Abstract
Insulin-producing cells normally occur only in the pancreas and thymus. Surprisingly, we found widespread insulin mRNA and protein expression in different diabetic mouse and rat models, including streptozotocin-treated mice and rats, ob/ob mice, and mice fed high-fat diets. We detected in diabetic mice proinsulin- and insulin-positive cells in the liver, adipose tissue, spleen, bone marrow, and thymus; many cells also produced glucagon, somatostatin, and pancreatic polypeptide. By in situ nucleic acid hybridization, diabetic, but not nondiabetic, mouse liver exhibited insulin transcript-positive cells, indicating that insulin was synthesized by these cells. In transgenic mice that express GFP driven by the mouse insulin promoter, streptozotocin-induced diabetes led to the appearance of GFP-positive cells in liver, adipose tissue, and bone marrow; the fluorescent signals showed complete concordance with the presence of immunoreactive proinsulin. Hyperglycemia produced by glucose injections in nondiabetic mice led to the appearance of proinsulin- and insulin-positive cells within 3 days. Bone marrow transplantation experiments showed that most of the extrapancreatic proinsulin-producing cells originated from the bone marrow. Immunoreactive proinsulin- and insulin-positive cells were also detected in the liver, adipose tissue, and bone marrow of diabetic rats, indicating that extrapancreatic, extrathymic insulin production occurs in more than one species. These observations have implications for the regulation of insulin gene expression, modulation of self-tolerance by insulin gene expression, and strategies for the generation of insulin-producing cells for the treatment of diabetes.
Keywords: pancreatic islets, hyperglycemia, ob/ob, obesity, bone marrow transplantation
Insulin is normally produced in highly specialized cells of the endocrine pancreas. The tissue-specific expression of insulin is tightly regulated at the transcriptional level, and the major regulatory elements are located in the 5′ flanking region of the insulin gene (1). Insulin expression is specific to β cells in the pancreatic islets, although proinsulin has also been detected in the fetal and postnatal thymus and spleen/lymphoid tissues (2, 3), a pattern of promiscuous expression of normally tissue-restricted “peripheral” proteins in these tissues (4, 5). Immuno-reactivity to insulin plays a key role in autoimmune diabetes in NOD mice (6, 7) and in type 1 diabetes in humans (7); expression of minute amounts of insulin in the thymus is believed to be important in the induction of tolerance to insulin and other “self-antigens” (8–11). In this study, we found insulin gene transcription and protein expression in multiple tissues outside the pancreas and thymus in mice and rats with diabetes. The widespread occurrence of extrapancreatic, extrathymic insulin gene products in diabetic animals is surprising; it suggests that we should revise our thinking on the regulation of insulin gene expression, the mechanism of immune tolerance to insulin, the pathophysiology of diabetes, and the strategies for producing insulin-producing cells in vitro for the treatment of diabetes.
Methods
Animals. WT and ob/ob mice in C57BL/6 background (The Jackson Laboratory), Wistar rats (Japan Clea, Hamamatsu, Japan), and MIP-GFP mice, a transgenic mouse line expressing GFP driven by the mouse insulin promoter (12), were maintained on regular chow. Diabetes was induced by i.v. streptozotocin (STZ, 100 mg/kg in mice and 40 mg/kg in rats) at 8–10 weeks of age, and diabetic animals were used for experiment 8 weeks later. We used commercial kits (Crystal Chem, Downers Grove, IL) to determine serum glucose and insulin. To generate high-fat diet-induced, obesity-related diabetes, we fed 8- to 10-week-old mice a high-fat/high-sucrose diet (per kg of feed containing 210 g of milk fat and 314 g of sucrose) from Harlan Teklad (TD 88137, Madison, WI) for 24 weeks. ob/ob mice were analyzed at 62 weeks. To induce hyperglycemia, we injected 25% glucose solution or saline i.p. (four times daily every 2 h with first injections at 8 a.m.), and mice were analyzed on days 3 and 15. For bone marrow transfer (BMT), female C57BL/6-Ly-5.1 mice were irradiated (9 Gy) and injected with 4 million bone marrow cells isolated from male C57BL/6-Rosa-Ly-5.2 mice. Engraftment was measured 6 weeks after transplant by fluorescence-activated cell sorting analysis of peripheral blood using an antibody against Ly-5.2 (clone 104, Pharmingen). Peripheral blood engraftment of bone marrow cells ranged from 61% to 81%. Eight weeks after BMT, we induced diabetes in one group of recipients by STZ treatment, and mice were analyzed 2 weeks later; we induced hyperglycemia with 25% glucose (or saline) in another group and studied it at day 15.
Analysis of Tissue mRNA Expression. We homogenized mouse and rat tissues in acid guanidinium-phenol-chloroform (TRIzol, GIBCO/BRL) and extracted and analyzed the total RNA by RT-PCR. We detected islet-specific transcripts by RT-PCR using 32 PCR cycles. PCR products were first confirmed by direct sequencing. Appropriate controls included DNase pretreatment and omission of reverse transcriptase in the reaction. The sequences of individual PCR primers were as described (13, 14).
