Abstract
Histone acetylation of promoters precedes activation of many genes. In addition, long-range histone acetylation patterns can be established over many kilobases of the chromatin of linked families of genes that are under common transcriptional control. It is not known whether establishment of long-range histone acetylation patterns is limited to gene families or is a common feature of many genes. The Ifng gene is not known to be a member of a gene family but exhibits complex strain-, cell lineage-, and stimulus-dependent regulation. For example, stimulation of naive T cells through their antigen receptor does not initiate Ifng gene transcription. However, stimulation of naive T cells through their antigen and IL-12 receptors initiates differentiation programs that yield effector cells with 100-fold greater rates of transcription of the Ifng gene after stimulation through the antigen receptor. Here, we demonstrate that these differentiation programs establish long-range histone hyperacetylation patterns that extend at least 50 kb in both upstream and downstream directions of the Ifng gene. Establishment of these histone acetylation patterns and Ifng gene expression is relatively IL-12-independent in T cells from autoimmuneprone nonobese diabetic mice. These results indicate that gene expression programs that mediate T cell differentiation are regulated by long-range histone acetylation patterns and that defective control of these patterns may contribute to development of autoimmunity.
Assembly of an active transcriptional complex at the promoter is an essential feature of eukaryotic gene expression (1–3). Histone acetylation of the promoter precedes the activation of many genes and is thought to establish a chromatin environment suitable for the assembly of the transcriptional complex (4–7). Recent evidence indicates that genes that are members of gene families, are located in relative close proximity to each other, and are regulated in a coordinated fashion exhibit long-range histone acetylation patterns that extend over many kilobases of DNA (8–11). Establishment of long-range histone acetylation patterns appears to permit communication between regulatory regions and the genes they regulate over a distance of many kilobases. In contrast, it is not known whether genes that are not members of gene families but exhibit complex forms of regulation exhibit long-range histone acetylation patterns or whether histone acetylation is limited to the promoter or gene region.
Naive T cells do not efficiently transcribe the Ifng gene in response to activation of the T cell receptor. Under appropriate conditions, naive T cells will differentiate into effector T helper 1 (Th1)/T cytotoxic 1 (Tc1) cells that transcribe the Ifng gene at 100-fold greater rates than naive T cells in response to stimulation of their antigen receptor. A limiting step in the differentiation of naive T cell precursors into effector cells that produce IFN-γ is the requirement for both an antigen stimulus to drive cell division and an inflammatory signal to stimulate IL-12 production that drives T cell differentiation toward the Th1/Tc1 lineage (12, 13). Epigenetic events, including chromatin remodeling of promoter and introns, histone acetylation of the promoter, and nuclear positioning of the Ifng locus, contribute to successful completion of this differentiation program (7, 14–19). Using a transgenic approach, we have also shown that distal regulatory elements are required to achieve both high-level and lineage-specific expression of the Ifng gene (20). Therefore, long-range communication between distinct genomic regions is most likely a critical feature of Th1/Tc1 differentiation.
Studies in tissue culture models, by us and others, demonstrate that, in contrast to other murine strains, CD4+ T cells from the autoimmune prone nonobese diabetic (NOD) strain are genetically programmed to use IL-2 as a driving cytokine to differentiate into IFN-γ-producing effector T cells (21–23). Thus, an antigen stimulus is sufficient to drive both T cell proliferation and Th1 differentiation. We believe this property represents a liability that may contribute to tissue inflammatory responses and development of autoimmune diabetes.
The purpose of studies presented here was to determine whether induction of histone hyperacetylation patterns during T cell differentiation was limited to the Ifng gene or exhibited an extended long-range pattern. We also wanted to examine lineage- and strain-dependent regulation of histone acetylation. We find that histone hyperacetylation of the Ifng promoter and introns 1 and 3 depends on stimulation of C57BL/6 CD8+ T cells through both antigen and IL-12 receptors. In contrast, NOD CD8+ T cells require stimulation only through their antigen receptors to achieve a similar level of histone acetylation. Surprisingly, histone hyperacetylation of the Ifng gene region extends far beyond the gene itself, and a discrete pattern of histone hyperacetylation is evident as far as ≈50 kb upstream and downstream of the Ifng gene. This finding argues that proper regulation of Ifng gene expression requires extended histone acetylation patterns across relatively large chromatin domains. Inhibition of histone deacetylases is sufficient to induce IFN-γ production by C57BL/6 CD8+ T cells in the presence of only a T cell receptor stimulus. Therefore, the major function of IL-12 in T cell differentiation may be to coordinate the establishment of long-range histone hyperacetylation patterns in the Ifng gene region.
