Abstract
During the differentiation of naïve CD4+ precursors to T helper 1 (Th1) or Th2 effector cells, several epigenetic changes occur in a lineage-specific manner at the IFN-γ or IL-4/IL-13 loci. These changes result in alterations in the chromatin structure of these loci and, hence, lineage-restricted expression of the corresponding cytokines. Intergenic transcripts have recently been shown to regulate the expression of genes in the β-globin locus; therefore, we have examined the Th2 cytokine gene cluster during human Th1/Th2 differentiation and in a transgenic mouse line containing the human IL-4/IL-13 genes for intergenic transcripts. We show for the first time that intergenic transcription of this locus is restricted to tissues and lineages in which IL-4 and IL-13 are expressed. We also show that intergenic transcription in the IL-4/IL-13 locus is up-regulated after Th2 differentiation. Furthermore, we demonstrate that the Th2 cytokines and intergenic transcripts are detectable in the thymus. We propose that intergenic transcription is tightly associated with transcriptional competence for the Th2 cytokines and may play a role in their regulation. These results support a progressive differentiation model of T cell lineage commitment.
The differentiation of naïve CD4+ T cells into T helper 1 (Th1) or Th2 cells has been the subject of intense study in both humans and mice (1, 2). Several lineage-specific transcription factors have been identified that are key regulators of murine Th1- or Th2-specific cytokines. GATA3 and c-Maf are required for expression of the Th2 cytokines IL-4 and IL-13, whereas T-bet is a key regulator of the Th1 cytokine IFN-γ (3, 4). We have shown that these transcription factors are expressed in a broadly analogous manner during human Th1/Th2 differentiation (5). Several elegant studies have demonstrated that inappropriate expression of these transcription factors is sufficient to skew the cytokine expression profile of developing or committed Th1 or Th2 cells and that differentiation of Th1/Th2 cells is accompanied by lineage-specific alterations in the chromatin structure of the cytokine loci (6–10). These changes include DNase I hypersensitive site formation and epigenetic changes such as DNA demethylation and histone hyperacetylation (reviewed extensively in ref. 11). This work has led to a model in which the “closed” chromatin of the cytokine loci in naïve precursors “opens” during differentiation in a lineage-specific process to allow the appropriate cytokine genes to be active upon stimulation.
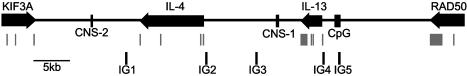
The genes encoding IL-4 and IL-13 are located in tandem within a cluster of cytokine genes on human chromosome 5q (12). The cytokine genes are interspersed with several unrelated genes; IL-4 and IL-13 are flanked by RAD50 and kinesin superfamily 3A (KIF3A) (Fig. 1). The genomic organization of this region is highly conserved on mouse chromosome 11. We and others have shown that the Th2 cytokines are coordinately expressed in individual Th2 cells (5, 13). The genomic organization and coordinated expression pattern of Th2 cytokines has led to considerable interest in the possible mechanisms that may control the lineage-restricted transcriptional competence of this locus.
Fig. 1.
Human IL-4/IL-13 locus. The position and orientation of genes are indicated by arrows, and exons are indicated by gray boxes below arrows. The positions of the known regulatory regions CNS-1 and CNS-2 are indicated. The positions of intergenic transcription amplimers (IG1–5) examined in this study are also shown.
Intergenic transcription was first identified in nonimprinted genes as a feature of the erythroid-specific β-globin gene cluster (14). Recent work has indicated that intergenic transcription is associated with developmental alterations in the chromatin structure of the β-globin locus (15). Fraser and colleagues (15) have demonstrated that the level of intergenic transcription correlates with the DNase I sensitivity of the β-globin cluster, and they have suggested that it demarcates differentially regulated chromatin subdomains of this cluster. We have shown that several intergenic regions (IG) of the human IL-4/IL-13 locus are transcribed in CD4+ T cells (16). To investigate intergenic transcription further we have generated a transgenic mouse line containing 118 kb of the human IL-4/IL-13 locus. We demonstrate that human IL-4 and IL-13 are transcribed in an activation-, lymphoid-, and Th2-specific manner at physiological levels in these transgenic mice. Examination of IL-4/IL-13 intergenic transcription reveals that it is specific to cells that have the potential to express IL-4 and IL-13. Furthermore, the intergenic transcripts are specifically up-regulated during Th2 differentiation. Our results indicate that intergenic transcription of this locus reflects the ability of a particular cell lineage to express the Th2 cytokines.
