Abstract
Antimicrobial peptides have established an important role in the defense against extracellular infections, but the expression of cationic peptides within macrophages as an antibacterial effector mechanism against intracellular pathogens has not been demonstrated. Macrophage expression of the murine cathelicidin-related antimicrobial peptide (CRAMP) was increased after infection by the intracellular pathogen Salmonella typhimurium, and this increase required reactive oxygen intermediates. By using CRAMP-deficient mice or synthetic CRAMP peptide, we found that CRAMP impaired Salmonella cell division in vivo and in vitro, resulting in long filamentous bacteria. This impaired bacterial cell division also depended on intracellular elastase-like serine protease activity, which can proteolytically activate cathelicidins. Macrophage serine protease activity induced filamentation and enhanced the activity of CRAMP in vitro. A peptide-sensitive Salmonella mutant showed enhanced survival within macrophages derived from CRAMP-deficient mice, indicating that Salmonella can sense and respond to cationic peptides in the intracellular environment. Although cationic peptides have been hypothesized to have activity against pathogens within macrophages, this work provides experimental evidence that the antimicrobial arsenal of macrophages includes cathelicidins. These results show that intracellular reactive oxygen intermediates and proteases regulate macrophage CRAMP expression and activity to impair the replication of an intracellular bacterial pathogen, and they highlight the cooperativity between macrophage antibacterial effectors.
Macrophages comprise an essential part of the innate immune response to bacterial infections (1). Because macrophages are highly phagocytic and are actively targeted by pathogenic bacteria, they must have effective mechanisms either for killing bacteria or controlling their replication to avoid becoming a reservoir of infection. Salmonella typhimurium is a bacterial pathogen that resides within murine splenic and liver macrophages (2, 3), causing in susceptible mice systemic disease resembling typhoid fever in humans. This pathogen counters macrophage antibacterial effectors with acid tolerance and perturbation of endosomal trafficking, to avoid oxidative and nitrosative damage and phagolysosomal degradation (4, 5). We have observed that macrophages impair cell division of intracellular S. typhimurium, resulting in the formation of filamentous bacteria with arrested septation (6). This morphology indicates a bacterial stress response and has been observed in bacteria responding to damage from low doses of antibiotics, starvation, and reactive oxygen and nitrogen species (7–9). We have shown previously that macrophages use a phagocyte oxidase (phox)-dependent and inducible NO synthase-independent mechanism to impair Salmonella cell division. Because S. typhimurium diverts the phox to minimize direct damage by reactive oxygen intermediates (ROIs) (4), we hypothesized that these ROIs impair bacterial cell division by regulating a previously uncharacterized antimicrobial effector mechanism(s).
Macrophages possess a variety of intracellular proteases, some of which are secreted whereas others show activity within a phagolysosomal compartment. Studies using knock-out mice have shown that the proteases neutrophil elastase and cathepsin G have important antibacterial activities within neutrophils (10, 11), but a role for macrophage proteases during bacterial infection has not been reported. In addition to a direct microbicidal role (12), proteases with elastase-like specificities proteolytically activate members of the cathelicidin family of cationic peptides, which are synthesized as inactive proproteins (13, 14). Mice express cathelicidin-related antimicrobial peptide (CRAMP), a cationic α-helical peptide with antimicrobial activity against Gram-positive and Gram-negative bacteria in vitro (reviewed in ref. 13). CRAMP expression by keratinocytes mediates control of bacterial skin infection by group A Streptococcus (15). Whereas alveolar macrophages express defensins, another class of antimicrobial peptides, macrophage expression of cathelicidins, has not been reported (16, 17).
S. typhimurium resists damage by cationic peptides in vitro by modifying their lipopolysaccharide (LPS) structure by using members of the PhoP–PhoQ regulon (refs. 18–21 and reviewed in ref. 22). PhoPnull mutants, which are more susceptible to peptides in vitro (20), exhibit decreased virulence in mice (23). It is of interest to determine whether murine macrophages use cationic peptides such as CRAMP to control Salmonella replication because these cells provide an intracellular niche for S. typhimurium within secondary lymphoid organs. This study examined whether macrophages use proteases and cationic peptides to limit replication of an intracellular bacterial pathogen.
Materials and Methods
Growth Conditions of Bacterial and Macrophage Cells. The Salmonella enterica serovar Typhimurium strains SL1344 and 14028s (Salmonella Genetic Stock Center, Calgary, AB, Canada), 14028s Pho24 PhoPconstitutive mutant (F. Heffron, Oregon Health and Sciences University, Portland), 14028s phoP::Tn10 MS7953s PhoPnull mutant (S. Miller, University of Washington, Seattle), SL1344 phoP::Tn10 (L. Knodler, Rocky Mountain Laboratories, Hamilton, MT), SL1344 ΔaroA (B. Stocker, Stanford University, Stanford, CA), and strains expressing pFPV25.1-GFP (S. Meresse, Marseille, France, and S. Falkow, Stanford University) were cultured as described (6). Strains in a SL1344 background were used unless indicated otherwise in the text. Bone marrow was isolated from the femurs of BALB/c (The Jackson Laboratory), 129/SVJ, or Cnlp–/– CRAMP-deficient mice in the 129/SVJ background (15), cultured for 7 days in DMEM (HyClone) supplemented with 20% FBS (Invitrogen)/2 mM l-glutamine/1 mM sodium pyruvate/30% L cell-conditioned media at 37°C in 5% CO2. Bone marrow-derived macrophages (BMDM) treated with chemical inhibitors were examined for cell death by propidium iodide, 7-amino-actinomycin D, and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling staining and death was <10%.