Materials and Methods
Mice. NOD, nonobese diabetes-resistant (NOR), and C57BL/6 mice were obtained from The Jackson Laboratory, bred in the Vanderbilt University animal facilities, and used between 4 and 6 wk of age.
Cell Purification and Cultures. Splenic T cells were purified by negative selection by using specific mAbs essentially as described (24). A magnetic-activated cell sorting system (Miltenyi Biotec, Auburn, CA) was used to separate memory and naive CD8+ T cells (anti-CD62L). Purified T cells were stimulated with immobilized anti-CD3 (2C11, American Type Culture Collection) as outlined in the text essentially as described in ref. 24. IFN-γ assays were performed by ELISA with mAbs recommended by BD Pharmingen.
Chromatin Immunoprecipitation (ChIP) Assays. For ChIP assays (acetyl-histone H4 ChIP assay kit, Upstate Biotechnology, Lake Placid, NY), ≈1 × 107 T cells were processed following the manufacturer's protocol. Briefly, cells were fixed with 1% paraformaldehyde and lysed in 1% SDS/10 mM EDTA/50 mM Tris-HCl, pH 8.1. Lysates were sonicated to achieve an average length of genomic DNA of ≈500 bp. Protein–DNA complexes were immunoprecipitated overnight at 4°C with anti-acetylhistone H4 Ab or normal rabbit IgG as control. Ab–histone–DNA complexes were purified with protein A agarose. Aliquots (20 μl) were processed without immunoprecipitation as input controls. DNA was purified after reversal of crosslinks at 65°C.
PCR primers for the ChIP assay spanned nonrepetitive conserved (mouse and human) genomic sequences from ≈60 kb upstream to ≈60 kb downstream of the Ifng gene. Sequence conservation was identified by using the Pipmaker web server (25). PCR primers were as follows: promoter, ctgtgctctgtggatgagaaat and aagatggtgacagataggtggg (PCR product, 254 bp); intron 1a, ggtccaaggtacaaagatgct and gaactttgcctcccattacttta (184 bp); intron 1b, cctgcagctaaaagaatgtaaca and ttccacatctatgccacttgag (223 bp); intron 3, tgtggcctaattactcatgctc and atggaaaggcagaagcaaagtc (161 bp); –53 kb, gcccacagatgccagtttaga and agggccacggttgtcaga (144 bp); –40 kb, gagggctttgggtgaactgtt and cccctattaaacatggtctcaaga (188 bp); –28 kb, aactgcttatgctggatttgagat and ctcctatgcttattggctggtcta (137 bp); –6 kb, cccagtgagtgctttaaaatttct and ctggatggttttgaaggataatgt (184 bp); –0.4 kb, cggggctgtctcatcgtc and ctcgggattacgtattttcacaa (165 bp); +10 kb, gacgcctagcagacaccatact and tcccagggctcccacaa (130 bp); +30 kb, cctggctgtgtgctgactca and gagcgatacggtcacagtgttt (153 bp); +40 kb, ggtcgtaagcacactgaggtca and tggctaaagatgggaacaagaata (176 bp); +59 kb, gccagtggagatgacgtgatta and ttcctgcaagtggttctctgatt (142 bp); Actb, tggcaccacaccttctacaat and ctcggtcaggatcttcatgag (310 bp).
Standard PCR conditions were used for 25, 30, or 35 cycles. PCR products were resolved by agarose gel electrophoresis, and image intensities were obtained by using the Stratagene Eagle Eye imaging and software packages. For quantitative comparisons, we used two procedures to normalize levels of histone acetylation. First, we divided the yield of the anti-H4 product by the input control product to normalize for varying efficiencies of different primer pairs. Second, we normalized to levels of histone acetylation of the Actb gene to control for variations among different experiments.