Materials and Methods
Generation of hIL-4/IL-13tg Mouse. A human genomic DNA library in the P1-derived artificial chromosome (PAC) vector pCYPAC-2 (17) was screened by PCR by using a primer pair immediately upstream of IL-13 (5′-GAAACTCTGCCCTGGACCCTTCTCAATAAG-3′ and 5′-AGGCAAGTGAGAGCAATGACCGTGGTCAAC-3′) and a primer pair immediately downstream of IL-4 (5′-CACTTAGCTGTGACACACTTCTCGAGAGAC-3′ and 5′-CAAGAAGTTTTCCAACGTACTCTGGTTGGC-3′). A clone (designated PAC 169.13.O) containing human IL-4 and IL-13 was obtained. The fragment was analyzed by restriction digestion, Southern hybridization, and sequencing and was found to be 118 kb (18). The ends of the fragment correspond to MboI position 32369 and MboI position 151147 in a contig of sequences from GenBank entries AC004237, AC004039, AC004041, and AC004042 numbered with reference to the start of clone AC004237 as position +1 (ref. 18 and data not shown). The IL-4/IL-13 genomic insert was liberated from the vector by NotI digestion and purified for pronuclear injection as described (19). Transgenic animals were generated by pronuclear injection of the purified PAC insert into the fertilized eggs of (CBA×C57BL/6)F1 mice. Screening for transgenic founders was performed by PCR on tail DNA by using the same primer pairs as were used in the library screening. Of 29 animals screened, 1 was transgenic and bred through the germ line (hIL-4/IL-13tg). The transgenic founder was crossed with wild-type (CBA×C57BL/6)F1 mice, and heterozygous progeny were used throughout the study. Analysis by Southern hybridization showed that the hIL-4/IL-13tg line contained three copies of the fragment (ref. 18 and data not shown). All mice used in this study were maintained under specified pathogen-free conditions. All of the animal experiments were approved by the institutional ethical review panel of King's College London.
Cells and in Vitro Culture. Murine CD4+ T cells were positively selected from splenocytes by using Dynabeads Mouse CD4 magnetic beads (Dynal, Great Neck, NY) according to the manufacturer's instructions. Purity of murine CD4+ cells was ≥98% in all experiments as assessed by flow cytometry (data not shown). Murine CD4+ T cells were cultured in RPMI medium 1640 supplemented with 10% FCS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Where indicated, cells were stimulated with 100 ng/ml phorbol 12,13-dibutyrate (Calbiochem) and 1 μg/ml ionomycin (Calbiochem) for 18 h.
For murine Th1 cultures, 1 × 106 cells per ml were stimulated with 1 μg/ml plate-bound anti-CD3ε (clone 2C11, BD Pharmingen) in the presence of 10 ng/ml recombinant mIL-2 (R & D Systems), 10 ng/ml recombinant mIL-12 (R & D Systems), and 10 μg/ml anti-IL-4 (clone 11B11, BD Pharmingen). To create murine Th2-polarizing conditions, 1 × 106 cells per ml were stimulated with anti-CD3ε in the presence of IL-2 with 1 μg/ml anti-CD28 (clone 37.51, BD Pharmingen), 100 ng/ml IL-4 (R & D Systems), and 10 μg/ml anti-IFN-γ (clone XMG1.2, BD Pharmingen). After 4 days, cultures were expanded by adding cytokines and antibodies as above but without anti-CD3ε or anti-CD28. After 7 days of culture, cells were restimulated in fresh medium with 1 μg/ml plate-bound anti-CD3ε for 5 h and then harvested for RT-PCR assay.
Peripheral blood mononuclear cells were isolated from human venous blood by separation over Lymphoprep (Nycomed, Oslo), and human CD4+ cells were purified by positive selection by using magnetic beads (Dynal) according to the manufacturers' instructions. Human Th1/Th2 cultures were performed as described (5). Normal human skin fibroblasts were obtained from the European Collection of Cell Cultures and cultured as described (8). Where indicated, fibroblasts were activated with IL-1β, tumor necrosis factor α, and IFN-γ for 24 h as described (8). The local ethical committee approved all experiments using material from human subjects.