Immunohistochemical Analysis. For the detection of insulin- or proinsulin-positive cells, we fixed 20-μm-thick frozen sections or 5-μm-thick paraffin sections and processed them for immuno-histochemical staining by using the avidin–biotin peroxidase complex (ABC) method and diaminobenzidine (DAB)-nickel reactions as described (13, 15). Briefly, for insulin or proinsulin staining, frozen sections were incubated for 2 days with antibody against insulin (guinea pig polyclonal, Linco Research, St. Charles, MO) or proinsulin (guinea pig polyclonal, Progen, Heidelberg) diluted 1:5,000 in 0.1% PBS containing 0.3% Triton X-100 (PBST) at 4°C. Paraffin sections were incubated for 2 h with antibody against proinsulin diluted 1:500 in PBST at room temperature. After the DAB-nickel reaction, the sections were counterstained with 0.1% neutral red solution. The image was transferred to light microscope (Olympus DX51, Olympus, Tokyo) and charge-coupled device camera (Olympus DP12). The density of proinsulin- and insulin-positive cells was calculated from the mean number of positive cells per unit area in 20 randomly selected fields at ×200 magnification under light microscopy.
For the immunofluorescence overlap staining of islet hormones, sections were processed for fluorescence triple staining with insulin/somatostatin/proinsulin, insulin/glucagon/pancreatic polypeptide (Ppy), proinsulin/somatostatin/glucagon, or proinsulin/Ppy/insulin (13). Antibodies used were specific for insulin (guinea pig polyclonal), somatostatin (rabbit polyclonal, Yanaihara, Shizuoka, Japan), proinsulin (mouse monoclonal, HyTest, Turku, Finland), glucagon (rabbit polyclonal, Biogenesis, Kingston, NH; or guinea pig polyclonal, Progen), or Ppy (mouse monoclonal or rabbit polyclonal, Yanaihara). For overlap staining of proinsulin/CD45 in the liver of STZ mice, liver sections were processed for fluorescence double immunostaining using antibodies against proinsulin (guinea pig polyclonal) and CD45 (rat monoclonal, BD Biosciences, San Diego). For overlap staining of proinsulin/β-galactosidase in STZ-treated or glucose-injected bone marrow transplant recipient mice, sections from bone marrow, liver, and pancreas were processed for fluorescence double staining using antibodies against proinsulin (guinea pig polyclonal) and β-galactosidase (rabbit polyclonal, Biogenesis). For overlap localization of GFP/proinsulin in MIP-GFP mice, sections from STZ-induced diabetic and nondiabetic liver, adipose tissue, and bone marrow were incubated with antibody against proinsulin (guinea pig polyclonal). After reaction with the first antibodies, sections were incubated with fluorescence-labeled second antibodies as described (13). All fluorescence photomicrographs were obtained from 1-μm-thick slices of the specimens under the confocal laser scanning microscope (LSM 510, Zeiss).
The specificity of the liver staining was examined by adsorption. Frozen and paraffin sections from STZ mice were incubated with proinsulin antibody diluted 1:5,000 (for frozen sections) or 1:500 (for paraffin sections) preadsorbed with 10 μM proinsulin (Biogenesis). The antigen and antibody mixture was incubated for 24 h at 4°C before application to the sections.
In Situ Hybridization. We used mouse insulin-2 cDNA (274 bp) and albumin cDNAs (805 bp) cloned in plasmid pGEMT as templates for in vitro transcription to produce riboprobes with a digoxigenin-RNA labeling kit (Roche Diagnostics) and performed hybridization with liver sections as described (16).
Results
Insulin mRNA and Protein-Producing Cells Are Present in the Liver of Diabetic Mice. We applied rigorous criteria to ensure that the insulin expression we observed in the liver of STZ-induced diabetic mice (13) was not an artifact. First, we determined that, by RT-PCR and with 40 cycles of amplification, we could not detect any insulin transcripts in samples of total liver RNA isolated from nondiabetic mice. We then arbitrarily set a limit of 32 cycles to assay for the presence of insulin mRNA in diabetic tissue samples and sequenced the RT-PCR products to ensure that the amplification band was authentic insulin cDNA. Using this approach, we established the presence of insulin transcripts in the RNA isolated from the liver of STZ-induced diabetic C57BL/6 mice (Fig. 1A) (13).
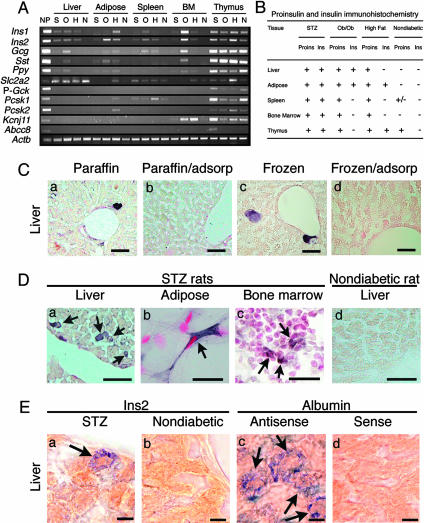
Fig. 1.