Results
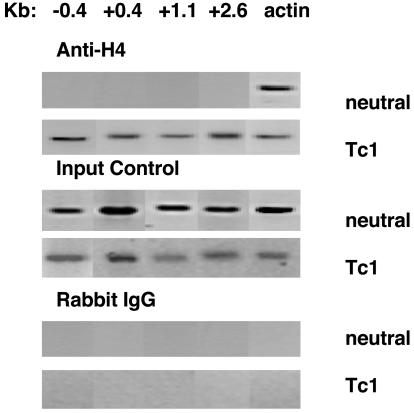
Long-Range, Lineage-Dependent Histone Hyperacetylation of the Ifng Gene Region. Activation of resting C57BL/6 T cells through their antigen and IL-12 receptors initiates differentiation programs that result in high levels of Ifng transcription within 72 h. In contrast, activation through their antigen receptor alone activates minimal Ifng transcription. To investigate changes in patterns of histone acetylation during T cell differentiation, we stimulated purified CD8+ T cells with plate-bound anti-CD3 with or without IL-12 (5 ng/ml) and anti-IL-4 mAb (11B11, 10 μg/ml). After 72 h, cells were harvested and processed for levels of H4-acetyl histone by ChIP assay. Cultures fluids were also harvested and analyzed for levels of IFN-γ protein by ELISA. Cultures from cells stimulated with or without IL-12 contained 288 ± 18 or 12 ± 8 ng/ml IFN-γ, respectively. First, we analyzed levels of acetylated histone H4 at the promoter, two sites in the first intron, and one site in the third intron of the Ifng gene by ChIP assay. These regions were relatively H4 histone-hypoacetylated in cells cultured without IL-12 and anti-IL-4 mAb (neutral conditions) (Fig. 1). In contrast, these regions were H4 histone-hyperacetylated in cells cultured with IL-12 and anti-IL-4 mAb (Tc1 conditions).
Fig. 1.
Lineage-dependent H4 histone hyperacetylation of the Ifng gene reflects gene activity. C57BL/6 CD8+ T cells were purified and stimulated with anti-CD3 under neutral or Tc1 conditions (5 ng/ml IL-12 and 10 μg/ml anti-IL-4 mAb). Cells were harvested on day 3 and processed for ChIP assay. The approximate positions of the PCR primers used in the ChIP assay are shown at the top. The Actb gene served as a positive control. All PCR products were of the expected size (see Materials and Methods).
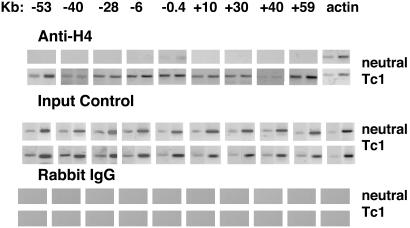
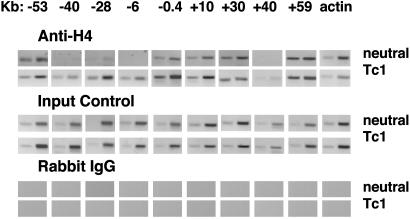
Using a transgenic approach, we demonstrated previously that distal regulatory elements are required to achieve both high-level and lineage-dependent expression of the Ifng gene (20). Therefore, we next wanted to determine whether lineage-dependent histone hyperacetylation extended beyond the Ifng gene. To do so, we designed primers at ≈10-kb intervals that spanned ≈50 kb upstream and downstream of the Ifng gene. ChIP analysis demonstrated extensive lineage-dependent histone H4 hyperacetylation across this genomic region from –53 kb to +59 kb 5′ and 3′ of the Ifng gene in T cells cultured under Tc1 conditions (Fig. 2). In contrast, histone H4 acetylation across this region was essentially undetectable in CD8+ T cells cultured under neutral conditions.
Fig. 2.
Long-range histone acetylation of the Ifng gene region during T cell differentiation. C57BL/6 CD8+ T cells were purified, stimulated under neutral or Tc1 conditions, and harvested and processed for ChIP assays as described in Fig. 1. The approximate positions of the different primer pairs relative to the Ifng gene are shown at the top. (Top) Anti-H4 histone immunoprecipitates. (Middle) Aliquots of purified total genomic DNA were amplified by PCR without immunoprecipitation. (Bottom) A control immunoprecipitation was performed with a nonspecific rabbit IgG, and samples were processed for ChIP assay.