RNA Isolation and RT-PCR. RNA isolations were performed by using a RNA/DNA kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA was treated with RQ1 RNase free DNase (Promega) to remove contaminating DNA. Reverse transcription reactions and control reactions without enzyme were performed as described (16). The conditions for RT-PCR amplification have been described (16). Species-specific primers were designed in the exons of human and murine β-actin, IL-4 and IL-13, human granulocyte/macrophage colony-stimulating factor (GM-CSF), and murine IFN-γ; each primer pair spanned an intron so that the amplimers derived from spliced mRNA could be distinguished from those amplified from DNA. Primer pairs used were as follows: human β-actin, 5′-CACCACACCTTCTACAATGAGCTGC-3′ and 5′-ACAGCCTGGATAGCAACGTACATGG-3′; human IL-4, 5′-ACATTGTCACTGCAAATCGACACC-3′ and 5′-TGTCTGTTACGGTCAACTCGGTGC-3′; human IL-13, 5′-GCAATGGCAGCATGGTATGG-3′ and 5′-AAGGAATTTTACCCCTCCCTAACC-3′; human GM-CSF, 5′-GCCAGCCACTACA AGCAGCAC-3′ and 5′-CAAAGGGGATGACAAGCAGAAAG-3′; murine β-actin, 5′-ATCGTGCGTGACATCA A AGAGA AGC-3′ and 5′-CCACAGGATTCCATACCCAAGAAGG-3′; murine IFN-γ, 5′-AACTCAAGTGGCATAGATGTGG-3′ and 5′-CAGGTGTGATTCAATGACGC-3′; mIL-4, 5′-CTGTAGGGCTTCCA AGGTGCT TCG-3′ and 5′-CCAT T TGCATGATGCTCTTTAGGC-3′; mIL-13, 5′-ATCTACAGGACCCAGAGGATAT TGC-3′ and 5′-CTGATGTGAGA A AGGAAAATGAGTCC-3′. The primers for human RAD50 and KIF3A and IG1–5 have been described (16). Where indicated, the intensity of the amplimer on an ethidium bromide-stained agarose gel was quantitated by using imagequant software (Molecular Dynamics). As shown in Fig. 2C, semiquantitative RT-PCR was carried out on RNA from Th1 and Th2 cells in parallel with identical control PCRs containing cytokine cDNA templates in increasing concentrations. To demonstrate that the amplification was in the linear range of the PCR, a standard curve was created for each primer pair. The intensity of the amplimer from the Th1 and Th2 samples was then calculated, and the quantity of starting cDNA was interpolated from the graph and expressed as copies per μg of RNA used in the reverse transcription reaction.
Fig. 2.
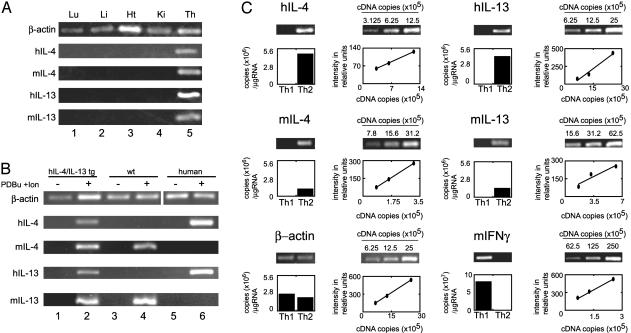
Th2-specific expression of human IL-4 and IL-13 in transgenic mice. (A) Expression of IL-4 and IL-13 is restricted to lymphoid tissues. RNA was isolated from transgenic lung (Lu), liver (Li), heart (Ht), kidney (Ki), and thymus (Th), reverse-transcribed, and analyzed by PCR for human and murine IL-4 and IL-13. Number of PCR cycles: β-actin, 18; hIL-4, 28; mIL-4, 28; hIL-13, 24; mIL-13, 28. (B) IL-4 and IL-13 are transcribed in activated CD4+ cells. CD4+ cells were purified from human blood and transgenic and wild-type murine spleens and were either activated with phorbol 12,13-dibutyrate and ionomycin for 18 h or left unactivated. RNA was isolated from each cell population, reverse-transcribed, and analyzed by PCR. Number of PCR cycles: β-actin, 18; hIL-4, 32; mIL-4, 32; hIL-13, 32; mIL-13, 32. (C) Th2-specific expression of human IL-4 and IL-13. CD4+ cells were purified from transgenic and nontransgenic spleens and cultured for 7 days under Th1 or Th2 conditions. The cells were then stimulated for 5 h with anti-CD3ε, and the RNA from each population was reverse-transcribed and used for PCR. The linear range of the PCR for each primer pair was determined by using increasing copies of cDNA as template and is shown in the line graphs. The levels of mRNA expression are shown in the histograms below the gels and are expressed as copies of cDNA per μg of RNA used in the reverse transcription reaction. Number of PCR cycles: β-actin, 18; hIL-4, 22; mIL-4, 22; hIL-13, 22; mIL-13, 22; murine IFN-γ, 16.
Results
Lineage-Specific Transcription of Human IL-4 and IL-13 in a Transgenic Mouse Line. A 118-kb genomic DNA fragment containing IL-4 and IL-13 was isolated from a human library in a PAC vector, pCYPAC-2 (17). The fragment encompasses the 3′ half of KIF3A, IL-4, and IL-13 and the 3′ half of RAD50 (Fig. 1). We generated transgenic mice harboring the fragment (see Materials and Methods). One transgenic line (hIL-4/IL-13tg) was identified that contained three copies of the IL-4/IL-13 fragment (data not shown).