Reagents. After infection, macrophages were treated with inhibitors at the following concentrations: 4 μM diphenyleneiodonium (DPI; Sigma), 250 μM acetovanillone (apocynin; Aldrich), 30 mM N-acetyl cysteine (Sigma), 100 μM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF; Calbiochem), 25 μM elastatinal (Calbiochem), 100 μM methoxysuccinyl (MeOSuc)-Ala-Ala-Pro-Ala-chloromethylketone (CMK; human neutrophil elastase inhibitor; Calbiochem), 100 μM MeOSuc-Ala-Ala-Pro-Val-CMK (human leukocyte elastase inhibitor; Calbiochem). The complete mini EDTA-free protease inhibitor mixture was used according to the manufacturer's instructions (Roche Diagnostics, Laval, Quebec). Control cells were treated with equivalent volumes of DMSO per ml of media. Human neutrophil elastase (Calbiochem) and MeOSuc-Ala-Ala-Pro-Val-para-nitroaniline (Colorimetric elastase substrate I; Calbiochem) were used for zymograms. CRAMP peptide, corresponding to the mature C-terminal 34-aa peptide, was synthesized according to methods published by the Protein Services Laboratory at the University of British Columbia.
Infection Conditions. For immunof luorescence and colony-forming unit (cfu) experiments, BMDM were seeded in 24-well plates (1 × 105 cells per well) and infected with S. typhimurium for 20 min, as described (6). Monolayers were washed with PBS, incubated in media containing 100 μg/ml gentamicin (Sigma) for 1.5 h, and maintained with 10 μg/ml gentamicin. Intracellular survival and/or replication of S. typhimurium was determined by using the gentamicin-resistance assay (6).
Immunofluorescence. Immunofluorescence staining was performed as described (6) by using rabbit anti-S. typhimurium LPS Ab (1:200) (Difco) without permeabilization to detect extracellular bacteria, a specific polyclonal rabbit anti-CRAMP Ab (1:150 dilution) (24) in the presence of 0.2% saponin, and an Alexa 568 mouse anti-rabbit Ab (1:200 to 1:400 dilution) (Molecular Probes). Bacteria were scored as “filamentous” when they were intracellular and more than three times longer than a typical bacterium (approximately >5 μm). To assess protease activity, BMDM were incubated with 10 μM carbobenzyloxy-Ala-Ala-Ala-Ala-rhodamine 110 (Calbiochem) for the last 2 h of infection. Confocal sections of 0.1-μm thickness were assembled into flat projections by using image software (Version 1.63; National Institutes of Health, Bethesda).
Flow Cytometry. BMDM were blocked and permeabilized on ice with 0.2% saponin (Sigma) and 2% FBS, and they were stained sequentially with rabbit anti-CRAMP Ab (1:150) and goat anti-rabbit phycoerythrin secondary Ab (1:200). Flow cytometry was performed by using a FACSCalibur fluorescence-activated cell sorter with cellquest software (BD Biosciences).
RT-PCR. RNA was isolated from BMDM by using TRIzol reagent, and cDNA was synthesized from equivalent quantities of RNA by using SuperScript II and oligo(dT) (Invitrogen). PCR was performed by using the following oligonucleotide primers: CRAMP, 5′-GCTGATGTCAAAAGAATCAGCG-3′ and 5′-TCCCTCTGGAACTGCATGGTTCC-3′; and GAPDH, 5′-AGAACATCATCCCTGCATCC-3′ and 5′-CTGGGATGGAAATTGTGAGG-3′.
In Vitro Filamentation Assay. Stationary-phase bacteria were cultured in N-minimal media (pH 7.4 or 5.8) containing 0.1% Casamino acids, 0.3% (vol/vol) glycerol, and 8 μM magnesium chloride, as well as concentrations of synthetic active CRAMP peptide or macrophage lysates, as indicated. Cultures were incubated overnight at 37°C with shaking and examined by microscopy. Filamentous bacteria induced by CRAMP were subsequently incubated with chemical inhibitors for 24 h, as indicated. To quantify the extent of bacterial filamentation, culture OD560 was measured with a SpectraFluor Plus spectrophotometer (Tecan, Groedig, Austria). Macrophage lysates were prepared by mechanical disruption, and postnuclear supernatants were processed as described (25).
Statistics. Significance was determined by calculating P values by using an unpaired two-tailed Student t test.