(A) RT-PCR of tissue RNA for islet-specific hormones Ins1 (insulin-1), Ins2 (insulin-2), Gcg (glucagon), Sst (somatostatin), and Ppy, and various β cell-specific transcripts including Slc2a2 (glucose transporter 2), P-Gck (glucokinase, pancreatic type), Pcsk1 (protein convertase 1/3), Pcsk2 (protein convertase 2), Kcnj11 (also known as Kir6.2), and Abcc8 (sulfonylurea receptor-1). Actb (β-actin) was used as control. BM, bone marrow; NP, nondiabetic pancreas; S, STZ-induced diabetic; O, ob/ob diabetic; H, high-fat diet-induced diabetic; N, nondiabetic. (B) Proinsulin (Proins)-positive and insulin (Ins)-positive cells as determined by immunohistochemical staining in diabetic and nondiabetic mice. +, Cells positive for proinsulin or insulin by immunohistochemistry. (C) Proinsulin-staining of the liver cells in the paraffin-embedded (a and b) and frozen (c and d) sections from STZ mice. (b and d) Immunoadsorption study for proinsulin. Proinsulin-positive cells seen in a and c were completely blocked in b and d. (D) Proinsulin-positive cells (arrows) in the liver (a), adipose tissue (b), and bone marrow (c) from STZ-induced diabetic rats and those in the liver from nondiabetic rats (d). (E) In situ hybridization. (a and b) Insulin-2 mRNA expression in the liver from STZ (a) and nondiabetic (b) mice. (c and d) Albumin mRNA expression detected with antisense probe (c) and sense probe control (d) in the liver of STZ mice. (Scale bars: 25 μm, C and D;10 μm, E.)
We next analyzed tissue sections of STZ mice by immunohistochemical staining for proinsulin and insulin (Fig. 1B). We first detected proinsulin-positive cells by using paraffin-embedded sections. The staining was specific for proinsulin because immunoadsorption blocked the staining. Because the finding was unexpected and nonspecific staining sometimes occurs with paraffin-embedded specimens, we repeated the experiment, using frozen sections and free-floating methods for immunohistochemistry. This technique had previously been shown to reduce nonspecific staining (17–20). The results confirmed the presence of proinsulin-producing cells in STZ-induced diabetic mouse livers, but not in nondiabetic controls (Figs. 1C and 2A). A few of the proinsulin-staining cells also stained positive for insulin, but the vast majority did not, indicating that the proinsulin was not processed to mature insulin in these cells, corroborating the lack of expression of the proinsulin-processing proteases Pcsk1 and Pcsk2 as assayed by RT-PCR (Fig. 1 A) in the liver of STZ mice. By RT-PCR, we also detected the presence of Pdx-1, neurogenin3, and beta2/neuroD transcripts in the liver RNA of diabetic but not of nondiabetic mice (data not shown).
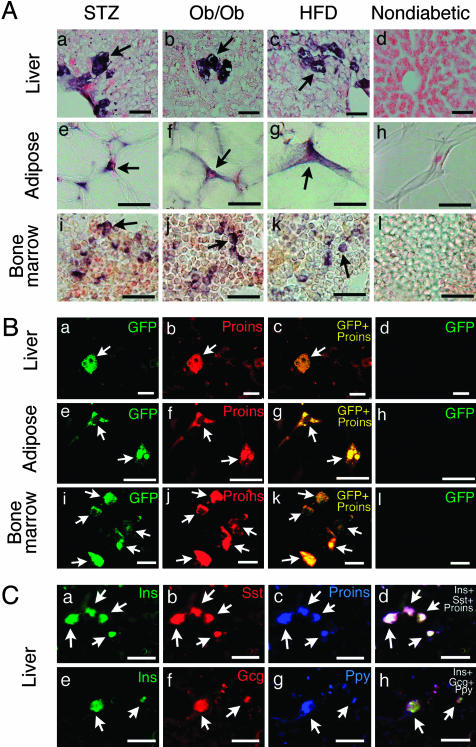
Fig. 2.
(A) Proinsulin-positive cells (arrows) in the liver (a–d), abdominal adipose tissue (e–h), and bone marrow (i–l) of STZ (a, e, and i), ob/ob (b, f, and j), and high-fat diet (HFD, c, g, and k) diabetic mice and nondiabetic mice (d, h, and l). In nondiabetic mice, proinsulin-positive cells were not found in the liver (d), adipose tissue (h), or bone marrow (l). (Scale bars, 25 μm.) (B) Overlap images of GFP/proinsulin in the liver (a–d), adipose tissue (e–h), and bone marrow (i–l) from STZ-induced diabetic (a–c, e–g, and i–k) and nondiabetic (d, h, and l) MIP-GFP mice. GFP and proinsulin signals completely overlap (arrows). (Scale bars: 25 μm, a–d; 20 μm, e–h; 10 μm, i–l.) (C) Overlap staining of insulin/somatostatin/proinsulin (a–d) and insulin/glucagon/Ppy (e–h) in the liver of ob/ob mice. Insulin, proinsulin, somatostatin, glucagon, and Ppy colocalize in most of the cells (arrows). (Scale bars, 50 μm.)
Because proinsulin expression in mouse liver cells was unexpected, we determined whether diabetes has the same effect in another species. We produced diabetes in rats by STZ and detected the presence of insulin transcripts in diabetic, but not nondiabetic control, liver RNA by RT-PCR (data not shown). By immunohistochemistry we found that diabetic, but not nondiabetic, rats displayed proinsulin-positive cells in the liver (Fig. 1Da).
We next used in situ nucleic acid hybridization and found the presence of insulin transcripts in select liver cells in diabetic mice (Fig. 1Ea) but not nondiabetic mice (Fig. 1Eb). In contrast, albumin transcripts were detected in essentially all liver cells in both diabetic and nondiabetic mice (Fig. 1Ec). Use of sense probes produced no hybridization signals (Fig. 1Ed).
