all the other Pt NPs, (P3, P4 and P5) showed bacterial counts similar to or in some cases even more than the control. Even very high (almost saturated) concentrations of NPs (15 mg/L) did not demonstrate bactericidal activity.
Figure 2.
(a), Graph showing the growth curve of bacteria in the presence of 8.5 mg/L concentrations of P1, P2, P3, P4, P5 Pt NPs and control, based on the total viable counts obtained by plate count method. (b), Graph showing the growth curve of bacteria in the presence of 15 mg/L concentrations of P1, P2, P3, P4, P5 Pt NPs and control, based on the total viable counts obtained by plate count method.
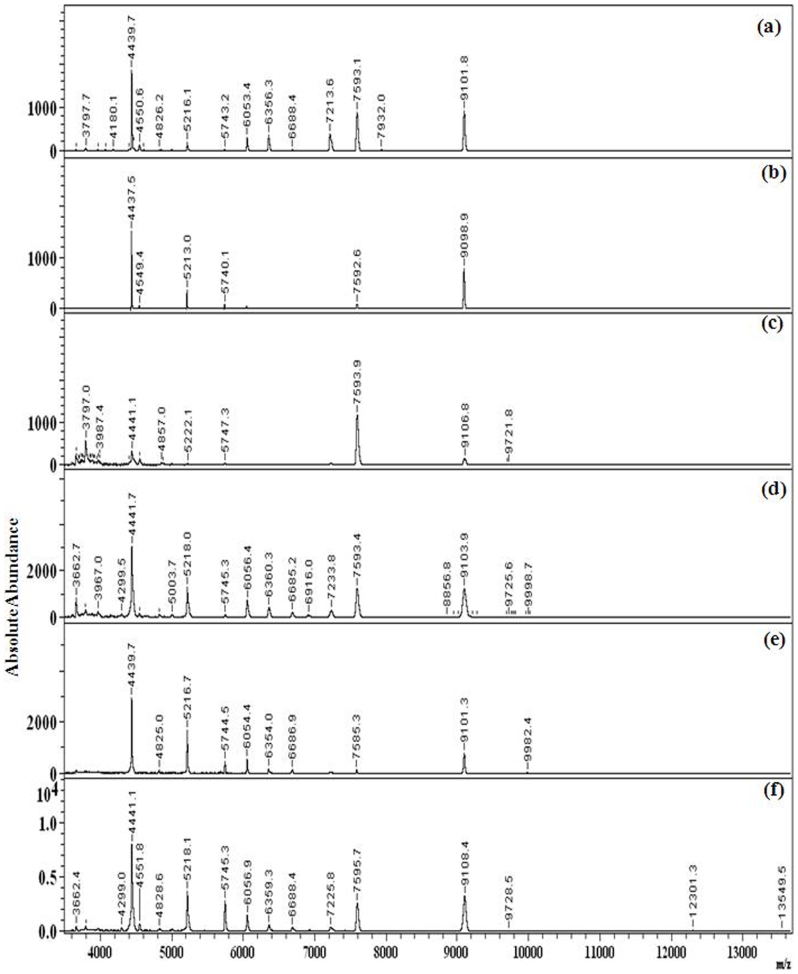
MALDI-MS studies of the cells after incubation with Pt NPs (15 mg/L) revealed that compared to the control (Fig.S4A(a)), P1(Fig. S4A(b)) and P2 (Fig. S4A(c)) produced lower number of bacterial peaks. Similar to the plate count results, the P3, P4 and P5 Pt NP interaction lead to enhanced bacterial signals compared to the control at incubation periods of 3 h (Fig. S4A(d–f)), 6 h (Fig.S4B(d–f)) and 24 h (Fig. 3(d–f). P1 and P2 did not show any distinct difference in the results up to 3 h (Fig. S4A(b–c)), but beyond 6 h (Fig. 4(b–c)) even up to 24 h (Fig. 3(b–c)), distinct suppression on the bacterial signals were observed. Such suppression of bacterial peaks, following incubation with NPs has been reported earlier by our group43,44. Thus, the MALDI-MS results indicate that the P1 and P2 Pt NPs showed bacterial toxicity, while the P3, P4 and P5 NPs showed bacterial compatibility.
Figure 3. MALDI-MS spectra of P. aeruginosa incubated with (a) no Pt NPs (control) and 15 mg/L concentrations of (b) P1 , (c) P2 , (d) P3, (e) P4, (f) P5 Pt NPs for 24 h, showing distinctly lesser peaks in P1 and P2.