Results
Macrophages Impair Salmonella Cell Division by Using an Oxidase-Dependent Mechanism. When primary macrophages are infected with S. Typhimurium, there is a decrease in the number of viable intracellular bacteria within the first few hours of infection, and the size of this surviving population then remains relatively stable over the next 20 h (26, 27). Many of these surviving bacteria exhibited impaired cell division and were unable to complete septation, resulting in filamentous bacteria after 24 h (Fig. 1, DMSO). BMDM, isolated splenic and peritoneal macrophages, and macrophages within infected mouse spleen were all capable of causing bacterial stress and filamentation (Fig. 1 and data not shown). We described previously that, by using RAW 264.7 murine macrophage-like cells, macrophage impairment of bacterial cell division is ROI-dependent and reactive nitrogen intermediate-independent (6). This phenotype was observed also in BMDM (Fig. 1 and data not shown). A significant decrease in the number of filamentous bacteria occurred in cells treated with DPI, a chemical inhibitor of ROIs (Fig. 1). A similar inhibition of bacterial filamentation was observed by using two other antioxidants, N-acetyl cysteine and acetovanillone (data not shown). S. typhimurium delivers proteins into infected macrophages by using a secretion apparatus encoded on the Salmonella pathogenicity island 2 (SPI2), which inhibit functional assembly of phox on the Salmonella-containing vacuole (4). Bacterial filamentation within macrophages was determined not to depend on SPI2 because no significant difference in the number of cells containing filamentous bacteria was observed when cells were infected with wild-type bacteria, which avoid colocalization with phox, or a SPI2 secretion mutant, which does not divert phox (data not shown). Therefore, we hypothesized that the stressor was not direct oxidative damage but rather an effector regulated by ROIs.
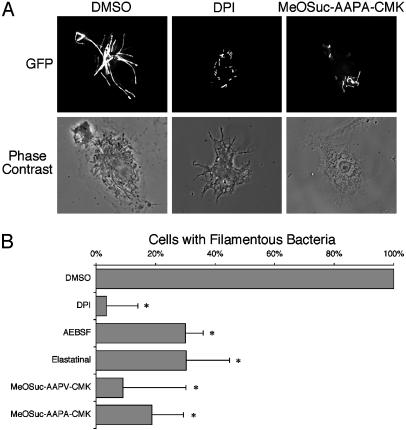
Fig. 1.
Serine protease inhibitors decrease bacterial filamentation in BMDM. (A) Immunofluorescence. BMDM were pretreated with inhibitors, infected with S. typhimurium expressing GFP for 24 h, and then examined by fluorescence microscopy. Mock-treated cells contained filamentous bacteria (GFP, DMSO). There was a decrease in the number of filamentous bacteria in cells treated with chemical inhibitors of ROIs (DPI) or elastase-like serine proteases (MeOSuc-AAPA-CMK). n ≥ 4. (B) Quantification. Cells were treated as described in A, and the percentage of infected macrophages containing filamentous bacteria were quantified in at least 100 infected cells per experiment. The mean percentage of infected DMSO-treated cells containing filamentous bacteria (24 ± 5%; n = 6) was normalized to 100%. *, P < 0.01; n ≥ 4.
Intracellular Protease Activity Mediates Impaired Salmonella Cell Division. Neutrophil elastase and cathepsin G activity are regulated by ROIs within early neutrophil phagosomes, and these proteases are important for killing phagocytosed Gram-negative bacteria (11). We investigated whether protease activity could be the ROI-regulated effector mechanism that causes bacterial filamentation within macrophages. We treated BMDM with a mixture of protease inhibitors and observed a significant decrease in the number of infected cells containing filamentous bacteria after protease inhibitor treatment (data not shown). To determine the pharmacological characteristics of the protease activity or activities correlating with impaired bacterial cell division, cells were treated with individual specific inhibitors. Macrophages treated with the protease inhibitors pepstatin, TLCK, leupeptin, or aprotinin contained filamentous bacteria and were indistinguishable from mock-treated cells (data not shown). In contrast, a significant decrease in the number of macrophages containing filamentous bacteria was observed in cells treated with the broad serine protease inhibitor AEBSF as well as three more specific inhibitors of elastase-like serine protease activity (elastatinal, MeOSuc-AAPA-CMK, and MeOSuc-AAPV-CMK) (Fig. 1B).
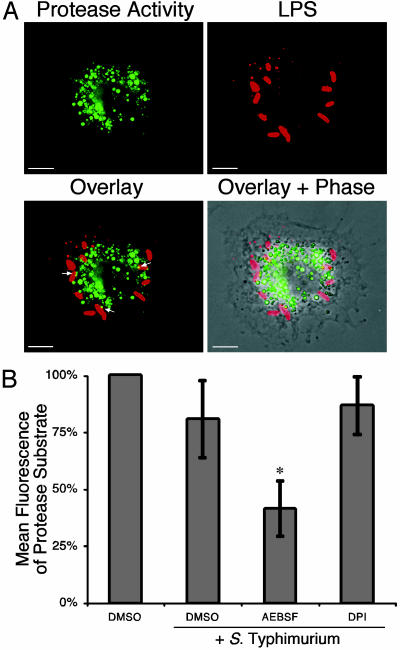
Because macrophages can store and secrete proteases, we examined the location of this protease activity relative to Salmonella-containing vacuoles. BMDM infected with S. typhimurium were examined 8 h after infection, which is immediately before the onset of filamentation and when we hypothesized that a putative effector mechanism would be active in impairing bacterial cell division. Cells were incubated with a cell-permeable elastase substrate carbobenzyloxy-AAAA-rhodamine 110 that fluoresces on cleavage. Fig. 2A shows that infected BMDM contain a protease activity capable of cleaving this substrate in a vesicular distribution with a perinuclear location similar to intracellular Salmonella. This fluorescence pattern was observed also in uninfected cells and cells infected for 24 h, which contain filamentous bacteria (data not shown). Macrophages treated with the serine protease inhibitor AEBSF, which contain fewer filamentous bacteria (Fig. 1), exhibit significantly decreased substrate proteolysis when the fluorescence of cleaved substrate was quantified by flow cytometry (Fig. 2B). Treatment with the antioxidants DPI (Fig. 2B) or N-acetylcysteine or acetovanillone (data not shown) did not significantly reduce the substrate cleavage, which is different from the regulation of neutrophil elastase by ROIs within neutrophil phagosomes (11). To begin to characterize the protease(s), total cell proteins were resolved in a gel containing MeOSuc-Ala-Ala-Pro-Val-para-nitroaniline, a substrate of elastase-like proteases that yields a white precipitate when cleaved. BMDM contain multiple activities that are capable of cleaving this substrate with apparent molecular weights distinct from neutrophil elastase (data not shown).