RT-PCR was used to analyze the tissue-specificity of IL-4 and IL-13 transcription in a range of tissues from the hIL-4/IL-13tg mice (Fig. 2 A). Transcription of both the human and murine IL-4 and IL-13 genes was detected in the thymus (lane 5) but was absent from the lung, liver, heart, and kidney. CD4+ T cells were isolated by positive selection from transgenic animals, control littermates, and humans. RT-PCR was carried out on RNA isolated from both resting and activated CD4+ T cells (Fig. 2B). Both the human and murine IL-4 and IL-13 cytokine genes were transcribed in the activated population of transgenic CD4+ cells (lane 2) but not in resting T cells (lane 1). In all cases, the PCRs yielded products that were both of the correct size and species-specific (compare lanes 2, 4, and 6). These results indicate that the human transgenes are expressed only in lymphoid tissues in which expression of the endogenous murine genes is also detected. Furthermore, the results demonstrate that, like the murine genes, the transgenes are expressed in an activation-dependent manner in CD4+ splenocytes.
To determine whether the hIL-4 and hIL-13 genes were specifically expressed in transgenic Th2 cells, we cultured CD4+ splenocytes for 7 days in Th1- or Th2-polarizing conditions. For Th1 conditions, cells were stimulated with anti-CD3 together with IL-2, IL-12, and anti-IL-4. For Th2 conditions, cells were stimulated with anti-CD3 and anti-CD28 together with IL-2, IL-4, and anti-IFN-γ. Polarized cells were restimulated with anti-CD3 for 5 h, and the steady-state levels of cytokine mRNA in the Th1/Th2 cells were quantified by using a semiquantitative RT-PCR based assay (Fig. 2C). RT-PCR was carried out on RNA from Th1 and Th2 cell populations in parallel with control PCRs containing known quantities of cDNA, which demonstrated that the PCRs were in the linear amplification range. As expected, the murine IL-4 and IL-13 genes were specifically expressed in Th2 cells; 1.1 × 106 and 1.4 × 106 copies of cDNA were detected per μg of RNA in the reverse transcription reaction, respectively. The Th1-specific gene IFN-γ was expressed in Th1 cells at 8 × 107 copies per μg. The human IL-4 and IL-13 genes were also specifically expressed in Th2 cells and at slightly higher levels than the murine genes at 4.8 × 106 and 4.4 × 106 copies per μg, respectively. These data demonstrate that the human transgenes are correctly regulated during murine Th2 differentiation and are transcribed at physiological levels. The data also show that the human IL-4 and IL-13 genes are not expressed in cells differentiated toward a Th1 phenotype. Hence, the hIL-4/IL-13tg line provides a suitable experimental model in which to further examine the regulation of the human Th2 cytokine cluster.
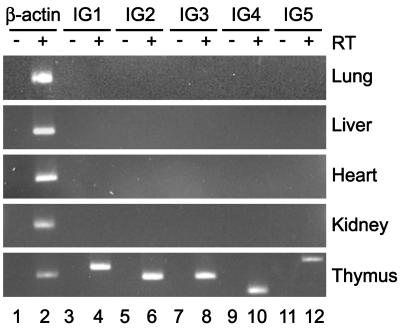
Tissue-Specific Intergenic Transcription in the Human Th2 Cytokine Locus. In previous work we have shown that intergenic transcription occurs at several points throughout the human Th2 cytokine cluster (labeled IG1–5 in Fig. 1) in human CD4+ T cells (16). We demonstrated that the intergenic transcripts are constitutively present in resting CD4+ cells even in the absence of detectable transcription of IL-4 or IL-13 and the intergenic transcripts were not up-regulated upon acute activation (16). To investigate the tissue-specificity of these intergenic transcripts, we isolated RNA from various tissues from the hIL-4/IL-13tg mice. To eliminate any contaminating DNA, the RNA samples were treated with DNase I. RT-PCR analysis for intergenic transcripts corresponding to IG1–5 was performed (Fig. 3). IG1 is between the 3′ end of IL-4 and the conserved noncoding sequence 2 (CNS-2), IG2 is in the IL-4 promoter, IG3 is between IL-4 and IL-13 near CNS-1, IG4 is in the IL-13 promoter, and IG5 is between IL-13 and the 3′ end of RAD50 near a CpG island. To control for DNA contamination, the RT-PCRs were performed in parallel with identical samples that had no enzyme added during the reverse transcription step (Fig. 3, –RT). Intergenic transcripts corresponding to each of the regions IG1–5 were detected in hIL-4/IL-13tg thymus (Fig. 3, thymus, lanes 4, 6, 8, 10, and 12). We did not detect intergenic transcription in these regions in any of the other tissues examined (Fig. 3, lung, liver, heart, and kidney). Furthermore, we did not detect intergenic transcripts in any of the regions IG1–5 in transgenic CD19+ splenocytes, which do not express IL-4 or IL-13 (data not shown). The presence of detectable intergenic transcripts in thymus alone is consistent with the observation that IL-4 and IL-13 transcription was detected only in thymus (Fig. 2). This indicates that the presence of Th2 cytokine locus intergenic transcripts in a tissue is closely associated with competence of that tissue to express Th2 cytokines.