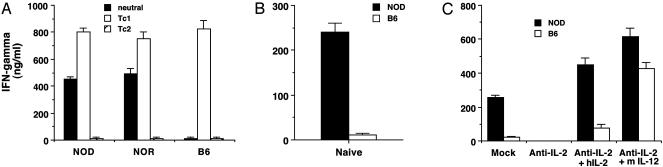
Strain-Dependent Variations in Histone Hyperacetylation of the Ifng Gene Region. In contrast to most strains, CD4+ T cells from autoimmune-prone NOD mice do not require IL-12 to differentiate into IFN-γ producers (21–23); they retain a cytokine requirement for this differentiation process. However, IL-2, which T cells produce, is sufficient to direct CD4+ T cells toward the Th1 lineage of IFN-γ-producing cells. We compared cytokine requirements for differentiation of NOD, NOR, or B6 CD8+ T cells into producers of IFN-γ. NOD or NOR CD8+ T cells, but not B6 CD8+ T cells, differentiated into strong producers of IFN-γ in the absence of IL-12 (Fig. 3A). Addition of IL-4 (Tc2 conditions) suppressed IFN-γ production by NOD or NOR T cells. Purified naive NOD CD8+ T cells produced markedly greater levels of IFN-γ than did naive B6 CD8+ T cells after stimulation by plate-bound anti-CD3 alone (Fig. 3B), although cells from both strains exhibited similar levels of cell proliferation (data not shown). This indicates that previous activation is not required for increased IFN-γ production by NOD T cells. In culture, NOD and B6 T cells produced approximately equivalent amounts of IL-2 (data not shown). Levels of IL-12 and IL-18 in these cultures were under the limit of our detection (<30 pg/ml). When neutralizing mAb against mouse IL-2, IL-12, or IFN-γ were added to the primary cultures, IFN-γ production by NOD CD8+ T cells in secondary culture was markedly inhibited only by anti-IL-2 mAb but not by other mAbs (Fig. 3C; anti-IL-2 mAb is shown, and others are not shown). This amount of anti-IL-2 did not inhibit T cell proliferation (data not shown). Either human IL-2 or mouse IL-12 abrogated anti-IL-2 mAb-induced inhibition of IFN-γ production (Fig. 3C). Therefore, IL-2 functioned as a potent IFN-γ inducer for NOD T cells but not C57BL/6 T cells.
Fig. 3.
Strain-dependent variations in IFN-γ production. (A) CD8+ T cells were purified from the indicated strains and stimulated with anti-CD3 mAb under neutral, Tc1, or Tc2 culture conditions. IFN-γ levels produced by NOD, NOR, and C57BL6 CD8+ T cells are shown. (B) IFN-γ production by purified NOD or C57BL/6 naive T cells after anti-CD3 stimulation. (C) NOD or C57BL/6 CD8+ T cells were stimulated with plate-bound anti-CD3 in the presence or absence of anti-murine IL-2 mAb, human IL-2, or murine IL-12.
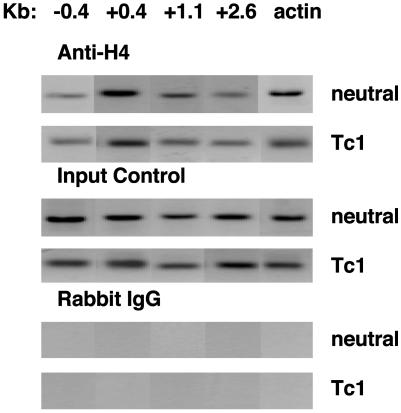
To characterize the state of histone H4 acetylation of the Ifng gene in NOD CD8+ T cells, we performed ChIP assays after in vitro stimulation. Under neutral or Tc1 conditions (IL-12 and anti-IL-4 mAb), NOD CD8+ T cells exhibited histone hyperacetylation of the promoter and introns 1 and 3 of the Ifng gene (Fig. 4). This finding is in contrast to C57BL/6 CD8+ T cells, where culture under Tc1 conditions was required to achieve histone hyperacetylation of the Ifng gene (Fig. 2). Levels of histone hyperacetylation were comparable in NOD and C57BL/6 CD8+ T cells after culture under Tc1 conditions. This correlation suggests that histone hyperacetylation plays a critical role in regulation of Ifng gene expression and contributes to increased IFN-γ production by NOD T cells after culture under neutral conditions.
Fig. 4.
Strain-dependent differences in H4 histone hyperacetylation of the Ifng gene reflect gene activity. NOD CD8+ T cells were purified and stimulated with anti-CD3 under neutral or Tc1 conditions. Cells were harvested on day 3 and processed for ChIP assay. The approximate positions of the PCR primers used in the ChIP assay are shown at the top. The Actb gene served as a positive control.
Given the above results, we wanted to determine whether histone hyperacetylation was limited to the Ifng gene promoter and introns or spread beyond the boundaries of the Ifng gene. To address this question, we repeated the above ChIP analysis from –53 kb upstream to +59 kb downstream of the Ifng gene using NOD CD8+ T cells cultured under neutral or Tc1 conditions. Levels of histone hyperacetylation across this region were indistinguishable between NOD and C57BL/6 T cells cultured under Tc1 conditions (Fig. 5, compare with Fig. 2). Sites at –53, –40, –28, –6, –0.4, +10, +30, and +59 kb were hyperacetylated. Histone H4 acetylation was very weak at the +40-kb site. Histone H4 hyperacetylation was also very evident in NOD T cells cultured under neutral conditions. This finding is in marked contrast to C57BL/6 CD8+ T cells, in which the chromatin across this region exhibited undetectable levels of histone H4 acetylation.