Fig. 2.
Localization and regulation of macrophage protease activity. (A) Confocal microscopy. Macrophages were infected with S. typhimurium for 8 h and incubated with the cell-permeable elastase substrate carbobenzyloxy-AAAA-R110 for the last 2 h. Cleavage of the protease substrate yields fluorescence at 488 nm, which is observed in a punctate perinuclear pattern (green). Intracellular S. typhimurium was detected by using an Ab specific for S. typhimurium LPS (red). White arrows in the overlay indicate an overlap of elastase activity with intracellular bacteria (yellow). (Scale bar, 5μm; n = 4.) (B) Flow cytometry. Elastase activity is not blocked by antioxidants. Cells were pretreated with AEBSF (serine protease inhibitor), DPI (antioxidant), or DMSO (control); infected with S. typhimurium for 8 h; incubated with the elastase substrate carbobenzyloxy-AAAA-R110, as described in A; and fluorescence-quantified by flow cytometry. The mean ± SD of the flow cytometry histogram for each infected population was compared with the fluorescence observed in uninfected cells (set at 100%). *, P < 0.01, compared with infected DMSO-treated cells; n ≥ 3.
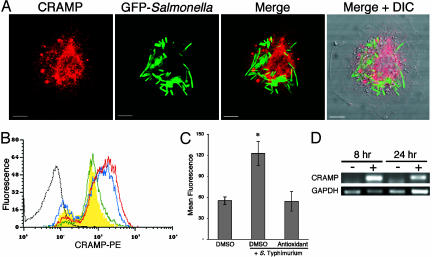
Macrophages Express the Cathelicidin CRAMP. Mechanisms by which proteolytic activity could cause this replication defect include damaging bacteria directly and activating cationic antimicrobial peptides (28). We tested the hypothesis that a cathelicidin mediates Salmonella filamentation within macrophages because a synthetic active peptide of indolicidin, a bovine cathelicidin, has been reported to induce bacterial filamentation in vitro (29). Mice produce a single cathelicidin, named CRAMP. When activated by proteolytic cleavage, the peptide has microbicidal activity against S. typhimurium (24), although its expression or biological function in macrophages has not been reported. By using confocal microscopy and an Ab specific for both the inactive proprotein and proteolytically active CRAMP peptide, BMDM (Fig. 3A) and splenic macrophages infected in vivo or in vitro (data not shown) express CRAMP in a punctate pattern, with more intense staining in the perinuclear region. A similar pattern was observed in uninfected macrophages and was not observed in unpermeabilized cells, indicating that CRAMP is intracellular (data not shown). S. typhimurium resides in a similar perinuclear location, with some overlap with CRAMP (Fig. 3A). Flow cytometry confirmed that CRAMP is expressed by uninfected macrophages (Fig. 3B, yellow curve) compared with cells incubated with secondary Ab alone (Fig. 3B, gray dotted line) or unpermeabilized cells (data not shown). CRAMP expression was significantly increased in macrophages after Salmonella infection (Fig. 3B, red line), and this increase was abrogated in infected macrophages pretreated with the antioxidants N-acetyl cysteine (Fig. 3B, blue dashed line, and quantified in Fig. 3C) or acetovanillone (data not shown). Elevated CRAMP protein in infected cells correlates with an increased abundance of CRAMP mRNA (Fig. 3D).
Fig. 3.
Macrophages express the antimicrobial cathelicidin CRAMP. (A) Confocal microscopy. Macrophages were infected with S. typhimurium expressing GFP (green) for 24 h and processed for immunofluorescence by using a CRAMP-specific Ab (red). CRAMP and GFP are flat projections of 50 optical sections, and the overlay is composed of single confocal sections. (Scale bar, 5 μm; n = 4.) (B) Flow cytometry. CRAMP expression in macrophages is up-regulated by S. typhimurium infection and modulated by ROIs. Macrophages were infected and processed as described in A and then analyzed by flow cytometry. CRAMP expression was detected in uninfected macrophages (yellow curve) when compared with cells incubated with secondary Ab alone (gray dotted line). CRAMP expression was elevated in infected macrophages (red line) and was decreased in infected cells treated with the antioxidant N-acetylcysteine (blue dashed line). A representative experiment is shown. (C) Quantification of flow cytometry. The histogram mean ± SD is shown. *, P < 0.01, compared with uninfected cells (n = 4). (D) RT-PCR. CRAMP mRNA is up-regulated in Salmonella-infected cells at 8 and 24 h after infection, relative to GAPDH abundance. –, uninfected; +, infected; n = 3.