Fig. 3.
Lymphoid-specific intergenic transcription in the human Th2 cytokine cluster. RNA was isolated from transgenic lung (Lu), liver (Li), heart (Ht), kidney (Ki), and thymus (Th), reverse-transcribed, and analyzed by PCR for human intergenic transcripts IG1–5. –RT indicates control reverse transcription without enzyme (lanes 1, 3, 5, 7, 9, and 11). Number of PCR cycles: β-actin, 20; IG1–5, 34.
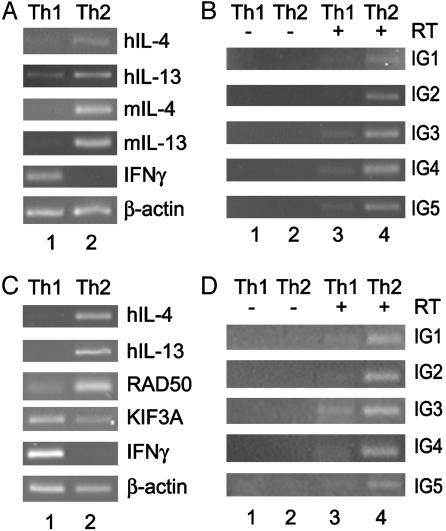
Intergenic Transcription in the Human Th2 Cytokine Locus After Th1/Th2 Differentiation. To examine whether the intergenic transcripts that we observe are selectively expressed in Th2 cells after differentiation, we compared the levels of IG1–5 transcripts in CD4+ hIL-4/IL-13tg T cells differentiated toward Th1 or Th2 phenotypes (Fig. 4). Fig. 4A shows that both the human and murine Th2 cytokine genes were selectively transcribed in Th2 cells (lane 2) but not in Th1 cells (lane 1). A Th2-specific up-regulation of intergenic transcription was observed at each of the regions examined when compared with Th1 cells (Fig. 4B, compare lanes 3 and 4).
Fig. 4.
Intergenic transcription after Th1 or Th2 differentiation. (A and B) Expression of IL-4 and IL-13 and intergenic transcripts in murine Th1/Th2 cells. CD4+ cells were purified from transgenic spleens and cultured for 7 days under Th1 or Th2 conditions. The cells were then stimulated for 5 h with phorbol 12-myristate 13-acetate (PMA) and ionomycin, and the RNA from each population was reverse-transcribed and used in PCR for the genic (A) and intergenic (B) transcripts indicated. Number of PCR cycles: β-actin, 18; hIL-4, 22; mIL-4, 24; hIL-13, 22; mIL-13, 24; murine IFN-γ, 16; IG1–5, 34. (C and D) Expression of IL-4 and IL-13 and intergenic transcripts in human Th1/Th2 cells. CD4+ cells were purified from peripheral blood and cultured for 14 days under Th1 or Th2 conditions. The cells were then stimulated for 5 h with PMA and ionomycin, and the RNA from each population was reverse-transcribed and used in PCR for the genic (C) and intergenic (D) transcripts indicated. Number of PCR cycles: β-actin, 22; hIL-4, 28; hIL-13, 22; RAD50, 28; KIF3A, 28; IFN-γ, 18; IG1–5, 34.
We also examined the Th2-specificity of intergenic transcription in this locus in human CD4+ T cells using an in vitro differentiation assay to generate Th1 or Th2 cells from naïve precursors (5). RT-PCR showed that, after 14 days in culture, we had highly polarized human Th1 and Th2 cell populations (Fig. 4C). The Th1 cells transcribed high levels of IFN-γ when compared with the Th2 cells, and the Th2 cells transcribed high levels of IL-4 and IL-13 compared with the Th1 cells (Fig. 4C, compare lanes 1 and 2). Once again, we observed a specific up-regulation of the intergenic transcripts in the Th2 cells compared with the Th1 cells at all of the regions examined (Fig. 4D, compare lanes 3 and 4). These data clearly show that the intergenic transcripts observed in this locus are selectively up-regulated in both human and transgenic Th2 cells. This finding provides further evidence that the level of intergenic transcripts is intimately associated with the competence of cells to transcribe the Th2 cytokines.