Fig. 5.
Long-range histone acetylation of the Ifng gene region during T cell differentiation reflects strain- and lineage-dependent differences in transcription. NOD CD8+ T cells were purified, stimulated under neutral or Tc1 conditions, and harvested and processed for ChIP assays as described in Fig. 1. The approximate positions of the different primer pairs relative to the Ifng gene are shown at the top. (Top) Anti-H4 histone immunoprecipitates. (Middle) Aliquots of purified total genomic DNA amplified by PCR before immunoprecipitation. (Bottom) A control immunoprecipitation.
The pattern of histone hyperacetylation observed in NOD CD8+ T cells cultured under Tc1 conditions was not completely identical to that observed in NOD CD8+ T cells cultured under neutral conditions. Sites at –28 and –6 kb were relatively hypoacetylated in T cells cultured under neutral conditions but were hyperacetylated in T cells from both C57BL/6 and NOD strains cultured under Tc1 conditions. These results argue that culture of NOD T cells culture under neutral conditions does not entirely recapitulate the differentiation program induced in NOD or C57BL/6 T cells by culture under Tc1 conditions.
Kinetic Analysis of Long-Range Histone Acetylation. Next we wanted to examine kinetics of histone acetylation of the Ifng gene region after activation of B6 CD8+ T cells cultured under neutral, Tc1, or Tc2 conditions. To control for interexperimental variations and efficiencies of different primers, we performed semiquantitative analysis of levels of histone H4 hyperacetylation by measuring band intensity obtained from PCR products after 25, 30, or 35 cycles of amplification selecting the linear range of amplification. We also included in our analysis results from Tc2 cultures (IL-4 and anti-IFN-γ mAb). The immediate Ifng gene region displayed low levels of histone acetylation within 1 day after activation that was quantitatively similar in cells from neutral or Tc1 cultures (Table 1). Between days 2 and 3, levels of histone acetylation progressively increased and spread across the 100-kb region in Tc1 cultures. In contrast, histone H4 acetylation in neutral cultures failed to spread and diminished to undetectable levels by day 3. The entire 100-kb region was not histone-acetylated in Tc2 cultures throughout this time period. We also examined levels of histone H3 acetylation by ChIP assay at select sites (–53, +0.4, and +59 kb) and found that levels of histone H3 acetylation were comparable to levels of H4 acetylation under the different strain and culture conditions (data not shown). In NOD T cells cultured under neutral conditions, the –40- to –6-kb region was relatively hypoacetylated, whereas the 0- to +30-kb region was relatively hyperacetylated. In contrast, both of these regions were hyperacetylated in both NOD and C57BL/6 CD8+ T cells cultured under Tc1 conditions. Taken together, these data suggest that there may be distinct chromatin subdomains in this region, whose histone acetylation is controlled by distinct regulatory mechanisms.
Table 1. Kinetics of long-range histone H4 acetylation across the ≈100-kb Ifng gene region during T helper cell differentiation.
| -53 | -40 | -28 | -6 | -0.4 | +0.4 | +1.1 | +2.6 | +10 | +30 | +40 | +59 | IFN-γ, ng/ml | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d1 (B6) | |||||||||||||
| Neutral | 0 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | <0.03 |
| Tc1 | 2 | 0 | 1 | 3 | 3 | 1 | 2 | 1 | 2 | 0 | 1 | 0 | <0.03 |
| Tc2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.03 |
| d2 (B6) | |||||||||||||
| Neutral | 0 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 0 | 0 | 1 | <0.03 |
| Tc1 | 3 | 0 | 2 | 5 | 5 | 2 | 3 | 3 | 3 | 3 | 1 | 4 | 2 ± 2 |
| Tc2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.03 |
| d3 (B6) | |||||||||||||
| Neutral | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 ± 8 |
| Tc1 | 7 | 2 | 6 | 8 | 9 | 6 | 6 | 6 | 6 | 7 | 1 | 12 | 288 ± 18 |
| Tc2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 ± 2 |
| d3 (NOD) | |||||||||||||
| Neutral | 8 | 0 | 0 | 0 | 4 | 5 | 6 | 6 | 4 | 8 | 0 | 12 | 260 ± 20 |
| Tc1 | 8 | 2 | 4 | 5 | 8 | 7 | 6 | 7 | 6 | 8 | 1 | 12 | 430 ± 35 |
| Tc2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 25 ± 4 |
Relative levels of histone acetylation at the indicated positions (kb) relative to the Ifng start site were determined by ChIP assay and are expressed in relative terms scaled from 0 to 10 (see Materials and Methods). Levels of IFN-γ in culture fluids were measured by ELISA (±SEM). ChIP analyses were performed a minimum of three times. SEM for each primer pair used for the ChIP analysis were <1 and are not shown.