CRAMP Mediates Filamentation in Vitro. An in vitro assay was developed to determine whether chemically synthesized active CRAMP peptide can impair S. typhimurium cell division in a way that is similar to the stressed bacterial morphology (filamentation) within macrophages. As shown in Fig. 4A, 10 μM CRAMP induced filamentous bacteria with arrested septum formation at pH 7.4, and this morphology was unaffected by subsequent incubation with protease inhibitors or antioxidants (Fig. 4A and data not shown). To quantify this increase in bacterial length, wild-type S. typhimurium was cultured with increasing concentrations of CRAMP peptide. An increase in culture OD was observed between 3 and 10 μM (Fig. 4b), which correlated with increasing bacterial length (Fig. 4C, Wild type), whereas cfu remained constant (data not shown). Filamentous bacteria of increasing length were observed at CRAMP concentrations of up to 15 μM (Fig. 4C), but this dose also had a partial bacteriostatic effect, resulting in decreasing culture OD (Fig. 4B) and cfu (data not shown). A concentration of 20 μM CRAMP was bacteriostatic, resulting in nonturbid cultures containing small, round bacteria (Fig. 4C). The majority of filamentous bacteria were viable (exclusion of propidium iodide), and many of them were motile (microscopic analysis; data not shown). Bacteria incubated in minimal media (pH 7.4) in the presence of the chemical inhibitors used in this study were not impaired in growth or cell division (data not shown). We have observed that filamentous S. Typhimurium, induced by incubation with synthetic CRAMP in vitro, are phagocytosed by macrophages and are rapidly killed (data not shown), suggesting that induction of filamentation could interrupt the cycle of infection.
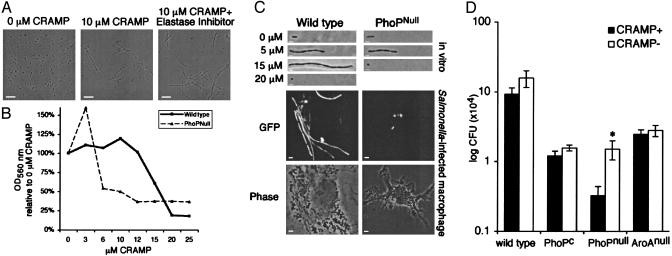
Fig. 4.
CRAMP mediates Salmonella filamentation in vitro and in vivo.(A) Fluorescence microscopy of bacteria in vitro. S. typhimurium was cultured overnight in a minimal media with or without 10 μM active synthetic CRAMP peptide. Filamentation is unaffected by culture with the protease inhibitor MeOSuc-AAPV-CMK for 24 h. (Scale bar, 10 μm; n = 5.) (B) Quantification of filamentation. PhoPnull mutant has increased susceptibility to CRAMP. Wild-type and PhoPnull bacteria were cultured as described in A in the presence of increasing concentrations of CRAMP, and the extent of bacterial filamentation and growth arrest were quantified by OD560.OD560 at 16 h were normalized to the absorbance of bacteria cultured in parallel without CRAMP (100%) (n = 4). (C) Fluorescence microscopy of bacteria in vitro and in vivo. PhoP mediates resistance to CRAMP. (Top) Bacteria expressing GFP were cultured in the presence of increasing concentrations of CRAMP in vitro as described in B, and illustrative individual filamentous bacteria were photographed. (Bottom) BMDM were infected for 24 h with the indicated bacterial strains and infected cells were examined by microscopy. (Scale bar represents 1 μm for in vitro images and 2 μm for infected macrophage images.) (D) Quantification of bacterial cfu. Significant increase in the number of PhoPnull bacteria at 24 h in CRAMP– macrophages. BMDM derived from CRAMP+ and CRAMP– mice were infected with S. typhimurium 14028 wild type, PhoPC, PhoPnull, or AroAnull mutants. Shown are the mean ± SEM of three independent experiments performed in duplicate, with each sample plated in triplicate. *, P = 0.03; n = 6.
Salmonella Minimizes Filamentation by Using a PhoP–PhoQ-Dependent Mechanism. Because the PhoP–PhoQ two-component regulatory system mediates Salmonella resistance to other cationic antimicrobial peptides in vitro, PhoP mutants were exposed to CRAMP in vitro. A PhoPnull mutant, which is more sensitive to damage from other antimicrobial peptides (19), exhibited filamentation and the corresponding increase in OD when cultured with 3–5 μM CRAMP (Fig. 4 B and C). Moreover, it was highly susceptible to lower CRAMP concentrations compared with wild-type S. typhimurium, preventing an increase in culture turbidity and yielding small, round bacteria at CRAMP concentrations of 10 μM or higher (Fig. 4 B and C). In contrast, a PhoPC constitutive active mutant, which can modify its LPS structure and decrease its sensitivity to antimicrobial peptides in vitro (20), does not undergo filamentation (data not shown). Paralleling what was observed in vitro, many wild-type bacteria become filamentous within BMDM, whereas the PhoPnull mutant does not (Fig. 4C, macrophage). A PhoPnull mutant in the 14028s strain background demonstrates the same increased susceptibility to CRAMP (data not shown).