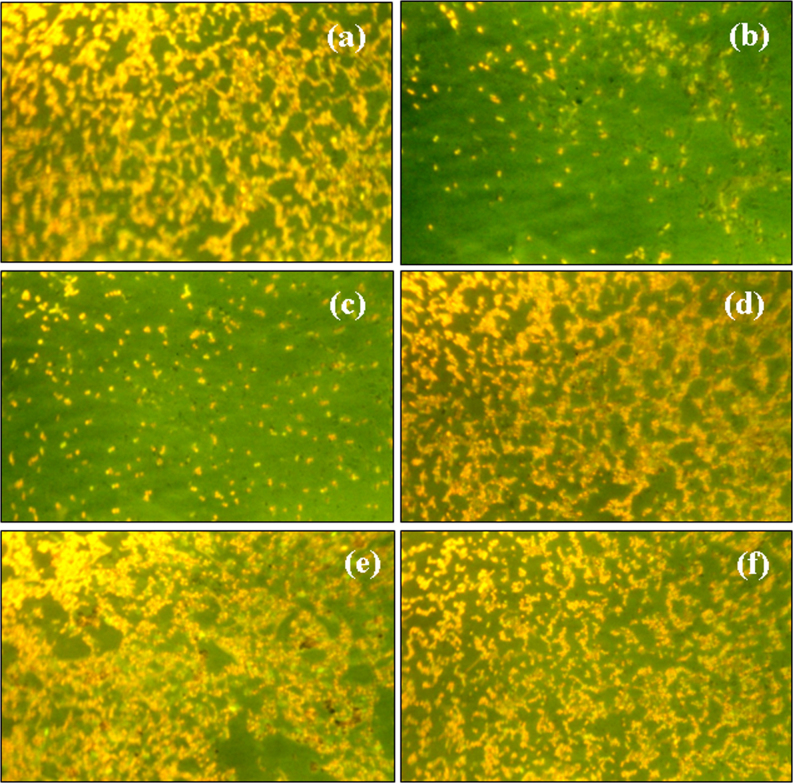
Figure 4. Epifluorescence micrographs showing the bacteria present in 10 μL of the bacterial culture suspension in (a) control and after incubating with 8.5 mg/L of (b) P1 (c) P2 (d) P3 (e) P4 (f) P5 Pt NPs for 9 h.
To further confirm the results obtained from MALDI-MS, we also applied the Fluorescence microscope for direct observation of the bacteria in 10 μL of the bacteria-NP suspension collected after an incubation period of 9 h. The plate count method has one limitation owing to the fact that the bacteria were grown on nutrient agar plates for 24 h before counting them. This time period and nutrition would assist the damaged bacteria to repair and survive45. Thus, the plate count method does not really signify the total viability of bacteria. Therefore, we applied direct microscopic studies which presented the current status of the bacteria after incubation with Pt NPs. Fig. 4 gives the epifluorescence micrographs of the bacteria after incubation with different sizes of Pt NPs at concentrations of 8.5 mg/L. Fig. 4(a) shows that numerous cells were present in the 10 μL smear in the control. P.aeruginosa incubated with P1 (Fig. 4(b)) and P2 (Fig. 4(c)) Pt NPs showed significant reduction in bacterial cells, supporting the trend reported in the plate count method and MALDI-MS studies too. However, as observed so far, P3, P4 and P5 Pt NPs did not show any decrease in bacterial counts (Fig. 4(d–f)); they appeared to even increase the bacterial counts compared to the control.
