Summary
The RV144 HIV-1 trial of the canary pox vector (ALVAC-HIV) plus the gp120 AIDSVAX B/E vaccine demonstrated an estimated efficacy of 31%, that correlated directly with antibodies to HIV-1 envelope variable regions 1 and 2 (V1–V2). Genetic analysis of trial viruses revealed increased vaccine efficacy against viruses matching the vaccine strain at V2 residue 169. Here, we isolated four V2 monoclonal antibodies from RV144 vaccinees that recognize residue 169, neutralize laboratory-adapted HIV-1, and mediate killing of field isolate HIV-1-infected CD4+ T cells. Crystal structures of two of the V2 antibodies demonstrated residue 169 can exist within divergent helical and loop conformations, which contrasted dramatically with the beta strand conformation previously observed with a broadly neutralizing antibody PG9. Thus, RV144 vaccine-induced immune pressure appears to target a region that may be both sequence variable and structurally polymorphic. Variation may signal sites of HIV-1 envelope vulnerability, providing vaccine designers with new options.
INTRODUCTION
Development of a safe and effective HIV-1 vaccine is a global priority. After several failed efficacy trials, in 2009 the HIV-1 field was encouraged by an estimated 31.2% vaccine efficacy in the RV144 Thai HIV-1 vaccine efficacy trial that used a canarypox virus vector (ALVAC) prime and a combination of clades B and E gp120 (AIDSVAX gp120 B/E) proteins as a boost (Rerks-Ngarm et al., 2009). This trial provided hope that a vaccine could induce protective immune responses to HIV-1 (Rerks-Ngarm et al., 2009). In 2012 an immune correlates study of the RV144 trial revealed that antibodies against the Env gp120 V1–V2 region were associated with lower risk of infection (Haynes et al., 2012a). Epitope mapping of plasma V1–V2 antibody responses showed that within V2, vaccine-induced antibodies targeted a region of HIV-1 Env, amino acid (aa) residues at positions163–178 (Karasavvas et al., 2012; Zolla-Pazner et al., 2011). There is considerable sequence variability in V1–V2, ~75% of the residues are conserved or demonstrated to be only conservative changes (Zolla-Pazner and Cardozo, 2010). Whereas the demonstration that V1–V2 antibody responses directly correlated with decreased infection risk was suggestive of their protective role in the trial, this association was not sufficient for proving causation of protection (Plotkin and Gilbert, 2012). Indeed further studies are needed to evaluate the ability of such responses to mediate immune pressure on HIV-1. Viral genetic (sieve) analyses, isolation of V1–V2 antibodies and understanding their effector function in vitro and in vivo, as well as validation of correlates of infection risk in future vaccine trials are needed.
By comparing sequences of breakthrough infections that occur in vaccinees versus placebo recipients, genetic or sieve analysis of sequences of viruses that caused breakthrough infections in a vaccine trial can demonstrate sites of immune pressure (Rolland et al., 2011). A recent genetic analysis of breakthrough HIV-1 infections in the RV144 trial demonstrated 48% (CI: 18 to 68%, p=0.0036) vaccine efficacy against viruses matching the CRF_01AE vaccine immunogens with a lysine (K) at position 169 (Rolland et al., 2012). Thus, it is critical to determine the binding site and effector functions of RV144-induced V1–V2 antibodies. Antibody effector function candidates for mediation of protection from HIV-1 transmission include the ability of V1–V2 antibodies to neutralize those virus strains involved in HIV-1 transmission (i.e. transmitted-founder viruses) (Keele et al., 2008), or to mediate other effector functions such as antibody-dependent cellular cytotoxicity (ADCC) (Haynes et al., 2012a). Herein, we have probed the specificities and effector functions of four V2 monoclonal antibodies (mAbs) isolated from RV144 ALVAC-AIDSVAX vaccine recipients, and determined the crystal structures of two of these mAbs with V2 peptides containing position 169.
We show that V2 residue 169 was in a structurally polymorphic region of HIV-1, thus revealing the structural heterogeneity of the V2 region to which Env immune responses correlated with decreased transmission. We go on to show that the V2 antibodies isolated from RV144 vaccinees mediated ADCC against RV144 trial breakthrough Env-target cells, and this ADCC activity was dependent on position 169 in breakthrough Envs. These data directly demonstrate the plausibility of these types of V2 antibodies to mediate immune pressure targeted at position169 of Env V2.