To ensure that induction of insulin was not a toxic side effect of STZ, we extended our study to the ob/ob mice and the high-fat diet-induced obese diabetic mice. The ob/ob mice had a fasting blood glucose of 176 ± 32 mg/dl (means ± SD, n = 7), and the diet-induced obese diabetic mice had a fasting blood glucose of 182 ± 18 mg/dl (n = 7). Both had elevated fasting plasma insulin concentrations of 17.81 ± 4.92 ng/ml (n = 7) and 2.54 ± 1.08 ng/ml (n = 7), respectively. The fasting plasma glucose and insulin levels for age-matched nondiabetic mice were 98 ± 12 mg/dl and 0.53 ± 0.14 ng/ml (n = 5), respectively. By RT-PCR, insulin transcripts were detected in the liver of both of these diabetic mouse models (Fig. 1 A). As in STZ mice (Fig. 2 Aa), we detected the presence of proinsulin-positive cells by immunohistochemistry in liver sections of diabetic ob/ob mice (Fig. 2 Ab) and mice fed a high-fat diet (Fig. 2 Ac).
Proinsulin mRNA and Protein Are Present in Multiple Extrapancreatic Tissues. We used RT-PCR to screen for insulin gene expression in tissues other than the liver and found insulin transcripts in RNA isolated from adipose tissue, spleen, bone marrow, and thymus of diabetic mice, but from only the thymus of nondiabetic animals (Fig. 1 A), although barely detectable levels were sometimes detected in the spleen of nondiabetic mice. Insulin gene transcripts were translated into proinsulin, which was detected by immunohistochemical staining in the liver, adipose tissue, and bone marrow of diabetic mice (Fig. 2A), the same tissues that harbored the insulin transcripts. In nondiabetic mice, we found proinsulin-producing cells almost exclusively in the thymus and rarely in the spleen (Fig. 4, which is published as supporting information on the PNAS web site). Compared to nondiabetic mice, the number of proinsulin-positive cells in the thymus and especially in the spleen was much higher in mice with diabetes (Table 1, which is published as supporting information on the PNAS web site). Again, we found insulin mRNA by RT-PCR (data not shown) and proinsulin by immunohistochemical staining in adipose tissue and bone marrow, in addition to the liver (Fig. 1D b and c), in diabetic STZ rats, indicating that diabetes-associated multitissue insulin gene expression occurs in more than one species.
The distribution of proinsulin- and insulin-positive cells in diabetic mice is summarized in Fig. 1B, and the density distribution of these cells can be found in Table 1. The presence of insulin in adipose tissue in the different diabetic models was accompanied by low-level expression of the processing enzyme mRNAs detectable by RT-PCR at 40 cycles (data not shown). We failed to detect any proinsulin or insulin-producing cells in kidney, intestine, or brain (data not shown).
Colocalization of Insulin Gene Transcription in Extrapancreatic, Extrathymic Proinsulin-Positive Cells in Diabetes. The presence of insulin gene transcripts (Fig. 1E) and immunoreactive proinsulin (Figs. 1C and 2 A) suggests that the transcripts were translated into protein. However, because these experiments were performed in different animals, we had to exclude the unlikely possibility that in some instances the presence of proinsulin was a consequence of cellular uptake, not expression (21). We examined MIP-GFP mice, a transgenic mouse line expressing GFP driven by the mouse insulin promoter (12), and found that STZ-induced diabetes in these mice led to the appearance of fluorescent signals (without staining) in select cells in the liver, adipose tissue, and bone marrow (Fig. 2B a, e, and i), but not in similar tissues of nondiabetic MIP-GFP controls (Fig. 2Bd, h, and l). By immunostaining there was complete concordance of proinsulin production with GFP expression in these tissues (Fig. 2Bc, g, and k), indicating that insulin gene transcripts in the extrapancreatic tissues in diabetes are translated into proinsulin and little, if any, of the proinsulin observed in these cells results from the uptake of proinsulin produced elsewhere. The fact that we failed to detect any proinsulin-positive cells that did not also display fluorescence further supports the specificity of the immunohistochemical staining.
Other Islet Hormones Are also Expressed in Extrapancreatic Tissues. By RT-PCR we found that the liver, adipose tissue, spleen, and bone marrow from the different diabetic mouse models express transcripts of the other islet hormones, glucagon, somatostatin, and Ppy (Fig. 1 A). The highest expression was detected in the bone marrow and liver of ob/ob mice (Fig. 1 A). Immunohistochemical analysis of ob/ob liver showed that proinsulin/insulin-harboring cells also contain glucagon, somatostatin, and Ppy. Observation and photomicrography of tissue planes at 1-μm intervals under a confocal laser scanning microscope confirmed that the cells simultaneously coexpressed all four hormones (Fig. 2C). We also detected cells that produce the four islet hormones in other extrapancreatic tissues (adipose tissue, spleen, and bone marrow), albeit at much lower frequencies (data not shown).