Because PhoP-regulated gene expression can protect S. typhimurium from CRAMP in vitro, we investigated whether PhoP regulates resistance to the concentration of CRAMP in macrophages. BMDM from Cnlp–/– CRAMP– and congenic wild-type mice were infected with S. typhimurium (14028s). The presence of CRAMP did not affect the number of bacteria that were internalized by macrophages (data not shown). Significantly more intracellular cfu was observed in CRAMP– macrophages after 24 h of infection with the S. typhimurium PhoPnull mutant (Fig. 4D). This mutant was more susceptible to impaired cell division and filamentation after in vitro incubation with CRAMP (Fig. 4 B and C). Similar results were observed by using wild-type and PhoPnull mutants in an SL1344 background (data not shown; P < 0.01; n = 6). In contrast, there was no difference in the number of intracellular cfu within CRAMP+ or CRAMP– macrophages infected with the peptide-resistant PhoPC mutant (Fig. 4D). Also, three other bacterial mutants attenuated for intramacrophage replication, the auxotroph ΔaroA and the virulence factor mutants ΔssaR and ΔsifA, are unaffected by the presence of CRAMP (Fig. 4D and data not shown) Therefore, S. typhimurium resists the bacteriostatic activities of CRAMP by a PhoP-dependent mechanism.
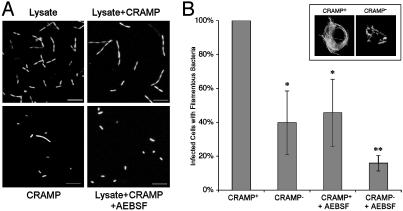
CRAMP and Proteases Cooperate to Impair Salmonella Cell Division in Vitro and in Vivo. To assess whether macrophage proteases could act in combination with CRAMP and impair bacterial cell division directly, macrophage lysates were incubated with S. typhimurium in vitro. Macrophage lysate impaired bacterial cell division and induced filamentation (Fig. 5A) in a dose-dependent manner (data not shown). Activity was observed at pH 5.8, the predicted pH of the SCV, and higher concentrations of CRAMP were required to induce filamentation at this lower pH (data not shown). Filamentation was more dramatic when bacteria were incubated with a combination of lysate and CRAMP, even at a concentration of CRAMP that induced little filamentation on its own. Filamentation was abrogated by addition of the serine protease inhibitor AEBSF (Fig. 5A), although this inhibitor alone had no effect on bacterial morphology (data not shown).
Fig. 5.
Cooperativity between CRAMP and protease effectors. (A) Confocal microscopy. S. typhimurium plus GFP was incubated for 16 h in minimal media (pH 5.8) with combinations of macrophage lysate (0.5 mg/ml protein), CRAMP (100 μM), and AEBSF (500 μM). Representative flat projections of 20 0.2-μm sections are shown (n = 3). (B) Fluorescence microscopy. CRAMP expression correlates with bacterial filamentation in vivo. BMDM derived from CRAMP+ and CRAMP– mice, with or without treatment with AEBSF, were infected with wild-type S. typhimurium plus GFP and examined 24 h after infection. The mean percentage of infected cells containing filamentous bacteria ± SD is shown relative to the number observed in CRAMP+ cells (14 ± 6%, normalized to 100%; n = 5). (Inset) Fluorescence microscopy of CRAMP+ or CRAMP– BMDM infected for 24 h with S. typhimurium plus GFP. *, P < 0.01, compared with CRAMP+ cells; **, significance relative to both CRAMP+ and CRAMP– BMDM; n = 4–5.
To determine whether CRAMP is required to impair bacterial cell division within macrophages, BMDM were derived from Cnlp+/+ and Cnlp–/– mice (15), and the bacterial morphology within infected cells was examined after 24 h. As shown in Fig. 5B, CRAMP– macrophages were impaired significantly in their ability to cause filamentation of intracellular S. typhimurium compared with cells expressing CRAMP. Because some filamentation was still observed in the absence of CRAMP, BMDM with or without CRAMP were treated with the serine protease inhibitor AEBSF before infection. The number of macrophages containing filamentous bacteria was reduced to a similar extent in CRAMP+ cells treated with protease inhibitor and CRAMP– cells with intact protease activity, relative to wild-type cells. CRAMP– macrophages treated with AEBSF were impaired further in their ability to interrupt bacterial cell division, indicating that protease activity also mediates filamentation independently of CRAMP activation.
Discussion
We have elucidated previously uncharacterized macrophage effector mechanisms for controlling the replication of a pathogenic bacterium. In addition, we observe interplay between these antibacterial effectors. S. typhimurium infection of macrophages increases intracellular ROIs, which are required for increased expression of the cationic antimicrobial protein CRAMP. Serine protease activity within macrophages impairs bacterial cell division both by proteolytic release of active CRAMP peptide and by a direct mechanism. Elastase-like proteases can be antimicrobial in their enzymatic (12) or nonenzymatic (30) activities. The cooperation between macrophage elastase-like proteases and CRAMP in vitro and in vivo is similar to synergy observed between lysozyme and defensins (31).
Cationic peptides play a central role in control of microbial infection, and deficiency of LL-37, the human homologue of CRAMP, may lead to susceptibility to chronic skin and oral infections (32, 33). High concentrations of antimicrobial peptides are stored by neutrophils and are expressed at lower levels by granulocytes, mast cells, keratinocytes, and epithelial cells (28). Although these antimicrobial peptides can be released, can be activated by extracellular proteases, and serve important roles in the killing of extracellular pathogens, their regulation within phagocytes and their activity against intracellular pathogens or phagocytosed bacteria are not well understood. Ectopic expression of foreign peptides has suggested that peptides can be important mediators of innate immune control of bacterial pathogens. Human LL-37 augments mouse lung defense against Pseudomonas aeruginosa (34), human defensin 5 in the intestine makes mice more resistant to oral challenge with S. typhimurium (35), and human defensin expression in macrophages controls replication of pathogens such as Mycobacterium tuberculosis (36, 37). Here, we show the expression, activation, and activity of an endogenous cationic peptide within macrophages, an important target cell of many significant intracellular pathogens. The residual filamentation observed in Cnlp–/– macrophages may suggest a contribution by other antimicrobial peptides.