RESULTS
Vaccine-Induced Antibodies
Using clonal memory B cell cultures and high throughput antigen-specific screening assays (Bonsignori et al., 2011), two mAbs, CH58 and CH59, were initially isolated and found to bind the RV144 vaccine immunogen AE.A244 gp120 and to a V2 peptide (residues 169–182, KKKVHALFYKLDIV) (Table S1; Figure S1A). Epitope mapping of mAbs CH58 and CH59 using alanine-scanning peptides with the sequence of AE.A244 Env from positions 165 to186 (LRDKKQKVHALFYKLDIVPIED) showed that the footprint for binding of mAb CH58 involved 10 residues (K168, K169, K171, V172, H173, F176, Y177, K178, D180, P183), while that of mAb CH59 involved 4 residues (K169, H173, F176, Y177) (Figure 1A). HIV-1 Env V1–V2 broadly neutralizing antibodies (bnAbs) have been described that bind both glycans as well as bind V1–V2 aa residues around and including position 169 (Bonsignori et al., 2011; Doria-Rose et al., 2012; McLellan et al., 2011; Walker et al., 2009). The epitope mapping results suggested that mAbs CH58 and CH59 (while themselves not bnAbs) bind at or near the site in V2 to which the HIV-1 V1–V2 bnAbs (e.g. PG9, CH01) bind (Doria-Rose et al., 2012; McLellan et al., 2011)
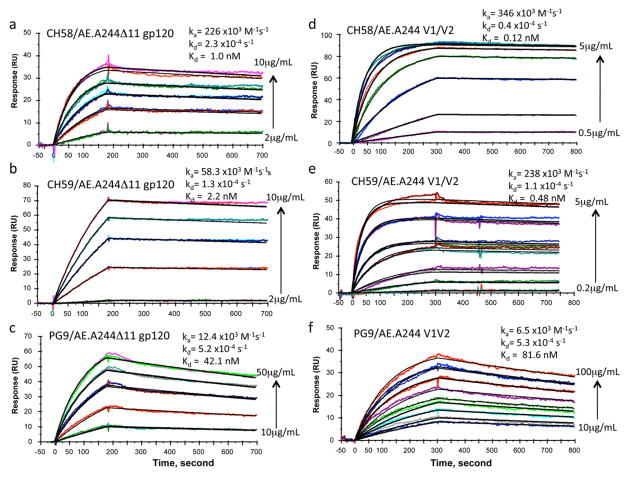
Figure 1. Binding of RV44 mAbs CH58 and CH59 to HIV-1-infected cells and to HIV-1 V2 peptides.
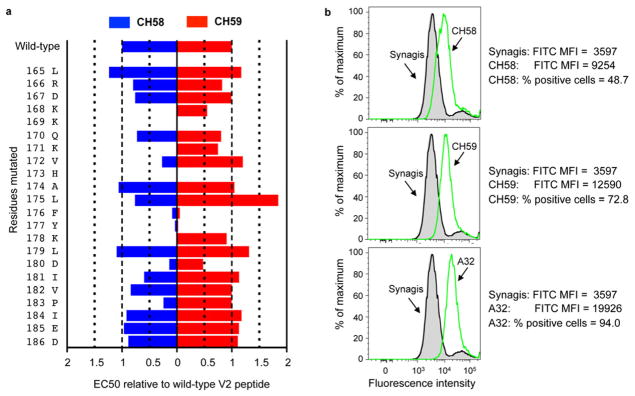
A. Effect of alanine point substituted mutations on the binding of CH58 (in blue) and CH59 (in red) to the HIV-1 V2 peptide. For each mutation (y axis), results were normalized as EC50 relative to wild-type V2 peptide. B. Shown is the flow cytometric analysis of binding of mAbs CH58 (upper left), CH59 (middle left), and A32 (lower left) to the activated PB CD4+ T cells infected IMCCM235. Synagis (anti-respiratory syncytial virus mAb) and mAb HIV-1 A32 were used as negative and positive controls, respectively. Mean fluorescence intensity (MFI) and % of positive cells are indicated next to the histograms. Data shown are representative of 3 independent experiments. See also Figure S1 and Table S1.