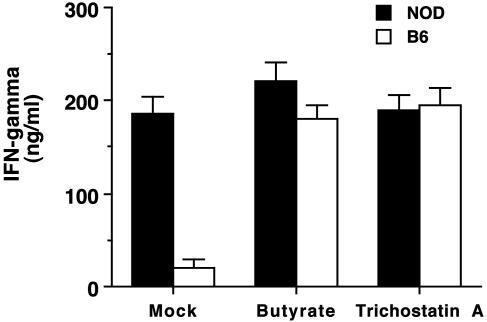
Inhibition of Histone Deacetylation Induces IFN-γ Production and Histone Acetylation of the Ifng Gene Region. The level of histone acetylation reflects a balance between the relative activities of histone acetyl transferases and histone deacetylases at a given genomic region (26, 27). To examine effects of inhibition of histone deacetylation on Ifng gene expression, we treated CD8+ T cells with two inhibitors of histone deacetylases (butyrate or trichostatin A) for 3 days under neutral conditions. Inhibition of histone deacetylases increased IFN-γ expression by C57BL/6 CD8+ T cells to levels comparable to that of NOD CD8+ T cells (Fig. 6). The amount of IFN-γ production by NOD T cells did not change substantially after culture with either trichostatin A or butyrate. The data suggest that, compared with C57BL/6 T cells, NOD T cells under neutral conditions may have impaired histone deacetylase activity at the Ifng gene region and that this may contribute to the elevated IFN-γ production by NOD CD8+ T cells cultured under neutral conditions.
Fig. 6.
Stimulation of IFN-γ production by histone deacetylase inhibitors. NOD and C57BL/6 CD8+ T cells were purified and stimulated with anti-CD3 in the presence or absence of the histone deacetylase inhibitors butyrate or trichostatin A. Cultures were harvested on day 4 and assayed for levels of IFN-γ by ELISA.
Next, we determined whether inhibition of histone deacetylase by trichostatin A caused an increase in histone acetylation across the Ifng gene region and whether IL-4 would inhibit the increase in both histone acetylation and IFN-γ production induced by trichostatin A. For these experiments, we did not add anti-IFN-γ mAb to the cultures to permit direct examination of the role of IL-4 in regulation of histone acetylation of the Ifng gene region. As we expected, trichostatin A induced increased histone acetylation across the Ifng gene region (Table 2). This increase in histone acetylation was inhibited by IL-4, as was IFN-γ production induced by trichostatin A. These results and those shown in Table 1 demonstrate that signaling pathways activated by IL-4 can interfere directly with histone acetylation of the Ifng region independent of the type of stimulus used to stimulate histone acetylation.
Table 2. Long-range histone acetylation across the ≈100-kb Ifng gene region is stimulated by trichostatin A and inhibited by IL-4.
| -53 | -40 | -28 | -6 | -0.4 | +10 | +30 | +40 | +59 | IFN-γ, ng/ml | |
|---|---|---|---|---|---|---|---|---|---|---|
| Trichostatin A | 9 | ND | 6 | 5 | 3 | 4 | 5 | ND | 0 | 220 ± 15 |
| Trichostatin A + IL-4 | 9 | ND | 4 | 0 | 0 | 0 | 0 | ND | 0 | 6 ± 2 |
Levels of histone acetylation were determined by ChIP assay in CD8+ T cell cultures stimulated with anti-CD3 and trichostatin A with or without IL-4 for 3 days. Levels of IFN-γ in culture fluids were measured by ELISA (±SEM). ChIP analyses were performed a minimum of three times. SEM were <1 for each analysis and are not shown. ND, not done.
Discussion
Histone H3 and H4 hypo- and hyperacetylation of promoters and enhancers are linked to the transcriptional silencing and activation of genes. Histone hyperacetylation is also involved in the function of genomic nuclear matrix attachment regions and other nuclear functions that regulate gene transcription (8, 28). For example, genomic regions that are involved in the regulation of transcription of linked genes under common control but are positioned kilobases away from the genes they regulate also exhibit histone hypo- and hyperacetylation patterns over extended chromatin domains (19–21). Histone H3 and H4 hyperacetylation of the Ifng promoter has been previously reported in T cells stimulated under Th1 differentiation conditions (6, 7). Here, we show that Ifng introns 1 and 3 are also histone H4-hyperacetylated in C57BL/6 CD8+ T cells stimulated under similar conditions.