Salmonella resistance to cationic peptides in vitro has been well described. PhoP–PhoQ is a two-component regulatory system that mediates pleiotropic changes in S. typhimurium gene expression (22). PhoPnull mutants are highly susceptible to growth inhibition by peptides in vitro, which is presumably caused by an inability to mediate LPS modifications that protect bacteria from membrane damage from peptides (20). Here, we show that S. typhimurium is exposed to cationic peptides within the endosomal compartment of macrophages. The increased cfu of Phonull mutants within CRAMP-deficient macrophages provides direct evidence that the PhoP regulon protects S. typhimurium from cationic peptides within macrophages. However, the PhoP-PhoQ regulatory system appears to mediate resistance to other peptides or regulates other virulence phenotypes because removal of CRAMP does not repair the attenuation of the PhoPnull mutant fully.
Perhaps the best-characterized antimicrobial effector is the oxidative burst. It has been shown in neutrophils that the influx of ROIs and potassium ions into the phagosome cooperate to activate the serine proteases neutrophil elastase and cathepsin G, and that protease activity is responsible for impaired bacterial survival (11). Neutrophil elastase can proteolytically cleave virulence proteins secreted by S. typhimurium (38). However, mature macrophages do not appear to express neutrophil elastase (39). We detected macrophage serine protease activities demonstrating activity and inhibitor sensitivity similar to elastases. This activity is regulated in a manner that is distinct from elastase in neutrophils because it is not sensitive to inhibitors of ROIs and potassium channels (Fig. 2 and data not shown). Rather, we show that an antibacterial effector regulated by ROIs within macrophages is the expression of CRAMP. Therefore, both macrophages and neutrophils possess indirect mechanisms of oxidase-dependent impairment of bacterial replication. In summary, these data indicate that ROIs and intracellular proteases can impair cell division of a bacterial pathogen within macrophages by means of complementary yet independent mechanisms by regulating the expression and activity of an antimicrobial peptide.
Acknowledgments
We thank Anna DiNardo for providing bone marrow from Cnlp+/+ and Cnlp–/– mice, Bob Hancock for providing synthetic CRAMP peptide, and the University of British Columbia BioImaging Facility and the Biomedical Research Centre MultiUser Flow Cytometry Facility for support. We also thank members of the Finlay laboratory, Bob Hancock, and Monisha Scott for insightful discussions and comments on the manuscript. This work was supported by the Canadian Institutes for Health Research and a Howard Hughes Medical Institute International Research Scholar award. C.M.R. was supported by Canadian Institutes for Health Research and Michael Smith Foundation for Health Research studentships. B.B.F. is the recipient of a Canadian Institutes for Health Research Distinguished Investigator award and is the University of British Columbia Peter Wall Distinguished Professor. R.L.G is supported by National Institutes of Health Grants AR45676 and AI52453 and a Veterans Affairs Merit Award.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AEBSF, 4-(2-aminoethyl)benzenesulfonyl fluoride; BMDM, bone marrow-derived macrophages; cfu, colony-forming unit; CMK, chloromethylketone; CRAMP, cathelicidin-related antimicrobial peptide; DPI, diphenyleneiodonium; LPS, lipopolysaccharide; MeOSuc, methoxysuccinyl; phox, phagocyte oxidase; ROI, reactive oxygen intermediate.
References
- 1.Rosenberger, C. M. & Finlay, B. B. (2003) Nat. Rev. Mol. Cell Biol. 4, 385–396. [DOI] [PubMed] [Google Scholar]
- 2.Richter-Dahlfors, A., Buchan, A. M. & Finlay, B. B. (1997) J. Exp. Med. 186, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salcedo, S. P., Noursadeghi, M., Cohen, J. & Holden, D. W. (2001) Cell. Microbiol. 3, 587–597. [DOI] [PubMed] [Google Scholar]
- 4.Vazquez-Torres, A., Xu, Y., Jones-Carson, J., Holden, D. W., Lucia, S. M., Dinauer, M. C., Mastroeni, P. & Fang, F. C. (2000) Science 287, 1655–1658. [DOI] [PubMed] [Google Scholar]
- 5.Chakravortty, D., Hansen-Wester, I. & Hensel, M. (2002) J. Exp. Med. 195, 1155–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberger, C. M. & Finlay, B. B. (2002) J. Biol. Chem. 277, 18753–18762. [DOI] [PubMed] [Google Scholar]
- 7.Davis, K. J., Vogel, P., Fritz, D. L., Steele, K. E., Pitt, M. L., Welkos, S. L., Friedlander, A. M. & Byrne, W. R. (1997) Arch. Pathol. Lab. Med. 121, 865–868. [PubMed] [Google Scholar]