CH58 and CH59 bound to two Env immunogens used in the ALVAC vCP1521-AIDSVAX B-E vaccine (AE.A244 and AE.92TH023) (Table S1). Both antibodies bound to the surface of CD4+ T cells infected with infectious molecular clone (IMC) of tier 2 (Seaman et al., 2010) HIV-1 AE.CM235 and mediated ADCC with CEM.NKRCCR5 cells as targets (Figure S1B). In Surface Plasmon Resonance (SPR) assays, mAbs CH58, CH59 and PG9 bound well to AE.A244 gp120Δ11 with Kds of 1.0 nM, 2.2 nM and 42.1 nM, respectively (Figures 2A–C, Figure S2), Similarly, CH58, CH59 and PG9 also bound well to AE.A244 V1–V2 tags protein with Kds of 0.12 nM, 0.48 nM, and 81.6 nM, respectively (Figures 2D–F). Binding kinetics of PG9 was distinct from CH58 and CH59 in having slower association rates (about 40-fold to AE.A244 V1–V2 tags and about 6–15 fold slower to AE.A244 gp120Δ11) (Figure 2). Thus, both RV144 V2 mAbs CH58 and CH59 and V1–V2 bnAb PG9 bound to recombinant HIV-1 Env proteins, A244 V1–V2 tags and AE.A244 gp120Δ11. HIV-1 gp120 has been demonstrated to interact with the α4β7 molecule on CD4+ T cells using a tripeptide (LDI) motif in V2 (Arthos et al., 2008; Nawaz et al., 2011). Both CH58 and CH59 mAbs inhibited interactions of V2 peptide with α4β7 (Table S2). When assayed for ability to capture infectious viruses, CH58 and CH59 captured only infectious virions of AE.92TH023 strain but not of HIV-1 AE.CM244 strain (Table S2). Thus, a prime candidate for vaccine-induced protective function is via antibody binding to virus-infected CD4+ T cells.
Figure 2. Binding of RV144 V2 and PG9 bnAbs to AE.A244 V1–V2 tags protein and AE.A244 gp120Δ11.
Each of the mAbs was captured on an anti-Fc antibody immobilized sensor surface to about 100–125 RU. For binding to A244gp120 Δ11, 2–10 ug/mL (CH58, A), 2–10 ug/mL (CH59, B), 10–50 ug per mL (PG9, C) of monomeric gp120 were injected over each of the mAbs. AE.A244 V1–V2 tags protein was injected at concentrations ranging from 0.5 – 5ug/mL (CH58, D), 0.1 – 5 ug/mL (CH59, E), 10–100 ug/mL (PG9, F). A negative control mAb (Synagis) was used to subtract non-specific binding. Each plot shows binding curves with increasing concentrations of gp120 or V1–V2 proteins (shown in different colors) injected over two independent flow cells immobilized with the same mAbs. For binding to CH58 and CH59 mAbs, A244 gp120Δ11 protein was injected at 2, 4, 6, 8, and 10ug/mL and AE.A244 V1–V2 protein at 0.2, 0.5, 1, 2, 3, 4 and 5 ug/mL. For PG9 mAb, A244 gp120Δ11 and AE.A244 V1–V2 proteins were injected at 10, 20, 30, 40 and 50ug/mL and 10, 25, 50, 75 and 100 ug/mL respectively. Global Curve fitting (shown in black) to a 1:1 Langmuir model was used to derive rate constants and Kd values following simultaneous fitting to binding data from two independent flow cells with the same mAb captured. A third flow cell with each of the mAbs gave similar rate constant values. See also Figure S2 and Tables S2.