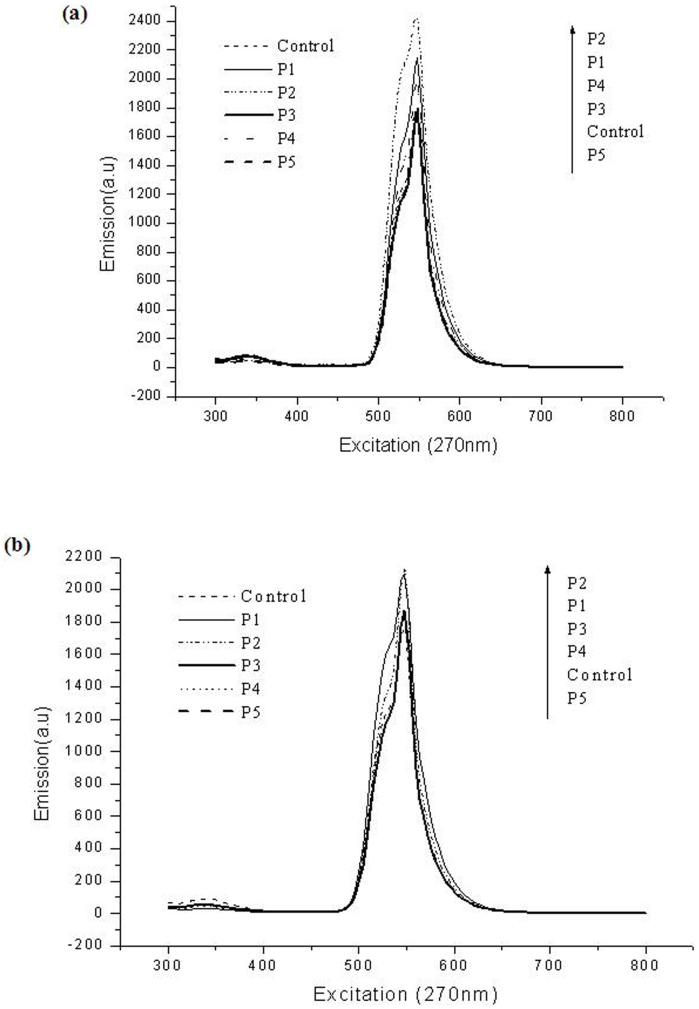
Finally, we also confirm the size dependant bacteriotoxic/compatible property of the Pt NPs using fluorescence spectroscopy. Recently, we have reported the use of acridine orange based techniques for differentiation of live/dead cells based on the 547 nm peak during fluorescence spectroscopic studies (unpublished data). Since the dead cells take up more green fluorescence, the intensity of this peak was higher in cell mixtures where dead cells dominated. We also used this technique to assay the cell viability of the P.aeruginosa cells after incubation with the different Pt NPs. As observed from Fig. 5a, the P1 and P2 Pt NPs showed highest 547 nm peaks compared to all the others at NPs concentration of 8.5 mg/L and also at 15 mg/L (Fig. 5b). The P3, P4, P5 and control showed lower intensity confirming their bacterio-compatibility. The same trend was observed after 9 h incubation (Fig. S5A) and even up to 24 h incubation (Fig. S5B) too. These results further confirmed the bacteriotoxic property exhibited by the P1 and P2 Pt NPs and the bacterio-compatible property exhibited by the P3, P4 and P5 Pt NPs.
Figure 5.
(a), Fluorescence spectra of the AO stained P.aeruginosa control cells and cells incubated with 8.5 mg/L concentrations of P1, P2, P3, P4 and P5 Pt NPs for 6 h. (b), Fluorescence spectra of the AO stained P.aeruginosa control cells and cells incubated with 15 mg/L concentrations of P1, P2, P3, P4 and P5 Pt NPs for 6 h.
HR TEM images (Fig. S6) were acquired in order to get some information on the interaction of the small and larger sized Pt NPs with P.aeruginosa cells. Although, in depth information was not obtained, the results signify that the smaller particles (P1) enter the cells (Fig. S6b) while the larger (P3 and P4) Pt NPs merely interact with the cell membranes (Fig. S6c and S6d) and therefore the localization of Pt NPs inside the cells was not observed.
Discussion
Of the five different Pt NPs sizes which were studied, the P1 showed the least size (1–2 nm) followed by P2 (2–3 nm); the rest of the Pt NPs, P3, P4 and P5 showed gradually increasing sizes with P5 attaining the largest size ( 16–18–20 nm). From all the results obtained using four different techniques, it can be concluded that the Pt NPs with particle sizes less than 3 nm have distinct bacterio-toxic property, while higher particle sizes show no toxic effect and in fact in most cases (P4 and P5) leading to enhanced growth of bacteria. Pelka et al.46 have investigated Pt NPs in the size range of 20 nm, 100 nm and > 100 nm for their influence on DNA; they reported that 20 nm Pt NPs showed pronounced effect on the DNA compared to larger particle sizes. Gehrke et al., demonstrated the size dependant effect of Pt NPs on DNA strand breaks in human colon carcinoma cells (HT29)47.