- 8.Imlay, J. A. & Linn, S. (1987) J. Bacteriol. 169, 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schapiro, J. M., Libby, S. J. & Fang, F. C. (2003) Proc. Natl. Acad. Sci. USA 100, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belaaouaj, A., McCarthy, R., Baumann, M., Gao, Z., Ley, T. J., Abraham, S. N. & Shapiro, S. D. (1998) Nat. Med. 4, 615–618. [DOI] [PubMed] [Google Scholar]
- 11.Reeves, E. P., Lu, H., Jacobs, H. L., Messina, C. G., Bolsover, S., Gabella, G., Potma, E. O., Warley, A., Roes, J. & Segal, A. W. (2002) Nature 416, 291–297. [DOI] [PubMed] [Google Scholar]
- 12.Belaaouaj, A., Kim, K. S. & Shapiro, S. D. (2000) Science 289, 1185–1188. [DOI] [PubMed] [Google Scholar]
- 13.Zaiou, M. & Gallo, R. L. (2002) J. Mol. Med. 80, 549–561. [DOI] [PubMed] [Google Scholar]
- 14.Cole, A. M., Shi, J., Ceccarelli, A., Kim, Y. H., Park, A. & Ganz, T. (2001) Blood 97, 297–304. [DOI] [PubMed] [Google Scholar]
- 15.Nizet, V., Ohtake, T., Lauth, X., Trowbridge, J., Rudisill, J., Dorschner, R. A., Pestonjamasp, V., Piraino, J., Huttner, K. & Gallo, R. L. (2001) Nature 414, 454–457. [DOI] [PubMed] [Google Scholar]
- 16.Selsted, M. E., Brown, D. M., DeLange, R. J. & Lehrer, R. I. (1983) J. Biol. Chem. 258, 14485–14489. [PubMed] [Google Scholar]
- 17.Lehrer, R. I., Selsted, M. E., Szklarek, D. & Fleischmann, J. (1983) Infect. Immun. 42, 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields, P. I., Groisman, E. A. & Heffron, F. (1989) Science 243, 1059–1062. [DOI] [PubMed] [Google Scholar]
- 19.Groisman, E. A., Parra-Lopez, C., Salcedo, M., Lipps, C. J. & Heffron, F. (1992) Proc. Natl. Acad. Sci. USA 89, 11939–11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, L., Lim, K. B., Gunn, J. S., Bainbridge, B., Darveau, R. P., Hackett, M. & Miller, S. I. (1997) Science 276, 250–253. [DOI] [PubMed] [Google Scholar]
- 21.Bader, M. W., Navarre, W. W., Shiau, W., Nikaido, H., Frye, J. G., McClelland, M., Fang, F. C. & Miller, S. I. (2003) Mol. Microbiol. 50, 219–230. [DOI] [PubMed] [Google Scholar]
- 22.Ernst, R. K., Guina, T. & Miller, S. I. (1999) J. Infect. Dis. 179, Suppl. 2, S326–S330. [DOI] [PubMed] [Google Scholar]
- 23.Miller, S. I., Kukral, A. M. & Mekalanos, J. J. (1989) Proc. Natl. Acad. Sci. USA 86, 5054–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallo, R. L., Kim, K. J., Bernfield, M., Kozak, C. A., Zanetti, M., Merluzzi, L. & Gennaro, R. (1997) J. Biol. Chem. 272, 13088–13093. [DOI] [PubMed] [Google Scholar]
- 25.Senior, R. M., Campbell, E. J., Landis, J. A., Cox, F. R., Kuhn, C. & Koren, H. S. (1982) J. Clin. Invest. 69, 384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buchmeier, N. A. & Heffron, F. (1989) Infect. Immun. 57, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vazquez-Torres, A., Jones-Carson, J., Mastroeni, P., Ischiropoulos, H. & Fang, F. C. (2000) J. Exp. Med. 192, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallo, R. L., Murakami, M., Ohtake, T. & Zaiou, M. (2002) J. Allergy Clin. Immunol. 110, 823–831. [DOI] [PubMed] [Google Scholar]
- 29.Subbalakshmi, C. & Sitaram, N. (1998) FEMS Microbiol. Lett. 160, 91–96. [DOI] [PubMed] [Google Scholar]
- 30.Del Rosso, M., Fibbi, G. & Schmitt, M. (2002) Biol. Chem. 383, 1–4. [DOI] [PubMed] [Google Scholar]
- 31.Hancock, R. E. & Scott, M. G. (2000) Proc. Natl. Acad. Sci. USA 97, 8856–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong, P. Y., Ohtake, T., Brandt, C., Strickland, I., Boguniewicz, M., Ganz, T., Gallo, R. L. & Leung, D. Y. (2002) N. Engl. J. Med. 347, 1151–1160. [DOI] [PubMed] [Google Scholar]
- 33.Putsep, K., Carlsson, G., Boman, H. G. & Andersson, M. (2002) Lancet 360, 1144–1149. [DOI] [PubMed] [Google Scholar]
- 34.Bals, R., Weiner, D. J., Moscioni, A. D., Meegalla, R. L. & Wilson, J. M. (1999) Infect. Immun. 67, 6084–6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzman, N. H., Ghosh, D., Huttner, K. M., Paterson, Y. & Bevins, C. L. (2003) Nature 422, 522–526. [DOI] [PubMed] [Google Scholar]
- 36.Kisich, K. O., Heifets, L., Higgins, M. & Diamond, G. (2001) Infect. Immun. 69, 2692–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couto, M. A., Liu, L., Lehrer, R. I. & Ganz, T. (1994) Infect. Immun. 62, 2375–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinrauch, Y., Drujan, D., Shapiro, S. D., Weiss, J. & Zychlinsky, A. (2002) Nature 417, 91–94. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, H., Nukiwa, T., Basset, P. & Crystal, R. G. (1988) J. Biol. Chem. 263, 2543–2547. [PubMed] [Google Scholar]