HIV-1 strains can be classified in tiers according to their relative ease or difficulty by which they are neutralized by Env antibodies (Seaman et al., 2010). Tier 1 HIV-1 strains are laboratory-adapted and neutralization-sensitive, whereas tier 2 HIV-1 strains are from field isolates and are more difficult to neutralize; no plasma tier 2 neutralizing activity was found in RV144 vaccinees (Montefiori et al., 2012). Both CH58 and CH59 mAbs neutralized the tier 1 virus pseudotyped with AE.92TH023 Env that was included in ALVAC as prime vaccine immunogen (Rerks-Ngarm et al., 2009) (Table S3). Neutralization of AE.92TH023 by both antibodies was abrogated when a K169Q mutation was introduced in the virus envelope by (Table S3). MAbs CH58 and CH59 also neutralized the tier 1 clade C SHIV1157ipEL-p, but not the tier 1 B.MN or the tier 2 AE.CM244 strains that comprised the gp120 components in AIDSVAX vaccine, nor did they neutralize other tier 1 or 2 HIV-1 strains tested (Table S3). Although both the tier 2 AE.CM244 and tier 1 AE.92TH023 HIV-1 strains have identical V1–V2 sequences spanning residues 165–184 that contain the CH58/CH59 epitopes, mAbs CH58 and CH59 neutralized only HIV-1 tier 1 strain, AE.92TH023. The differential neutralization of tier 1 AE.92TH023 but not tier 2 AE.CM244 can be explained by the failure of CH58 and CH59 to capture tier 2 AE.CM244 infectious virions, while both antibodies can capture tier 1 AE.92TH023 infectious virions (Table S2). Thus, neutralization of HIV-1 is not a likely candidate for protective effector function of vaccine induced antibodies.
Since bnAbs that target V1–V2 regions (Bonsignori et al., 2011; Walker et al., 2009) (e.g. PG9, PG16 and CH01) and V2 conformational mAbs (e.g. 697D) (Gorny et al., 2012) that also target epitopes in V2 are glycan-dependent, we next used peptides, recombinant gp120 and V1–V2 constructs to determine the dependence of CH58 and CH59 binding on adjacent N160, N156 glycan sites. Whereas 697D, PG9, PG16 and CH01 did not bind to linear and cyclic V2 peptides (not shown), both CH58 and CH59 bound well to V2 peptide (KKKVHALFYKLDIV) with EC50 of 0.06 μg/ml and 0.003 ug/ml, respectively (Figure S1A). Thus, the binding of CH58 and CH59 to V2 is glycan-independent.
We have previously described that bnAbs PG9, PG16 and CH01 bound well to recombinant HIV-1 Env AE.A244 gp120 (Bonsignori et al., 2011). We constructed N160K mutants of AE.A244 gp120 and found that whereas the binding of bnAbs PG9, PG16 and CH01 to AE.A244 gp120 was abrogated by the N160K mutation, the binding of CH58 and CH59 mAbs was not (Table S1). In addition, native deglycosylation (Ma et al., 2011) (that removes all glycans) of the AE.A244 V1–V2 tags protein (Figure S1C) abrogated the binding of V1–V2 bnAbs CH01, PG9 and PG16 and decreased the binding of mAb 697D six-fold but did not affect CH58 and CH59 Env binding (Table S1). Similarly, the N156Q, N160Q mutations had no effect on CH58 and CH59 binding to A244 V1–V2 tags protein, while the two mutations completely abrogated the binding of bnAbs PG9, PG16, and CH01 and markedly decreased the binding by mAb 697D, thus confirming the glycan independence of CH58 and CH59 V1–V2 binding (Figure S1D).