Surprisingly, histone hyperacetylation extends far beyond the Ifng gene. Regions as far as ≈50 kb upstream and downstream of the Ifng gene exhibit a common pattern of histone H4 hyperacetylation in both C57BL/6 and NOD Tc1 cells. In contrast, marked differences in the pattern of histone hyperacetylation are seen in C57BL/6 and NOD CD8+ T cells cultured under neutral conditions. C57BL/6 CD8+ T cells exhibit a pattern of histone hypoacetylation, but NOD CD8+ T cells exhibit a pattern of histone hyperacetylation. Moreover, closer inspection suggests that the histone hyperacetylation pattern may be divided into subdomains. As many as five domains may exist within this ≈110-kb region. Subdomains 1 (–53 kb), 3 (–0.4 to +4 kb), 4 (+30 kb), and 5 (+59 kb) exhibit equivalent levels of histone hyperacetylation in C57BL/6 CD8+ T cells cultured under Tc1 conditions and NOD CD8+ T cells cultured under either neutral or Tc1 conditions. In contrast, domain 2 (–28 to –6 kb) is histone-hypoacetylated in NOD CD8+ T cells cultured under neutral conditions and hyperacetylated in C57BL/6 and NOD CD8+ T cells cultured under Tc1 conditions. Regions at –40 kb and +40 kb are relatively histone-hypoacetylated in CD8+ T cells under all culture conditions. Broad, non-uniform acetylation patterns are known to occur in gene families, such as the growth hormone gene family and the β-globin gene family (29–31). However, to our knowledge, this is the first demonstration of these types of patterns surrounding a gene not known to be a member of an extended gene family. This is also, to our knowledge, the first demonstration of strain-dependent differences in the acetylation pattern of subdomains surrounding a gene that may account for strain-dependent differences in gene expression. Given their spatial separation and distinct acetylation patterns, it is likely that these subdomains possess unique functions. Taken together, these results suggest that broad non-uniform histone acetylation patterns may be a common property of genes that have complex expression profiles and may not be a unique property of gene families.
Our kinetic analysis demonstrates that initial histone acetylation of the immediate Ifng gene region is relatively IL-12- and IFN-γ-independent in B6 CD8+ T cells. However, establishment and maintenance of these extended domains of histone hyperacetylation depend on the presence of IL-12. Levels of histone acetylation reflect the activities of histone acetyl-transferases and histone deacetylases. Increased histone acetylation of the Ifng gene region may be a function of increased activity of histone acetyl-transferases or decreased activity of histone deacetylases in this region of genomic DNA. Our results demonstrate that inhibiting histone deacetylases stimulates both production of IFN-γ by B6 CD8+ T cells cultured under neutral conditions as well as establishment of extended domains of histone hyperacetylation. This supports the notion that differences in histone hyperacetylation of the Ifng gene locus resulting from altered activity of either histone acetyl-transferases or histone deacetylases account for strain and lineage differences in IFN-γ production by NOD and B6 CD8+ T cells. Interestingly, IL-4 effectively blocks both production of IFN-γ and histone hyperacetylation regardless of the stimulus or strain. This finding argues that signaling paths activated by IL-4 directly interfere with histone acetylation of the Ifng gene region rather than more proximal events in Th1/Tc1 differentiation.
Taken together, our results support a model in which NOD T cells differentiate into IFN-γ-producing effector cells by an autocrine pathway. NOD T cells clearly need a cytokine to induce Th1/Tc1 differentiation. However, the cytokine is IL-2, which T cells produce, rather than IL-12, which monocyte-lineage cells produce in response to inflammatory stimuli. Our results also suggest that these differences between NOD and other strains are genetic differences rather than reflections of the diabetic process because they are clearly seen in naive T cells and in the closely related NOR strain, which does not develop diabetes. These intrinsic differences in NOD T cells may represent a genetic trait that favors development of cell-mediated autoimmune diseases such as type I diabetes.
Acknowledgments
This work was supported by National Institutes of Health Grants RO1 DK 58765 and RO1 AI 44924.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Th1, T helper 1; Tc1, T cytotoxic 1; ChIP, chromatin immunoprecipitation; NOD, nonobese diabetic; NOR, nonobese diabetes-resistant.