Although such scattered reports do exist regarding the interaction of Pt NPs with cells, so far no report deals with assessing the size dependence on the toxicity/compatibility of Pt NPs on bacterial cells. We have systematically synthesized a series of close lying Pt NP sizes within the narrow range distribution of 1–20 nm and assessed their bacterio toxic/compatible properties using four powerful techniques. All the evaluations confirmed that the spherical 1–2 nm sized P1 and P2 NPs show high toxicity. Borm et al. and Oberdörster et al48,49. reported that, Pt particles accumulate at traffic hot spots and therefore might enter the food chain, and thus their study focused on the toxicological relevance of metallic Pt NP after oral uptake. While larger particles (10 μm) were absorbed by the mucus of the bronchi and trachea, smaller particles (2.5 μm) can invade the respiratory tract much deeper. It appears that based on their report too, it is possible that smaller sized Pt NPs would be able to enter and interact with the bacterial cells much easier than those of the larger sized particles. It is also possible that the porins on the bacterial cell membrane are also small in the size of few nanometers and hence the access of the small sized spherical nanoparticles into the bacterial cell is easier compared to the larger nanoparticles similar to the case of the Pt NPs reported by Borm et al. Once these NPs invade the cells then they are able to interact with the molecular machinery inside the cells, especially the DNA and lead to effective cell toxicity46.
Moreover, ultrafine particles always demonstrate enhanced properties compared to their larger counterparts. Pt NPs generally do not exhibit much cytotoxic effect; so far no such report on the bacterial toxicity of Pt NPs has been documented. The ultrafine sized Pt NPs in this report with sizes less than 3 nm show bactericidal property; this is the first report in this direction. The exact mode of action by these NPs is still unraveled. This work suggests the possibility of the existence of a size dependant tunability of the Pt NPs for their toxic or compatible properties. We report that Pt NPs with sizes > 4 nm were compatible. The P4 and P5 Pt NPs were observed to enhance the growth of bacteria as shown by the various results. This could be due to the fact that Pt NPs have been demonstrated to possess antioxidative capacity50,51. This may be associated with the enhanced growth exhibited by the bacteria in the presence of P3, P4 and P5 NPs.
The spherical morphology of the P1 and P2 Pt NPs may also play a role in the bactericidal property observed. Most of the ultra sized nanodots and quantum dots show extravagant toxicity to cells. It is possible that spherical morphologies can gain access into cells easily through membrane pores which are usually spherical too. The cuboidal (P3), oval (P4) and flowers(P5) could have tough time getting in, this may also additionally contribute towards them being biocompatible. Based on the application it is thus possible to choose the ideal particle size as far as Pt NPs are concerned. For inhibition of cells or killing the P1, P2 Pt NPs can be used and for drug delivery and cell compatible studies P3, P4 and P5 Pt NPs would prove effective. Figure S7 gives a schematic speculation of the possibility of the size and shape dependent toxicity of the Pt NPs based on their entry into the bacterial cells. As reported in the HR TEM results, the P1 NPs could enter the cells while the P3 and P4 larger NPs could not. So, maybe the spherical shape of the smaller Pt NPs facilitates their easy entry into the cells compared to the other complicated NPs. Entry into the cell would provide these smaller NPs access to the cell contents and thus they would be able to affect cell growth and viability. However, since the larger Pt NPs do not enter they do not gain entry into the cells, therefore they exhibit biocompatibility. Most of the Pt based toxic effects reported so far involve its interaction with cellular components inside the cytoplasm and inside the nucleus41. Therefore, the P1 and P2 small sized and spherical particles have a greater probability to interact as they enter the cells, while the larger P3, P4 and P5 particles having lesser active surface area and also restriction on entry remain compatible. Although, our speculation for the mechanism of interaction is preliminary, more detailed investigations in this direction could provide conclusive evidences to this speculation.
In conclusion, we report the synthesis of five different sized Pt NPs within the narrow size regime of 1–20 nm using an extended method from an earlier report. The existence of bacterio toxicity/compatibility of the Pt NPs was evaluated using P.aeruginosa a clinical pathogen. The results revealed by plate count method, direct microscopic imaging, fluorescence spectroscopy and MALDI-MS proved that the < 3 nm sized P1 and P2 Pt NPs showed bacterio-toxic properties and the larger P3, P4 and P5 Pt NPs exhibited bacterio-compatible properties. This study describes for the first time that varying sized and shaped Pt NPs can vary their bacterial toxicity/compatibility.
Methods
Chemicals
Dihydrogen hexachloroplatinate(IV)hexahydrate and 3.5-Dimethoxy-4-hydroxycinnamic acid were bought from Alfa Aesar, UK. Polyvinylpyrrolidone (PVP, K value 29–32) was purchased from Sigma Aldrich, USA. Ethylene glycol and sodium hydroxide were purchased from Riedël-de Haën, USA. Acetonitrile (HPLC grad 99.9%) and Acetone (GC grad 99.8%) were bought from J, T Baker, Phillipsburg, NJ, USA. Trifluoroacetic acid (TFA, 98%) was bought from Sigma chemical Co. (St. Louis, MO, USA).