Crystal Structures of Vaccine-Induced Antibodies and HIV-1 Env
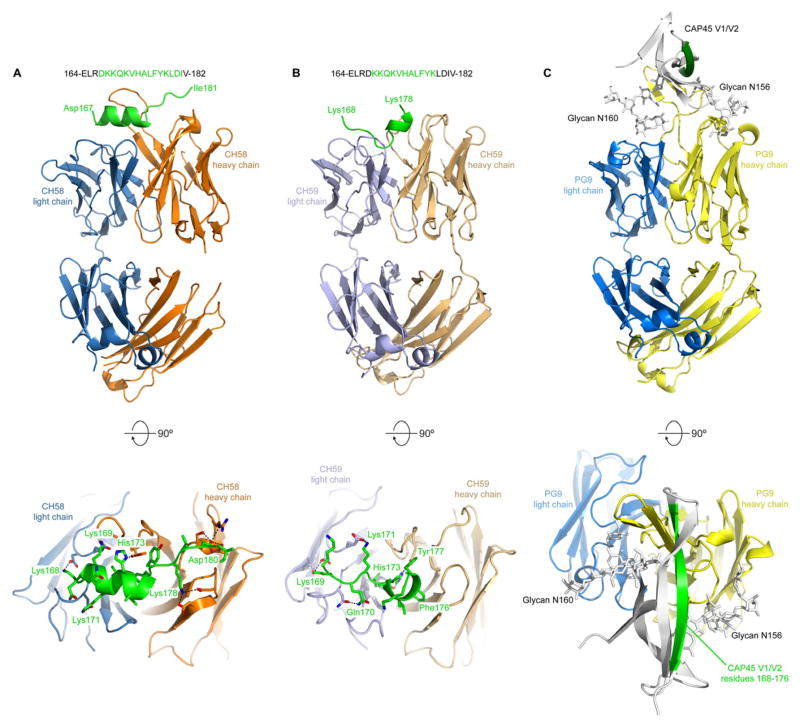
We determined a crystal structure of the CH58 antigen-binding fragment (Fab) alone, but several residues in the third complementarity determining region of the heavy chain (CDRH3) were disordered (Figure S3A). We next determined crystal structures of the CH58 or CH59 Fabs in complex with an AE.92TH023 Env V2 peptide 164-ELRDKKQKVHALFYKLDIV-182 to 1.7 Å and 1.5 Å, respectively. The CH58:peptide structure was refined to an Rcryst/Rfree of 0.185/0.211 (Table 1) and revealed that CH58 recognizes V2 residues 167–176 as an α-helix and residues 177–181 as an extended coil (Figure 3A). Surprisingly, the CH59:peptide structure, which was refined to an Rcryst/Rfree of 0.171/0.187 (Table 1), revealed that CH59 recognizes residues 168–173 as coil or turn, and residues 174–176 as a short 310 helix (Figure 3B). Thus, the two Fabs recognize similar V2 residues in completely different conformations. In general, the interactions observed in the crystal structures agreed well with the peptide mapping studies, and both Fabs made multiple hydrogen bond or salt bridge interactions with K169 and H173. Collectively, these results demonstrate antibody binding sites for both mAbs CH58 and CH59 at the position of imputed immune pressure (K169) induced by ALVAC-AIDSVAX B-E vaccination (Rolland et al., 2012).
Table 1.
Data Collection and Refinement Statistics (Molecular Replacement). See also Figure S2.
| PDB ID | CH58 Fab | CH58 w/V2 peptide | CH59 w/V2 peptide |
|---|---|---|---|
|
| |||
| 4HQQ | 4HPO | 4HPY | |
| Data collection | |||
| Space group | P212121 | C2 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 63.0, 70.4, 135.8 | 140.6, 75.8, 54.7 | 41.9, 79.2, 127.1 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 112.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0-2.4 (2.44-2.40)* | 50.0-1.7 (1.73-1.70)* | 20.0-1.5 (1.53-1.50)* |
| Rsym or Rmerge (%) | 13.6 (50.6) | 9.6 (38.7) | 11.8 (87.4) |
| I/σI | 18.2 (2.0) | 19.5 (1.9) | 21.2 (3.7) |
| Completeness (%) | 97.7 (82.8) | 90.7 (56.1) | 100 (100) |
| Redundancy | 6.3 (3.3) | 3.3 (2.2) | 7.0 (5.6) |
| Refinement | |||
| Resolution (Å) | 25.8-2.40 | 24.8-1.69 | 19.9-1.50 |
| No. reflections | 23,681 | 53,364 | 68,657 |
| Rwork/Rfree | 0.183/0.225 | 0.185/0.211 | 0.171/0.187 |
| No. atoms | |||
| Protein | 3,232 | 3,413 | 3,344 |
| Ligand/ion | - | 24 | 12 |
| Water | 191 | 403 | 521 |
| B-factors | |||
| Protein | 52.8 | 42.1 | 16.5 |
| Ligand/ion | - | 92.7 | 22.1 |
| Water | 49.1 | 42.4 | 32.1 |
| R.m.s. deviations | |||
| Bond lengths(Å) | 0.004 | 0.007 | 0.004 |
| Bond angles (°) | 0.87 | 1.11 | 1.00 |
Values in parentheses are for highest-resolution shell.
Figure 3. Structures of antibodies CH58 and CH59 bound to an HIV-1 gp120 V2 peptide.