Hyperglycemia Induces Extrapancreatic Expression of Insulin and Other Islet Hormones. The above observations indicate that extrapancreatic, extrathymic insulin gene expression occurs in multiple tissues of three different mouse models of diabetes mellitus and in a rat STZ diabetes model. The STZ diabetes model is one of insulin deficiency, whereas the diabetes in ob/ob and diet-induced obese mice is associated with hyperinsulinemia and insulin resistance. The plasma triglyceride level was normal in these different mouse models, and the cholesterol level was slightly elevated in the mice fed a high-fat diet, but not in the other models (data not shown). Therefore, hyperglycemia appears to be the common denominator in the three models. To test the hypothesis that hyperglycemia per se induces extrapancreatic insulin gene expression, we administered i.p. injections of 25% glucose to produce hyperglycemia in WT nondiabetic C57BL/6 mice (Fig. 3A) and assayed for insulin transcripts by RT-PCR on day 3 and again on day 15 of treatment. We injected control mice with 25% mannitol but unexpectedly found that mannitol led to hyperglycemia and appearance of trace amounts of insulin mRNA by RT-PCR (data not shown). We therefore used a saline control that produced no hyperglycemia and no insulin mRNA in extrapancreatic extrathymic mouse tissues (Fig. 5, which is published as supporting information on the PNAS web site). We found that 3 days of hyperglycemia induced by glucose injections induced insulin gene expression. We detected insulin-1 and insulin-2 transcripts in the five extrapancreatic, extrathymic tissues tested, being most abundant in fat (Fig. 5). Moreover, glucagon and somatostatin transcripts were also detected in adipose RNA of hyperglycemic mice, as were those for the proinsulin-processing enzymes Pcsk1 and Pcsk2. We observed a low level of somatostatin transcript in fat and liver. Ppy was either barely detectable or undetectable in tissues outside the pancreas or thymus. Interestingly, in the spleen, insulin and glucagon transcripts were often present, albeit at barely detectable levels, even in the absence of hyperglycemia. As expected, the thymus expressed transcripts for all of the genes tested in both saline- and glucose-injected mice.
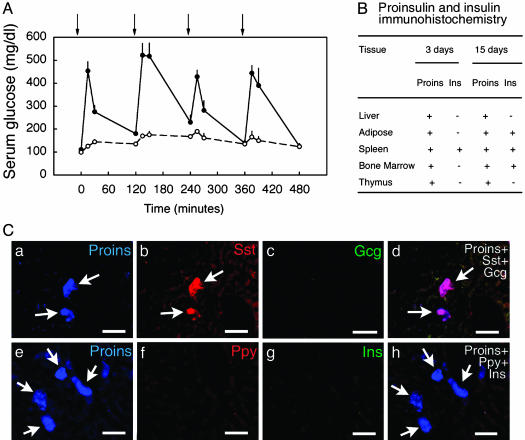
Fig. 3.
(A) Serum glucose levels in WT C57BL/6 mice treated with repeated i.p. injections (indicated by arrows) of 25% glucose (•) or saline (○); data are shown for day 7. (B) Proinsulin (Proins)-positive and insulin (Ins)-positive cells by immunohistochemical staining in mice receiving daily i.p. glucose after 3 and 15 days. (C) Proinsulin (Proins)-positive cells in the liver at day 15 of glucose injection. Proinsulin (Proins, a and e) and somatostatin (Sst) (b) colocalize in most of the cells (d, arrows); insulin (Ins) (g), glucagon (Gcg) (c), Ppy (f), and insulin (g) were not detected in these cells. (Scale bars, 25 μm.)
To determine whether the hyperglycemia-induced transcripts were translated into proteins, we studied the glucose-injected mice by immunohistochemistry. By confocal laser microscopy we found the coexpression of proinsulin and somatostatin protein within the same cells in the liver of these mice (Fig. 3Ca–d). However, we did not detect any mature insulin-, glucagon- or Ppy-producing cells in the liver (Fig. 3C). Proinsulin-producing cells were also present in fat, spleen, bone marrow, and thymus (Fig. 6, which is published as supporting information on the PNAS web site), and rare insulin-positive cells were detected in fat, spleen, and bone marrow after 15 days. The distribution of proinsulin- and insulin-positive cells in liver, fat, spleen, bone marrow, and thymus at days 3 and 15 of hyperglycemia is summarized in Fig. 3B, and the relative density distribution of these cells can be found in Table 2, which is published as supporting information on the PNAS web site. We failed to detect any proinsulin- or insulin-producing cells in kidney, intestine, or brain.
Bone Marrow Origin of Extrapancreatic Proinsulin-Producing Cells in Diabetes. Evidence thus far indicates that the hyperglycemia of diabetes induces proinsulin production in multiple tissues, including the bone marrow. The possible hematopoietic origin of some of these cells outside the pancreas or bone marrow was suggested by the coexpression of CD45 in some, although not all, of the proinsulin-producing cells in STZ mice (Fig. 7, which is published as supporting information on the PNAS web site). We performed BMT from nondiabetic mice that constitutively expressed β-galactosidase to nondiabetic C57BL/6 mice and induced diabetes in the recipient animals 8 weeks later by STZ treatment. By immunohistochemistry 2 weeks after STZ-induced diabetes, 23% of the bone marrow cells of recipient mice stained positive for proinsulin (Fig. 7B a–c). Most (88%) of the proinsulin-positive cells in the bone marrow also stained positive for β-galactosidase (Fig. 7B a–c). In the liver, 90% of proinsulin-positive cells were also positive for β-galactosidase (Fig. 7Bd–f).