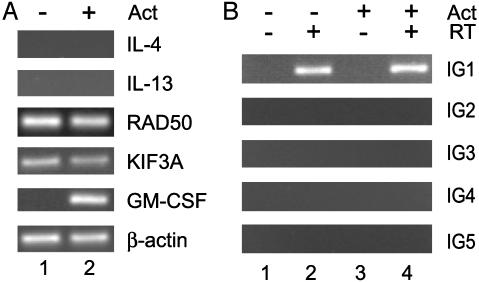
Intergenic Transcription in Fibroblasts. To further assess the lineage specificity of the Th2 cytokine locus intergenic transcripts, we examined resting and activated normal human skin fibroblasts. These cells constitutively transcribe the surrounding genes RAD50 and KIF3A but do not transcribe IL-4 or IL-13, even after activation (Fig. 5A). Fibroblasts readily express GM-CSF, another chromosome 5q cytokine, under these activation conditions (Fig. 5A, lane 2). When we examined these cells for the presence of intergenic transcripts, we observed constitutive transcription at region IG1 that was not affected by activation (Fig. 5B, lanes 2 and 4). However, intergenic transcripts were not detected in any of the other regions examined (IG2–5) in either resting or activated fibroblasts. These data provide further evidence for the lineage specificity of intergenic transcription in the Th2 cytokine locus. It is intriguing that we observe transcription in IG1 in these cells; because this region is between KIF3A and IL-4, it is possible that the presence of the IG1 transcripts is related to the expression of KIF3A in fibroblasts.
Fig. 5.
Transcription across the Th2 cytokine cluster in human fibroblasts. RNA was isolated from resting or activated normal skin fibroblasts and subjected to RT-PCR for the Th2 cytokine cluster genic (A) and intergenic (B) transcripts. Where indicated (+Act) cells were activated with IL-1β, tumor necrosis factor α, and IFN-γ for 24 h. Number of PCR cycles: β-actin, 22; hIL-4, 34; hIL-13, 34; RAD50, 27; KIF3A, 27; GM-CSF, 32; IG1–5, 34.
Discussion
In this study the human Th2 cytokine locus was examined by RT-PCR for the presence of intergenic transcripts in a variety of tissues and primary cell lineages from transgenic mice and humans. Our results clearly show that intergenic transcription is present throughout this locus in cell types that transcribe, or are competent to transcribe, IL-4 and IL-13. The intergenic transcripts are not detectable in cell lineages that do not express the Th2 cytokines. Furthermore, we have shown that the intergenic transcripts are up-regulated after Th2 differentiation. Interestingly, we also detected expression of IL-4, IL-13, and intergenic transcripts in the thymus, i.e., in early lymphoid tissue before effector T cell differentiation.
The transgenic mouse line that we generated contains a 118-kb fragment of the human Th2 cytokine locus containing IL-4 and IL-13 (Fig. 1). Recently Locksley and colleagues (20, 21) used a similar approach to begin to examine the regulation of human Th2 cytokines. They used two ≈400-kb fragments and reported Th2-specific expression of the human genes. The transgene reported here is considerably smaller than any previously described and, importantly, does not contain the promoters of either of the flanking genes RAD50 or KIF3A. Because we observe Th2 cell-specific, activation-responsive, physiological expression of human IL-4 and IL-13, it is likely that the 118-kb region described here contains all of the regulatory elements required for Th2-specific expression of these genes. The data also strongly suggest that expression of RAD50 or KIF3A is not a prerequisite for expression of Th2 cytokines. Notably, the transgene does not contain IL-5, and thus distant IL-5-proximal elements are not required for IL-4 or IL-13 expression. Our results also suggest that at least some of the DNA elements that control IL-4 and IL-13 expression and the transcription factors that regulate them are conserved between the human and mouse loci. A recent paper by Lee et al. (22) has identified a 25-kb region around the 3′ end of murine RAD50 that contains locus control region activity. The equivalent region of human RAD50 is contained within our transgene, and it will be interesting to determine whether it confers copy number-dependent expression on human IL-4 and IL-13. Because the transgenic mouse line that we have generated expresses the human Th2 cytokines in a physiological manner in mice, it could also be used to investigate the regulation of the human Th2 cytokine genes in murine models of allergic inflammation and asthma.