Vaccine-elicited antibodies CH58 and CH59 recognize alternative conformations of V2 compared to bnAb PG9. A. top: Ribbon representation of the CH58 antigen-binding fragment in complex with an A244 V2 peptide. Heavy chain is colored orange, light chain is blue, and peptide is green. The sequence of the peptide is shown, with modeled residues in green; bottom: Close-up of the top panel rotated 90° about a horizontal axis. The side-chains of residues involved in hydrogen bonds or salt bridges are shown as sticks, with the interactions depicted as dashed lines. B, Structure of CH59 in complex with peptide, depicted as in A. The heavy chain is tan, and the light chain is light blue. C, Structure of bnAb PG9 in complex with the V1–V2 domain from HIV-1 strain CAP45 (PDB ID: 3U4E)(McLellan et al., 2011). The PG9 structure is shown as ribbons with heavy and light chains (colored yellow and blue, respectively) in the same orientation as in A and B. The V1–V2 domain is shown as a grey ribbon with residues 168–176 colored green, and N-linked glycans attached to residues Asn156 and Asn160 shown as sticks. See also Figures S3 and Table S3.
CH58 and CH59 mAb binding sites also involved residues that are components of the bnAb PG9 binding site(Doria-Rose et al., 2012) (McLellan et al., 2011) (Figure 3C). In addition to binding to aa within 168 – 178 of a scaffolded-V1–V2 recombinant protein, mAb PG9 binds to N-linked glycans at N160 and N156 (Pancera et al., 2010; Walker et al., 2009). PG9 also recognizes positively charged V2 residues using a combination of sulfated tyrosines and acidic residues. MAbs CH58 and CH59, however, demonstrated no tyrosine sulfation (Figure S3B).
The crystal structures of CH58, CH59 and PG9 antibodies in complex with their epitopes reveal substantial differences in the conformations of V2 residues 168–176. These residues are bound to CH58 as an alpha helix, to CH59 as a coil and 310 helix, and to PG9 as a beta strand (McLellan et al., 2011) (Figure 3). While PG9 is a bnAb for tier 2 HIV-1 strains, PG9 and the related PG16 and CH01-CH04 bnAbs do not neutralize most tier 1 HIV-1 strains (Bonsignori et al., 2011; Pancera et al., 2010; Walker et al., 2009). Conversely, CH58 and CH59 neutralize only select tier 1 HIV-1 and no tier 2 strains. That PG9 binds to V1–V2 in a beta strand conformation whereas CH58 and CH59 recognize alternative V2 conformations raises several hypotheses. First, this V1–V2 region may exist in multiple conformations since CH58, CH59, and PG9 all bind to AE.A244 gp120 and V1–V2 tags proteins (Table S1, Figures 2A–F) despite their binding to different conformations in their respective crystal structures. Alternatively, the predominant conformation of V1–V2 on peptides and recombinant Env constructs may be different from that on virion trimeric Env and on virus infected CD4+ T cells. Third, the conformation of V2 aa 168–176 may take on alternate conformations in tier 1 versus tier 2 HIV-1 strains on virions and virus-infected cells.
Reactivity of RV144 Vaccinee Plasma with RV144 Vaccine Envs, RV144 Placebo Infection Breakthrough AE.Envs and gp70V1-V2 Case A2 Fusion Protein Mutants
An immune correlates study of RV144 vaccinees demonstrated that antibodies to a gp70V1-V2 fusion protein (gp70V1-V2 CaseA2) correlated with lowered infection risk (Haynes et al., 2012a). Whereas in the RV144 sieve analysis, vaccine efficacy was 48% (p=0.0036, 95% CI:18%, 66%) against viruses that matched the vaccine at V2 aa169K (Rolland et al., 2012), the gp70V1-V2 CaseA2 protein has a valine at aa169, and of CH58 and CH59, only CH58 binds to gp70V1-V2 CaseA2 fusion protein. Thus, there are two distinct but related antibody correlates of risk of infection identified in the RV144 trial that are relevant to CH58 and CH59 antibodies in this paper: 1) antibodies that bind to gp70V1-V2 CaseA2 scaffold (e.g. CH58-like antibodies) and 2) antibodies that bind to K169 and can potentially mediate immune pressure as found in the RV144 sieve analysis (e.g. CH58- and CH59-like antibodies).