References
- 1.Blackwood, E. M. & Kadonaga, J. T. (1998) Science 281, 60–63. [DOI] [PubMed] [Google Scholar]
- 2.Felsenfeld, G. Boyles, J., Chung, J., Clark, D. & Studisky, V. (1996) Proc. Natl. Acad. Sci. USA. 93, 9384–9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ptashne, M. & Gann, A. (1997) Nature 386, 569–577. [DOI] [PubMed] [Google Scholar]
- 4.Varga-Weisz, P. D. & Becker, P. B. (1998) Curr. Opin. Cell Biol. 10, 346–353. [DOI] [PubMed] [Google Scholar]
- 5.Wade, P. A. & Wolfe, A. P. (1999) Mol. Cell. Biol. 20, 2167–2175. [Google Scholar]
- 6.Brown, C. E., Lechner, T., Howe, L. & Workman, J. L. (2000) Trends Biochem. Sci. 25, 15–19. [DOI] [PubMed] [Google Scholar]
- 7.Struhl, K. (1998) Genes Dev. 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 8.Forsberg, E. C. & Bresnick, E. H. (2001) BioEssays 23, 820–830. [DOI] [PubMed] [Google Scholar]
- 9.Im, H., Grass, J. A., Christensen, H. M., Perkins, A. & Bresnick, E. H. (2002) Biochemistry 41, 15152–15160. [DOI] [PubMed] [Google Scholar]
- 10.Kiekhaefer, C. M., Grass, J. A., Johnson, K. D., Boyer, M. E. & Bresnick, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 14309–14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, K. D., Christensen, H. M., Zhao, B. & Bresnick, E. H. (2001) Mol. Cell 8, 465–471. [DOI] [PubMed] [Google Scholar]
- 12.Glimcher, L. H. & Murphy, K. M. (2000) Genes Dev. 14, 1693–1711. [PubMed] [Google Scholar]
- 13.O'Garra, A. (1998) Immunity 8, 275–283. [DOI] [PubMed] [Google Scholar]
- 14.Ansel, K. M., Lee, D. U. & Rao, A. (2003) Nat. Immunol. 4, 616–623. [DOI] [PubMed] [Google Scholar]
- 15.Agarwal, S. & Rao, A. (1998) Immunity 9, 765–775. [DOI] [PubMed] [Google Scholar]
- 16.Grogan, J. L., Mohrs, M., Harmon, B., Lacy, D. A., Sedat, J. W. & Locksley, R. M. (2001) Immunity 14, 205–215. [DOI] [PubMed] [Google Scholar]
- 17.Fields, P. E., Kim, R. M. & Flavell, R. A. (2002) J. Immunol. 169, 647–650. [DOI] [PubMed] [Google Scholar]
- 18.Avni, O., Lee, D., Macina, F., Szabo, S. J., Glimcher, L. H. & Rao, A. (2002) Nat. Immunol. 3, 643–651. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Balbas, M. A., Bannister, A. J., Martin, K., Haus-Seuffert, P., Meisterernst, M. & Kouzarides, T. (1998) EMBO J. 17, 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soutto, M., Zhou, W. & Aune, T. M. (2002) J. Immunol. 169, 6664–6667. [DOI] [PubMed] [Google Scholar]
- 21.Zhou, W., Zhang, F. & Aune, T. M. (2003) J. Immunol. 170, 735–740. [DOI] [PubMed] [Google Scholar]
- 22.Koarada, S., Wu, Y. & Ridgeway, W. M. (2001) J. Immunol. 167, 1693–1702. [DOI] [PubMed] [Google Scholar]
- 23.Koarada, S., Wu, Y., Olshansky, G. & Ridgway, W. M. (2002) J. Immunol. 169, 6580–6587. [DOI] [PubMed] [Google Scholar]
- 24.Soutto, M., Zhang, F., Enerson, B., Tong, Y., Boothby, M. & Aune, T. M. (2002) J. Immunol. 169, 4205–4212. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz, S., Zhang, Z., Frazer, K., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R. & Miller, W. (2000) Genome Res. 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger, S. L. (2002) Curr. Opin. Genet. Dev. 12, 142–148. [DOI] [PubMed] [Google Scholar]
- 27.Ahringer, J. (2000) Trends Genet. 16, 351–356. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez, L. A., Winkler, M. & Grosschedl, R. (2001) Mol. Cell. Biol. 21, 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elefant, F., Cooke, N. E. & Liebhaber, S. A. (2000) J. Biol. Chem. 275, 13827–13834. [DOI] [PubMed] [Google Scholar]
- 30.Elefant, F., Su, Y., Liebhaber, S. A. & Cooke, N. E. (2000) EMBO J. 19, 6814–6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shubeler, D., Francastel, C., Cimbora, D. M., Reik, A., Martin, D. I. & Groudine, M. (2000) Genes Dev. 14, 940–950. [PMC free article] [PubMed] [Google Scholar]