Synthesis of Pt NPs
Different sized Pt nanoparticles stabilized by PVP were prepared with some procedural extensions from a previous report42. Ethylene glycol (10 mL) was used as solvent and the reaction temperature was maintained at 120°C. To initiate the platinum nucleation-reduction reaction, NaOH (1 M, 50 uL) was added. Different amounts (5.0, 20.0, 50.0, 200.0, 400.0 and 600.0 mg) of the precursor, dihydrogen hexachloroplatinate(IV)hexahydrate was mixed in 1 mL PVP (91 mM in ethylene glycol, based on polymer monomer) solution and hot ethylene glycol was added drop wise. Slowly, a brown color appeared which finally turned to a grey tinted solution indicating the formation of Pt nanoparticles stabilized with PVP. Reaction mixture was stirred for 30 min more and cooled to room temperature. The Pt NPs were precipitated using acetone and centrifuged at 18000 rpm for 5 min and the supernatant discarded. Pt NPs were dispersed in deionized water for further use and stored at 4°C. The size and shape of the Pt NPs was confirmed using high resolution-transmission electron microscope (HR-TEM), JEOL TEM-3010, Tokyo, Japan at 75 keV. The particle size distribution of the various NPs was obtained using OPTIMAS 6.1 software based on the TEM images. The functionalization of PVP was confirmed using Fourier transform infra red (FTIR) spectroscopy.
Bacterial studies
Pseudomonas aeruginosa (BCRC 10303) standard culture was purchased from Bioresource Collection and Research Center (Hsin-Chu, Taiwan) and used for these studies. One mL of P.aeruginosa containing 1.05 × 109 cfu/mL of cells was inoculated into 20 mL of sterile nutrient broth and incubated with 0 mg/L, 8.5 mg/L and 15 mg/L concentrations of the five different types of Pt NPs. This setup was incubated at 37°C in an orbital shaker incubator (Firstek Inc., Taiwan) at 120 rpm. 2 mL of sample was taken from the reaction mixture at 3 h, 6 h, 9 h and 24 h intervals.
Evaluating the biotoxicity/biocompatibility of the Pt NPs
Plate count method
0.1 mL of the bacterial culture after incubation with P1, P2, P3, P4. P5 NPs and control at 3 h, 6 h, 9 h, 24 h intervals was serially diluted in sterile water and plated on nutrient agar by pour plate method for estimating the total viable count (TVC) of bacteria as per microbiological standard procedures.
Fluorescence microscopic studies
10 μL of the variously sized Pt NPs (8.5 mg/L) incubated with bacterial cells for 9 h was uniformly smeared using a sterile loop onto a clean glass slide and left to air dry under sterile condition in the laminar flow. The dry smears were flooded with acridine orange (0.1% solution in distilled water). After 2 min, the excess stain was drained off and the glass slides were washed in sterile water, dried and observed. Acridine orange, a fluorescent dye, differentially stains single stranded RNA and double stranded DNA, fluorescing orange when intercalated with the former and green while complexing with the latter when observed under a ESPA FI40(NIB-100F, ESPA systems Co. Ltd., Taiwan) inverted epifluorescence microscope (excitation filter BP 490; barrier filter O 515). Thus, the numbers of orange fluorescing cells (especially with respect to bacterial cells) depict the actively metabolizing cells51. Based on this technique it was possible to directly image the cells prior to exposure to the various Pt NPs and in the control.
Fluorescence spectroscopy
Fluorescence spectra were acquired using a Fluorescence spectrophotometer (Hitachi, F-2700, PerkinElmer, USA) operating with the FL solutions version 4.1 software. The spectrometer is equipped with a xenon discharge light source and a photomultiplier detector and includes a single position cell holder for measuring liquid samples. 1 mL of the bacterial culture was retrieved from the reaction mixture of various Pt NPs and from the control at 6 h, 9 h, 24 h intervals and centrifuged at 15,000 rpm for five mins. The supernatant was discarded and the bacterial cell pellet was resuspended in 1 mL of 0.1% acridine orange and incubated at room temperature for 15 min. Then the stained cells were separated by centrifugation at 15,000 rpm for five min. The stained bacterial pellet was resuspended in distilled water thrice to remove excess stain. The washed cells were finally suspended in 1 mL distilled water and placed in a high-grade standard (1 cm square) quartz cuvette (10 mm path length) with a screw cap and placed inside the cell holder. All measurements of the samples were made at room temperature (20–22°C).