Similarly, we analyzed mice by glucose injection-induced hyperglycemia after BMT (Fig. 7Ca–c). At 15 days after induced hyperglycemia, 29.8% of the bone marrow cells stained positive for proinsulin and 75% of the proinsulin-positive cells contained immunoreactive β-galactosidease (Fig. 7Ca–c). In the liver, 96% of the proinsulin-positive cells were also positive for β-galactosidase (Fig. 7C d–f). Conversely, in the liver of both STZ- and glucose-injected mice, 100% of β-galactosidase-positive cells were positive for proinsulin. We conclude that hyperglycemia induces the appearance of proinsulin-producing cells in the bone marrow. When these cells migrate outside the bone marrow and home to the liver, they do so with a markedly increased efficiency over bone marrow cells that do not produce proinsulin. The alternative explanation is that in hyperglycemic states essentially all bone marrow cells that migrate to the liver acquire the capacity to produce proinsulin after they reach the liver. It is possible that both scenarios happen to a varying degree.
To investigate whether similar mechanisms are operative in the pancreas, we performed immunohistochemical staining of the pancreas of BMT recipient mice that were treated with STZ or glucose injections. In STZ mice the area density of proinsulin-positive cells in the pancreas was 5.2 ± 0.4% that of normal mice. Some islet remnants were evident (Fig. 7Bg). In these remnants, none of the proinsulin-positive cells were positive for β-galactosidase, CD45, or Thy-1 (data not shown), suggesting that they were not of hematopoietic origin. Interestingly, we observed the presence of β-galactosidase-positive cells in the acinus, all of which also stained positive for proinsulin (Fig. 7B g–i) and CD45 (data not shown). For mice with glucose-induced hyperglycemia on both days 3 and 15 of injection, we failed to detect any significant islet hypertrophy or hyperplasia because islet sizes did not differ significantly from those of untreated nondiabetic mice. An extensive search failed to reveal any β-galactosidase-positive cells within the islets (Fig. 7Cg–i). However, as in the STZ model, we observed very rare β-galactosidase-positive cells in the acinus, all of which also stained positive for proinsulin (data not shown). Therefore, in mice with hyperglycemia caused by STZ-induced diabetes or glucose injections, all β-galactosidase-positive, and thus bone marrow-derived, cells in the pancreas also expressed proinsulin. Compared with those in the liver or adipose tissue, such cells are much rarer in the pancreas. However, these rare β-galactosidase-positive and proinsulin-positive cells seemed to have preferentially homed to the acinar, not the endocrine, pancreas of these animals.
Discussion
In this article, we made the surprising observation that hyperglycemia, with or without established diabetes, activates insulin gene transcription and proinsulin production in multiple extrapancreatic, extrathymic tissues. Bone marrow-derived cells (BMDC) appear to be a major source of proinsulin-expressing cells in these tissues. In the liver we also detected a few proinsulin-expressing cells that were negative for β-galactosidase, indicating their origin from non-bone marrow cells or from the progeny of BMDC fused with cells in the liver of the recipient mice, as was observed recently in a liver regeneration model (22, 23).
The contributions of BMDC to the pancreas vary according to the model studied. Ianus et al. (24) found that BMDC populate pancreatic islets in nondiabetic mice. In contrast, Hess et al. (25) found that BMT into STZ-induced diabetic mice stimulated islet neogenesis and the appearance of BMDC that stained positive for an endothelial cell marker. In our model, which involves BMT to nondiabetic mice followed by induction of diabetes in the recipients, proinsulin-positive BMDC appeared in the acinar pancreas. This location of BMDC paralleled that found by Li et al. (26) who performed BMT to E2f1-/-E2f2-/- mice that had diabetes and pancreatic acinar degeneration. They detected proinsulin-negative BMDC in the vicinity of pancreatic islets, presumably in the acinar region that had been replaced with scar tissue.
During embryonic development, extrapancreatic insulin gene expression is detectable in the yolk sac (27) and brain (28) of rodents. In adult mammals, apart from the thymus and other peripheral lymphoid tissues, which express many “peripheral proteins” (5), insulin is thought to be produced only in the β cells in the pancreas. The presence of insulin-related material in the brain and other extrapancreatic tissues of adult nondiabetic vertebrates was reported 20 years ago (reviewed in ref. 29). The nature of the insulin-related material was unclear, and by immunohistochemical staining we failed to detect any immunoreactive proinsulin or insulin-positive cells in brain or other extrapancreatic, extrathymic tissues in nondiabetic mice.
There are some important implications for the extrapancreatic, extrathymic insulin gene expression induced by hyperglycemia. Self-tolerance induced by low-level insulin expression in the thymus and lymphoid tissues is thought to be important in preventing the development of autoimmunity to insulin (ref. 8 and reviewed in refs. 5 and 30), and restoration of self-tolerance may be an approach to prevent, control, or alleviate β cell autoreactivity in type 1 diabetes (10). Administration of insulin either orally or s.c. in NOD mice leads to a decrease in the incidence of diabetes (reviewed in ref. 7). Pilot studies suggest that low-dose insulin therapy may also delay the onset of diabetes in individuals with insulin and other autoantibodies (31–33), although a recent clinical trial failed to delay the onset of diabetes in patients with a high risk for type 1 diabetes (34). The most likely mechanism for the prevention of diabetes in mice and people by low-dose insulin therapy is the induction of tolerance to insulin. Would the occurrence of extrathymic and extrapancreatic insulin expression during a brief period of hyperglycemia in humans who are predisposed to type 1 diabetes, e.g., in utero for a fetus, offer any protection, or will it accelerate or precipitate the onset of diabetes?