Intergenic transcription in this locus shows considerable lineage specificity and parallels the competence of a particular cell type to express the Th2 cytokines. We propose that intergenic transcription is tightly associated with transcriptional competence for the Th2 cytokines and may play a role in their regulation. This finding is highly reminiscent of the situation found in the β-globin gene cluster (14, 15). Recent work on β-globin has shown that some of the intergenic transcripts emanate from the locus control region whereas others appear to derive from promoters in the intergenic regions (15, 23, 24). Deletion of one of these promoters greatly affected expression of the adult β-globin genes, suggesting that intergenic transcription is required for the formation of active chromosomal domains. Another example of intergenic transcription has recently been described in another lineage-specific gene cluster, the MHC class II locus (25). This locus also contains a locus control region around which the intergenic transcripts appear to be centered. Although the precise role of intergenic transcription in controlling gene expression remains to be elucidated, it has been proposed that lineage-specific transcription factors binding to distal regulatory elements such as locus control regions recruit RNA polymerase II, which may then activate the surrounding chromosomal region by means of “looping,” “tracking,” or “linking” mechanisms in which nascent transcripts are generated (26–29). Further experiments will be required to determine the position of the intergenic promoters within the Th2 cytokine locus. Deletion of these regions in transgenic animals will provide further insight into the role of intergenic transcription in this locus.
In tissues that are not competent to express Th2 cytokines we did observe a difference in the pattern of intergenic transcripts between transgenic and human samples. In the transgenic tissues (Fig. 3) and B cells (data not shown) no intergenic transcripts were detected at any point in the cluster, whereas in human fibroblasts the region IG1 was transcribed (Fig. 5), which is similar to the pattern we have reported for HeLa cells (16). This difference could simply be due to the different lineages examined in the human and murine analyses. Alternatively, it is possible that the transcription at IG1, which is between IL-4 and KIF3A, is in some tissues associated with expression of the KIF3A gene, which is not intact in our transgenic model. This is an intriguing possibility, because it would suggest that there may be a boundary between the KIF3A and Th2 cytokine domains in this locus. High-resolution mapping of the limits of intergenic transcripts in this region may identify potential boundary elements or silencers.
It is noteworthy that we observe transcription of human and murine IL-4 and IL-13 in thymus tissue from transgenic mice and the murine genes in the wild-type controls (Fig. 2 A). It is possible that these genes are activated in thymocytes due to T cell receptor-mediated signals generated during positive or negative selection, which suggests that T cells, or a subset thereof, are competent to express these cytokines much earlier during lineage specification than previously thought. Further studies will be needed to determine at which stage during thymocyte development the Th2 cytokines are activated. It will also be interesting to determine whether the level of IL-4 expression in the thymus has any effect on the Th1/Th2 lineage choice of naïve T cells after they exit the thymus. The expression of Th2 cytokines in the thymus strongly supports a progressive model of T cell lineage commitment in which early progenitor cells are competent to express hallmark genes of several lineages that are progressively silenced during differentiation (30, 31).
Acknowledgments
We thank Lee Stuckey for performing microinjections, the staff the transgenic facilities of King's College London for excellent technical assistance, Paul Lavender for critical reading of the manuscript, and other members of our department for helpful discussions. This work was funded by a Medical Research Council program grant (to D.Z.S. and T.H.L.).
Abbreviations: KIF3A, kinesin superfamily 3A; PAC, P1-derived artificial chromosome; hIL, human IL; mIL, murine IL; IG, intergenic region; CNS, conserved noncoding sequence; GM-CSF, granulocyte/macrophage colony-stimulating factor; Th, T helper.