HR-TEM
In order to understand the interaction of the different Pt NPs, HR TEM was employed. P.aeruginosa cells incubated with 15 mg/L of P1, P3 and P4 Pt NPs, representing the small (P1) and bigger (P3, P4) Pt NPs were studied. The bacterial cells were collected after 9 h incubation and pelleted and fixed with glutaraldehyde overnight and then placed on the copper grids. Images were acquired using a JEOL TEM-3010 (Tokyo, Japan) transmission electron microscope at 75 keV.
MALDI-MS analysis
0.5 mL of the culture was taken for MALDI analysis after 3 h, 6 h, 9 h, and 24 h in Eppendorf tubes and centrifuged at 15,000 rpm for five min. The supernatant was removed and the pellet was washed in 1 mL of distilled water by vortexing for 1 min and 5 μl of this sample was spotted on a stainless steel target plate, each spot was overlaid with 5 μl of matrix (0.05 M sinapinic acid (SA) in 3:1 Acetonitrile:Water containing 0.1% TFA and air dried. All experiments were performed in triplicates. All mass spectra were obtained in positive ion mode using MALDI-TOF MS (Microflex, Bruker Daltonics, Bremen, Germany). The MALDI source was equipped with a nitrogen laser (337 nm) for irradiation of analytes and accelerating voltages were set at +20 kV. All experiments were performed under similar conditions in linear mode with laser energy of 63.2 μJ and 200 laser shots.
Author Contributions
J.G. performed the experiments and also wrote the paper, N.H. took care of the nanoparticle synthesis and characterization part, M.M. and H.F.W. participated in the organization of the experiments and also in the manuscript writing and reviewing. H.F.W. is the principle investigator of this NSC funded project and has contributed in the supervision for the project ideas, project direction, discussion and progress of all experiments, has revision for the manuscript and provided all materials, instruments and resources including post-doc funding.
Supplementary Material
Supporting Information
Acknowledgments
We thank National Science Council for financial support.
References
- Teranishi T., Hosoe M., Tanaka T. & Miyake M. Size Control of Monodispersed Pt Nanoparticles and Their 2D Organization by Electrophoretic Deposition. J. Phys. Chem. B 103, 3818–3827 (1999). [Google Scholar]
- Králik M. & Biffis A. Catalysis by metal nanoparticles supported on functional organic polymers. Journal of Molecular Catalysis A: Chemical. 177, 113–138 (2001). [Google Scholar]
- Rao C. N. R., Kulkarni G. U., Thomas P. J. & Edwards P. P., Metal nanoparticles and their assemblies. Chem. Soc. Rev. 29, 27–35 (2000). [Google Scholar]
- Lam D. M. K. & Rossiter B. W. Clear Advantage. Sci. Am. 265, 48–52 (1991).1862331 [Google Scholar]
- Lewis L. N. Chemical catalysis by colloids and clusters. Chem. Rev. 93, 2693–2730 (1993). [Google Scholar]
- Maier S. et al. Plasmonics-A Route to Nanoscale Optical Devices. Adv. Mater. 13, 1501–1505 (2001). [Google Scholar]
- Feldheim D. L. & Keating C. D. Self-assembly of single electron transistors and related devices. Chem. Soc. Rev. 27, 1–12 (1998). [Google Scholar]
- Kamat P. V. Photophysical, Photochemical and Photocatalytic Aspects of Metal Nanoparticles. J. Phys. Chem. B. 106, 7729–7744 (2002). [Google Scholar]
- Murray C. B., Sun S., Doyle H. & Betley T. Monodisperse 3D transition-metal (Co, Ni, Fe) nanoparticles and their assembly into nanoparticle superlattices. Mater. Res. Soc. Bull. 26, 985–991 (2001). [Google Scholar]
- Kim Y., Johnson R. C. & Hupp J. T. Gold Nanoparticle-Based Sensing of “Spectroscopically Silent” Heavy Metal Ions. Nano Lett. 1, 165–167 (2001). [Google Scholar]
- Nicewarner-Pena S. R. et al. Submicrometer metallic barcodes. Science, 294, 137–141 (2001). [DOI] [PubMed] [Google Scholar]