We next determined if CH58, CH59 and other V2 mAbs including conformational V2 mAb 607D and bnAbs CH01, PG9 and PG16 reacted with the vaccine Env A244 wildtype (WT), RV144 placebo (i.e. breakthrough) infection Envs (that did not match vaccine Envs), and gp70V1-V2 CaseA2 scaffold WT (V169), and evaluated the effect of mutations introduced in the CH58 and CH59 footprints (Table 2).
Table 2.
Effect of Amino Acid Substitutions in the V2 Region of HIV-1 Env on Ability of HIV-1 V2 mAbs to Bind. See Also Table S4.
| mAb | EC50, ug/ml
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A244 gp120 | AE.703359 gp120 | AE.427299 gp120 | gp70 B.CaseA2 V1–V2 | |||||||||
| WT | K169V | Mut3 | WT | Q169K | Mut3 | WT | Q169K | Mut4 | WT | V169K | Mut3 | |
| RV144V2 antibodies: | ||||||||||||
| CH58 | 0.03 | 0.05 | 7 | 0.38 | 0.04 | 0.07 | 22.5 | 3.4 | 0.03 | 1.2 | 0.025 | 0.03 |
| CH59 | 0.029 | 0.58 | NB | NB | 1.2 | 0.05 | NB | NB | 0.03 | NB | NB | 0.02 |
| Other V1V2 antibodies: | ||||||||||||
| 697D | 0.1 | 0.06 | 0.08 | 0.21 | 0.51 | 0.12 | 0.07 | 0.13 | NB | 0.04 | 0.14 | 0.04 |
| PG9 | 0.6 | 3 | >100 | >5 | 0.53 | 0.54 | 0.47 | 0.13 | >10 | NB | >100 | >10 |
| PG16 | >10 | >100 | NB | NB | >10 | >10 | >10 | 0.26 | NB | NB | NB | NB |
| CH01 | 0.35 | 0.13 | 0.93 | NB | NB | NB | >10 | >10 | NB | NB | NB | NB |
The indicated HIV-1 Env willdtype (WT), and Env mutants with a single a.a residue substitution at the position 169, 3 a. a. residue substitutions (Mut3) or 4 a. a. residue substitutions (Mut4) were assayed in ELISA.
AE.A244 gp120/Mut 3 = K169V/V172E/H173Y.
AE.703359 gp120/Mut 3 = Q169K/R170Q/Q173H.
AE.427299 gp120/Mut 4 = R168K/Q169K//Y173H/A174V.
gp70 B.CaseA2 V1–V2/Mut3 = V169K/E172V/E173H.
We found that CH58 and CH59 binding was high on WT A244 gp120 and either reduced for CH58 or abrogated for CH59 on A244 gp120 with K169V, V172E and H173Y mutations (mutations away from the sequence of the vaccine Envs) (Table 2). For Envs 703357 and 427299 gp120s derived from RV144 placebo breakthrough infection (that did not match the vaccine in V2), binding of CH58 was low on WT and increased by two logs on R168K, Q169K, Y173H, A174V Env mutants (mutations that changed the breakthrough infection Env sequences to match those of the Env vaccines). Similarly, CH59 did not bind to WT breakthrough infection Envs but did bind well to breakthrough Envs with Q169K, R170Q and Q173H mutations (Table 2). Finally, while CH58 but not CH59 bound to gp70V1-V2 CaseA2 fusion protein, the introduction of V169K, E172V and E173H substitutions (to match the vaccine Envs) within the CH58 and CH59 footprints on the V1–V2 fusion protein resulted in increased binding of CH58 and the appearance of CH59 binding (Table 2). The V2 conformational mAb 697D, and V1–V2 bnAbs CH01, PG9 and PG16 had either little or varying binding patterns to these Envs and their mutants (Table 2). The evidence that the same mutations to whole gp120 proteins and V2 peptides similarly abrogated the binding of mAbs CH58 and 59, lends to, but do not prove, the relevance of the structures obtained with V2 peptides.