Both transgenic expression of proinsulin in the thymus driven by the MHC class II promoter (35) and the intrathymic administration of insulin B chain (36) have been shown to inhibit diabetes development in NOD mice. Mice expressing subnormal, but not those expressing normal, levels of insulin display detectable T cell peripheral reactivity to insulin, underscoring the role of minor variation in intrathymic proinsulin expression in insulin-specific T cell self-tolerance (11). We showed that hyperglycemia increases the density of intrathymic proinsulin-producing cells (Table 1). Would a brief period of hyperglycemia upregulate intrathymic proinsulin production, affect the self-tolerance to insulin, and alter the clinical course of autoimmune diabetes in those who are predisposed to the disease?
The induction of insulin production from non-β cells, such as embryonic stem cells (37–39) and adult progenitor cells (40, 41), for the treatment of diabetes is an active area of investigation. The manipulations used include elaborate and prolonged (many months) exposure of the cells to high glucose concentration in a special medium (38). Herein, we showed that a mere 3-day exposure of mice to elevated blood glucose was sufficient to activate extrapancreatic insulin-1 and insulin-2 gene expression. The appearance of extrapancreatic insulin gene expression in response to a relatively brief period of hyperglycemia has important implications for the regulation of insulin gene expression in non-β cells and our continued effort to generate insulin-producing cells for the treatment of diabetes.
Supplementary Material
Acknowledgments
We thank Maiko Ninomiya, Atsunori Kashiwagi, and Ryuichi Kikkawa for advice; Margaret A. Goodell and Kathyjo A. Jackson for assistance with the bone marrow transplantation and critical readings of the manuscript; and Takefumi Yamamoto (Central Research Laboratory, Shiga University of Medical Science) for technical support in laser scanning microscopy. This research was supported by National Institutes of Health Grants DK61245 and DK20595 (to M.H.) and National Institutes of Health Grant HL16512/DK068037 and the Betty Rutherford Chair from St. Luke's Episcopal Hospital and Baylor College of Medicine (to L.C.).
Abbreviations: STZ, streptozotocin; BMT, bone marrow transfer; Ppy, pancreatic polypeptide; BMDC, bone marrow-derived cells.
References
- 1.Ohneda, K., Ee, H. & German, M. (2000) Semin. Cell. Dev. Biol. 11, 227-233. [DOI] [PubMed] [Google Scholar]
- 2.Vafiadis, P., Bennett, S. T., Todd, J. A., Nadeau, J., Grabs, R., Goodyer, C. G., Wickramasinghe, S., Colle, E. & Polychronakos, C. (1997) Nat. Genet. 15, 289-292. [DOI] [PubMed] [Google Scholar]
- 3.Pugliese, A., Zeller, M., Fernandez, A., Jr., Zalcberg, L. J., Bertlett, R. J., Ricordi, C., Pietropaolo, M., Eisenbarth, G. S., Bennett, S. T. & Patel, D. D. (1997) Nat. Genet. 15, 293-297. [DOI] [PubMed] [Google Scholar]
- 4.Derbinski, J., Schulte, A., Kyewski, B. & Klein, L. (2001) Nat. Immunol. 2, 1032-1039. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan, D. (1998) Curr. Opin. Immunol. 10, 656-662. [DOI] [PubMed] [Google Scholar]
- 6.Moriyama, H., Abiru, N., Paronen, J., Sikora, K., Liu, E., Miao, D., Devendra, D., Beilke, J., Gianani, R., Gill, R. G., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 10376-10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottlieb, P. A. & Eisenbarth, G. S. (2002) Clin. Immunol. 102, 2-11. [DOI] [PubMed] [Google Scholar]
- 8.Jolicoeur, C., Hanahan, D. & Smith, K. M. (1994) Proc. Natl. Acad. Sci. USA 91, 6707-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugliese, A., Brown, D., Garza, D., Murchison, D., Zeller, M., Redondo, M., Diez, J., Eisenbarth, G. S., Patel, D. D. & Ricordi, C. (2001) J. Clin. Invest. 107, 555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooke, A., Phillips, J. M. & Parish, N. M. (2001) Nat. Immunol. 2, 810-815. [DOI] [PubMed] [Google Scholar]
- 11.Chentoufi, A. A. & Polychronakos, C. (2002) Diabetes 51, 1383-1390. [DOI] [PubMed] [Google Scholar]
- 12.Hara, M., Wang, X., Kawamura, T., Bindokas, V. P., Dizon, R. F., Alcoser, S. Y., Magnuson, M. A. & Bell, G. (2003) Am. J. Physiol. 284, E177-E183. [DOI] [PubMed] [Google Scholar]