References
- 1.Glimcher, L. H. & Murphy, K. M. (2000) Genes Dev. 14, 1693–1711. [PubMed] [Google Scholar]
- 2.Murphy, K. M. & Reiner, S. L. (2002) Nat. Rev. Immunol. 2, 933–944. [DOI] [PubMed] [Google Scholar]
- 3.Glimcher, L. H. & Singh, H. (1999) Cell 96, 13–23. [DOI] [PubMed] [Google Scholar]
- 4.O'Garra, A. (2000) Curr. Biol. 10, R492–R494. [DOI] [PubMed] [Google Scholar]
- 5.Cousins, D. J., Lee, T. H. & Staynov, D. Z. (2002) J. Immunol. 169, 2498–2506. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal, S. & Rao, A. (1998) Immunity 9, 765–775. [DOI] [PubMed] [Google Scholar]
- 7.Takemoto, N., Koyano-Nakagawa, N., Yokota, T., Arai, N., Miyatake, S. & Arai, K. (1998) Int. Immunol. 10, 1981–1985. [DOI] [PubMed] [Google Scholar]
- 8.Santangelo, S., Cousins, D. J., Winkelmann, N. E. & Staynov, D. Z. (2002) J. Immunol. 169, 1893–1903. [DOI] [PubMed] [Google Scholar]
- 9.Messi, M., Giacchetto, I., Nagata, K., Lanzavecchia, A., Natoli, G. & Sallusto, F. (2003) Nat. Immunol. 4, 78–86. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita, M., Ukai-Tadenuma, M., Kimura, M., Omori, M., Inami, M., Taniguchi, M. & Nakayama, T. (2002) J. Biol. Chem. 277, 42399–42408. [DOI] [PubMed] [Google Scholar]
- 11.Ansel, K. M., Lee, D. U. & Rao, A. (2003) Nat. Immunol. 4, 616–623. [DOI] [PubMed] [Google Scholar]
- 12.Frazer, K. A., Ueda, Y., Zhu, Y., Gifford, V. R., Garofalo, M. R., Mohandas, N., Martin, C. H., Palazzolo, M. J., Cheng, J. F. & Rubin, E. M. (1997) Genome Res. 7, 495–512. [DOI] [PubMed] [Google Scholar]
- 13.Kelly, B. L. & Locksley, R. M. (2000) J. Immunol. 165, 2982–2986. [DOI] [PubMed] [Google Scholar]
- 14.Ashe, H. L., Monks, J., Wijgerde, M., Fraser, P. & Proudfoot, N. J. (1997) Genes Dev. 11, 2494–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gribnau, J., Diderich, K., Pruzina, S., Calzolari, R. & Fraser, P. (2000) Mol. Cell 5, 377–386. [DOI] [PubMed] [Google Scholar]
- 16.Rogan, D. F., Cousins, D. J. & Staynov, D. Z. (1999) Biochem. Biophys. Res. Commun. 255, 556–561. [DOI] [PubMed] [Google Scholar]
- 17.Ioannou, P. A., Amemiya, C. T., Garnes, J., Kroisel, P. M., Shizuya, H., Chen, C., Batzer, M. A. & de Jong, P. J. (1994) Nat. Genet. 6, 84–89. [DOI] [PubMed] [Google Scholar]
- 18.Rogan, D. F. (2002) Ph.D. thesis (Univ. of London, London).
- 19.Raguz, S., Hobbs, C., Yague, E., Ioannou, P. A., Walsh, F. S. & Antoniou, M. (1998) Dev. Biol. 201, 26–42. [DOI] [PubMed] [Google Scholar]
- 20.Loots, G. G., Locksley, R. M., Blankespoor, C. M., Wang, Z. E., Miller, W., Rubin, E. M. & Frazer, K. A. (2000) Science 288, 136–140. [DOI] [PubMed] [Google Scholar]
- 21.Lacy, D. A., Wang, Z. E., Symula, D. J., McArthur, C. J., Rubin, E. M., Frazer, K. A. & Locksley, R. M. (2000) J. Immunol. 164, 4569–4574. [DOI] [PubMed] [Google Scholar]
- 22.Lee, G. R., Fields, P. E., Griffin, T. J. & Flavell, R. A. (2003) Immunity 19, 145–153. [DOI] [PubMed] [Google Scholar]
- 23.Plant, K. E., Routledge, S. J. & Proudfoot, N. J. (2001) Mol. Cell. Biol. 21, 6507–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Routledge, S. J. & Proudfoot, N. J. (2002) J. Mol. Biol. 323, 601–611. [DOI] [PubMed] [Google Scholar]
- 25.Masternak, K., Peyraud, N., Krawczyk, M., Barras, E. & Reith, W. (2003) Nat. Immunol. 4, 132–137. [DOI] [PubMed] [Google Scholar]
- 26.Carter, D., Chakalova, L., Osborne, C. S., Dai, Y. F. & Fraser, P. (2002) Nat. Genet. 32, 623–626. [DOI] [PubMed] [Google Scholar]
- 27.Bulger, M. & Groudine, M. (1999) Genes Dev. 13, 2465–2477. [DOI] [PubMed] [Google Scholar]
- 28.Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F. & de Laat, W. (2002) Mol. Cell 10, 1453–1465. [DOI] [PubMed] [Google Scholar]
- 29.Hatzis, P. & Talianidis, I. (2002) Mol. Cell 10, 1467–1477. [DOI] [PubMed] [Google Scholar]
- 30.Fisher, A. G. (2002) Nat. Rev. Immunol. 2, 977–982. [DOI] [PubMed] [Google Scholar]
- 31.Lanzavecchia, A. & Sallusto, F. (2002) Nat. Rev. Immunol. 2, 982–987. [DOI] [PubMed] [Google Scholar]