- Nie S. & Emory S. R. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science. 275, 1102–1106 (1997). [DOI] [PubMed] [Google Scholar]
- Cao Y. C., Jin R. & Mirkin C. A. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 297, 1536–1540 (2002). [DOI] [PubMed] [Google Scholar]
- Dick L. A., McFarland A. D., Haynes C. L. & Van Duyne R. P. Metal Film over Nanosphere (MFON) Electrodes for Surface-Enhanced Raman Spectroscopy (SERS): Improvements in Surface Nanostructure Stability and Suppression of Irreversible Loss. J. Phys. Chem. B. 106, 853–860 (2002). [Google Scholar]
- Hirsch L. R., Jackson J. B., Lee A., Halas N. J. & West J. L. A whole blood immunoassay using gold nanoshells. Anal. Chem. 75, 2377–2383 (2003). [DOI] [PubMed] [Google Scholar]
- Hirsch L. R. et al. Nanoshell-mediated near-infrared thermal therapy of tumors under magnetic resonance guidance. Proc. Natl Acad. Sci. USA. 100, 13549–13554 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Woo H. J., Kim C. K. & Yoon C. S. The catalytic effect of Pt nanoparticles supported on silicon oxide nanowire. Nanotechnology. 20, 235306 (pp7) (2009). [DOI] [PubMed] [Google Scholar]
- Cheng H., Xi C., Meng X., Hao Y., Yu Y. &, Zhao F. . Polyethylene glycol-stabilized platinum nanoparticles: The efficient and recyclable catalysts for selective hydrogenation of o-chloronitrobenzene to o-chloroaniline. Colliod. Interface Sci. 336, 675–678 (2009). [DOI] [PubMed] [Google Scholar]
- Xu J. et al. Enhanced dehydrogenation of LiBH4 catalyzed by carbon-supported Pt nanoparticles. Chem. Commun. 44, 5740–5742 (2008). [DOI] [PubMed] [Google Scholar]
- Ahmadi T. S., Wang Z. L., Green T. C., Henglein A., El-sayed M. A. Shape-Controlled Synthesis of Colloidal Platinum Nanoparticles. Science. 272, 1924–1926 (1996). [DOI] [PubMed] [Google Scholar]
- Chen J., Xiong Y., Yin Y., Xia Y. Pt nanoparticles surfactant-directed assembled into colloidal spheres and used as substrates in forming Pt nanorods and nanowires. Small. 2, 1340–1343 (2006). [DOI] [PubMed] [Google Scholar]
- Chen Y., Johnson E. & Peng X. Formation of monodisperse and shape-controlled MnO nanocrystals in non-injection synthesis: self-focusing via ripening. J. Am. Chem. Soc. 129, 10937–10947 (2007). [DOI] [PubMed] [Google Scholar]
- Ford W. E., Harnack O., Yasuda A. & Wessels J. M. Platinated DNA as Precursors to Templated Chains of Metal Nanoparticles. Adv. Mater. 13, 1793–1796 (2001). [Google Scholar]
- Jana N. R. & Peng X. Single-Phase and Gram-Scale Routes toward Nearly Monodisperse Au and Other Noble Metal Nanocrystals. J. Am. Chem. Soc. 125, 14280–14281 (2003). [DOI] [PubMed] [Google Scholar]
- Narayanan R. & El-Sayed M. A. Shape-Dependent Catalytic Activity of Platinum Nanoparticles in Colloidal Solution. Nano Lett. 4, 1343–1348 (2004). [Google Scholar]
- Petroski J. M., Green T. C. & El-Sayed M. A. Self-Assembly of Platinum Nanoparticles of Various Size and Shape. J. Phys. Chem. A. 105, 5542–5547 (2001). [Google Scholar]
- Teng X. & Yang H. Synthesis of Platinum Multipods: An Induced Anisotropic Growth. Nano Lett. 5, 885–891 (2005). [DOI] [PubMed] [Google Scholar]
- Fenske D. et al. J. Colloidal Synthesis of Pt Nanoparticles: On the Formation and Stability of Nanowires. Langmuir. 24, 9011–9016 (2008). [DOI] [PubMed] [Google Scholar]
- Mafune F., Kohno J., Takeda Y. & Kondow T. Formation of Stable Platinum Nanoparticles by Laser Ablation in Water. J. Phys. Chem. B. 107, 4218–4223 (2003). [Google Scholar]
- Kinge S. & Bonnemann H. One-pot dual size- and shape selective synthesis of tetrahedral Pt nanoparticles. Appl. Organomet. Chem. 20, 784–787 (2006). [Google Scholar]
- Herricks T., Chen J. & Xia Y. Polyol Synthesis of Platinum Nanoparticles: Control of Morphology with Sodium Nitrate. Nano Lett. 4, 2367–2371 (2004). [Google Scholar]