- 13.Kojima, H., Fujimiya, M., Matsumura, K., Younan, P., Imaeda, H., Maeda, M. & Chan, L. (2003) Nat. Med. 9, 596-603. [DOI] [PubMed] [Google Scholar]
- 14.Kojima, H., Nakamuta, T., Fujita, Y., Kishi, A., Fujimiya, M., Yamada, S., Kudo, M., Nishio, Y., Maegawa, H., Haneda, M., et al. (2002) Diabetes 51, 1398-1408. [DOI] [PubMed] [Google Scholar]
- 15.Fujimiya, M., Okumiya, K., Yamane, T. & Maeda, M. (1997) Histochem. Cell Biol. 107, 105-114. [DOI] [PubMed] [Google Scholar]
- 16.Nishimura, T., Yamada, H., Kinoshita, M. & Ochi, J. (1994) Biomed. Res. 15, 291-298. [Google Scholar]
- 17.Kohler, C. (1983) J. Comp. Neurol. 221, 247-262. [DOI] [PubMed] [Google Scholar]
- 18.Yu, P. L., Fujimura, M., Okumiya, K., Kinoshita, M., Hasegawa, H. & Fujimiya, M. (1999) J. Comp. Neurol. 411, 654-665. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, H., McGeer, P. L., Peng, F. & McGeer, E. G. (1980) Science 208, 1057-1059. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, H., McGeer, P. L., Peng, J. H. & McGeer, E. G. (1981) J. Comp. Neurol. 200, 151-201. [DOI] [PubMed] [Google Scholar]
- 21.Rajagopal, J., Anderson, W. J., Kume, S., Martinez, O. I. & Melton, D. A. (2003) Science 299, 363. [DOI] [PubMed] [Google Scholar]
- 22.Wang, X., Willenbring, H., Akkari, Y., Torimaru, Y., Foster, M., al-Dhalimy, M., Lagasse, E., Finegold, M., Olson, S. & Grompe, M. (2003) Nature 422, 897-900. [DOI] [PubMed] [Google Scholar]
- 23.Vassllopoulos, G., Wang, P.-R. & Russell, D. W. (2003) Nature 422, 901-904. [DOI] [PubMed] [Google Scholar]
- 24.Ianus, A., Holz, G. G., Theise, N. D. & Hussain, M. A. (2003) J. Clin. Invest. 111, 843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hess, D., Martin, M., Sakano, S., Hill, D., Strutt, B., Thyssen, S., Gray, D. A. & Bhatia, M. (2003) Nat. Biotechnol. 21, 763-770. [DOI] [PubMed] [Google Scholar]
- 26.Li, F. X., Zhu, J. W., Tessem, J. S., Beilke, J., Varella-Garcia, M., Jensen, J., Hogan, C. J. & DeGregori, J. (2003) Proc. Natl. Acad. Sci. USA 100, 12935-12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giddings, S. J. & Carnaghi, L. (1989) J. Biol. Chem. 264, 9462-9469. [PubMed] [Google Scholar]
- 28.Deltour, L., Leduque, P., Blume, N., Madsen, O., Dubois, P., Jami, J. & Bucchini, D. (1993) Proc. Natl. Acad. Sci. USA 90, 527-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Roith, D., Hendricks, S. A., Lesniak, M. A., Rishi, S., Becker, K. L., Havrankova, J., Rosenzweig, J. L., Brownstein, M. J. & Roth, J. (1983) Adv. Metab. Disord. 10, 303-340. [DOI] [PubMed] [Google Scholar]
- 30.Pugliese, A. & Miceli, D. (2002) Diabetes/Metab. Res. Rev. 18, 13-25. [DOI] [PubMed] [Google Scholar]
- 31.Keller, R. J., Eisenbarth, G. S. & Jackson, R. A. (1993) Lancet 341, 927-928. [DOI] [PubMed] [Google Scholar]
- 32.Ziegler, A., Bachmann, W. & Rabl, W. (1993) Diabetes Metab. Rev. 9, 289-293. [DOI] [PubMed] [Google Scholar]
- 33.Fuchtenbusch, M., Rabl, W., Grassl, B., Bachmann, W., Standl, E. & Ziegler, A. G. (1998) Diabetologia 41, 536-541. [DOI] [PubMed] [Google Scholar]
- 34.Diabetes Prevention Trial–Type 1 Diabetes Study Group (2002) N. Engl. J. Med. 346, 1685-1691. [DOI] [PubMed] [Google Scholar]
- 35.French, M. B., Allison, J., Cram, D. S., Thomas, H. E., Dempsey-Collier, M., Silva, A., Georgiou, H. M., Kay, T. W., Harrison, L. C. & Lew, A. M. (1997) Diabetes 46, 34-39. [DOI] [PubMed] [Google Scholar]
- 36.Cetkovic-Cvrlje, M., Gerling, I. C., Muir, A., Atkinson, M. A., Elliot, J. F. & Leiter, E. H. (1997) Diabetes 46, 1975-1982. [DOI] [PubMed] [Google Scholar]
- 37.Hori, Y., Rulifson, I. C., Tsai, B. C., Heit, J. J., Cahoy, J. D. & Kim, S. K. (2002) Proc. Natl. Acad. Sci. USA 99, 16105-16110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, L., Li, S., Hatch, H., Ahrens, K., Cornelius, J. G., Petersen, B. E. & Peck, A. B. (2002) Proc. Natl. Acad. Sci. USA 99, 8078-8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soria, B., Roche, E., Berna, G., Leon-Quinto, T., Reig, J. A. & Martin, F. (2000) Diabetes 49, 157-162. [DOI] [PubMed] [Google Scholar]
- 40.Lechner, A. & Habener, J. F. (2003) Am. J. Physiol. 284, E259-E266. [DOI] [PubMed] [Google Scholar]
- 41.Ramiya, V. K., Maraist, M., Arfors, K. E., Schatz, D. A., Peck, A. B. & Cornelius, J. G. (2000) Nat. Med. 6, 278-282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.