- Chen J., Herricks T. & Xia Y. Polyol Synthesis of Platinum Nanostructures: Control of Morphology through the Manipulation of Reduction Kinetics. Angew. Chem. Int. Ed. 44, 2589-2592 (2005). [DOI] [PubMed] [Google Scholar]
- Lim B. et al. Facile Synthesis of Highly Faceted Multioctahedral Pt Nanocrystals through Controlled Overgrowth. Nano Lett. 8, 4043–4047 (2008). [DOI] [PubMed] [Google Scholar]
- Tsuji M. et al. Toward to branched platinum nanoparticles by polyol reduction: A role of poly(vinylpyrrolidone) molecules. Colloids Surf. A. 317, 23–31 (2008). [Google Scholar]
- Ahmadi T. S., Wang Z. L., Henglein A. & El-Sayed M. A. “Cubic” Colloidal Platinum Nanoparticles. Chem. Mater. 8, 1161–1163 (1996). [DOI] [PubMed] [Google Scholar]
- Lee H. et al. Morphological Control of Catalytically Active Platinum Nanocrystals. Angew. Chem. Int. Ed. 45, 7824–7828 (2006). [DOI] [PubMed] [Google Scholar]
- Kijima T. et al. Noble-Metal Nanotubes (Pt, Pd, Ag) from Lyotropic Mixed-Surfactant Liquid-Crystal Templates. Angew. Chem. Int. Ed. 43, 228–232 (2004). [DOI] [PubMed] [Google Scholar]
- Song Y. et al. Synthesis of Platinum Nanowire Networks Using a Soft Template. Nano Lett. 7, 3650–3065 (2007). [DOI] [PubMed] [Google Scholar]
- Konya Z. et al. A. Novel Two-Step Synthesis of Controlled Size and Shape Platinum Nanoparticles Encapsulated in Mesoporous Silica. Catal. Lett. 81, 137–140 (2002). [Google Scholar]
- Kundu S. & Liang H. Photoinduced formation of shape-selective Pt nanoparticles. Langmuir. 26, 6720–6727 (2010). [DOI] [PubMed] [Google Scholar]
- Asharani P. V., Xinyi N., Hande M. P. & Valiyaveettil S. DNA damage and p53-mediated growth arrest in human cells treated with platinum nanoparticles. Nanomedicine, 5, 51–64 (2010). [DOI] [PubMed] [Google Scholar]
- Koebel M. M., Jones L. C. & Somorjai G. A. Preparation of size-tunable, highly monodisperse PVP-protected Pt-nanoparticles by seed-mediated growth. J. Nanopart Res. 10, 1063–1069 (2008). [Google Scholar]
- Gopal J., Wu H. F. & Lee C. H. Photocatalyst Inhibition of Microbial Adhesion by Anodized Titanium. Analyst. 136, 5077–5083 (2011).22005882 [Google Scholar]
- Gopal J., Lee C. H. & Wu H. F. MALDI-TOF MS as a rapid and reliable technique for directly evaluating bactericidal activity: Probing the critical concentration of ZnO nanoparticles as affinity probes. Anal. Chem. 82, 9617–9621 (2010). [DOI] [PubMed] [Google Scholar]
- Pelka J. et al. Cellular uptake of platinum nanoparticles in human colon carcinoma cells and their impact on cellular redox systems and DNA integrity. Chem Res. Tox. 22, 649–659 (2009). [DOI] [PubMed] [Google Scholar]
- Gehrke H. et al. Platinum nanoparticles and their cellular uptake and DNA platination at non-cytotoxic concentrations. Arch Toxicol. 85, 799–812 (2011). [DOI] [PubMed] [Google Scholar]
- Borm P. J. A. et al. The potential risks of nanomaterials: a review carried out for ECETOC. E. Part. Fibre Toxicol. 3, 11, (pp 35) (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdörster G., Oberdörster E., Oberdörster J. Nanotoxicology: An Emerging Discipline Evolving from Studies of Ultrafine Particles. J. Environ Health Perspect. 113, 823–839 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M. et al. Platinum nanoparticle is a useful scavenger of superoxide anion and hydrogen peroxide. Free Radic. Res. 41, 615–626 (2007). [DOI] [PubMed] [Google Scholar]
- Watanabe A. et al. In vitro free radical scavenging activity of platinum nanoparticles. Nanotechnology. 20, 455105 (9pp), (2009). [DOI] [PubMed] [Google Scholar]
- Gopal J., George R. P., Muraleedharan P. & Khatak H. S. Photocatalyst Inhibition of Microbial Adhesion by Anodized Titanium. Biofouling. 20, 167–175 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information