We next studied the reactivity of plasma from 40 RV144 vaccinees to these Env proteins and their mutants (Figure S4A). Antibody binding decreased significantly on A244 gp120 K169V compared to WT A244 gp120 (p<0.0001, paired Student’s t test), and decreased further with the K169V, V172E and H173Y (Mut3) mutant of A244. For RV144 breakthrough Envs AE.703359 and AE.427299, binding was significantly less on WT Env than on the Envs containing mutations necessary for optimal CH58 and CH59 binding (AE703359 mut3 and AE427299 mut4) (p<0.0001 for both Envs, paired Student’s t test). Moreover, RV144 plasma antibodies bound AE.427299 Q169K Env significantly better than WT (p<0.0001, paired Student’s t test).
RV144 plasma antibodies bound to gp70V1-V2 B.CaseA2 protein (Figure S4A), with no significant improvement in binding to the V169K mutant scaffold, but dramatically improved vaccinee plasma binding to the gp70V1-V2 B.CaseA2 protein with 169K, E172V and E173H triple mutations (p<0.0001, paired Student’s t test). Thus, the RV144 vaccine clearly induced antibodies dependent on the CH58 and CH59 footprint aa residues found in the vaccine, and used this footprint for recognition of vaccine immunogen Env A244 gp120, and most importantly, for recognition of breakthrough CRF_AE01 infection Envs with CH58 and CH59 footprint mutations.
Reactivity of Unmutated Ancestor Antibodies of RV144 V2 mAbs and PG9, PG16 and CH01 V1–V2 bnAbs
The presence of multiple tyrosines in HCDR3s of CH58, CH01, and PG9 and their unmutated ancestor antibodies (UAs) (Figure S3C) led us to suspect that reactivity of UAs of RV144 V2 mAbs and these bnAbs may be similar. We constructed reverted UAs as models for naïve B cell receptors for CH58, CH59, PG9–PG16, CH01–CH04 and 697D, and compared their reactivity to 43 recombinant Envs or V1–V2 scaffold constructs (Table 3, Figures S5A–N, Table S4). We found that whereas the mature CH58 and CH59 antibodies reacted with 26 of 43 and 15 of 43 of the Env constructs, respectively, their UAs reacted with only 5 of 43 (Figures S5A–D). Remarkably, although CH58 and CH59 are from different VH families, their UAs reacted with the same Env constructs (AE.A244 gp120Δ11, AE.A244 V1–V2 tags, C.1086 gp120Δ7, C.1086 gp70 V1–V2 scaffold and C.1086 V1–V2 tags proteins.
Table 3.
Binding of Recombinant HIV-1 Envs and Select V1–V2 Recombinant Constructs By Mature Mutated RV144 V2, V2–V3 bnAbs and V2 Conformational mAb And Their Reverted Unmutated Ancestor Antibodies (UAs) in ELISA. See Also Figure S5 and Table S4.
| HIV-1 Env* | EC50, nM
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CH58 | CH59 | CH01 | 697D | PG9 | PG16 | CH58 UA | CH59 UA | CH01 UA3 | 697D UA | PG9 UA | PG16 UA | |
| AE. A244 V1–V2 tags | 0.02 | 0.014 | 8.9 | 0.1 | 1.1 | 3.7 | 4.9 | 0.13 | 18 | NB | NB | NB |
| AE.A244gp120Δ11 | 0.07 | 0.087 | 0.47 | 0.23 | 0.53 | 37.6 | >667 | 15.5 | 9.3 | >667 | NB | NB |
| A.Q23V1-V2 tags | >66.7 | NB | >66.7 | >66.7 | 14.94 | >66.7 | NB | NB | NB | >667 | NB | NB |
| B.6240 gp140C | NB | NB | >667 | 0.016 | 0.3 | 10.3 | NB | NB | NB | 0.19 | NB | NB |
| gp70 B.CaseA2 V1–V2 | 9.3 | NB | NB | 0.08 | NB | NB | NB | NB | NB | >667 | NB | NB |
| gp70 C.1086V1-V2 | 0.05 | 0.067 | NB | 0.067 | NB | NB | >66.7 | 11.3 | NB | NB | NB | NB |
| C.1086V1-V2 tags | 0.08 | 0.1 | NB | 0.23 | >667 | NB | >667 | 50.1 | NB | NB | NB | NB |
| C.1086C gp120Δ7 | 0.055 | 0.07 | NB | 0.02 | NB | NB | >667 | 66.7 | NB | NB | NB | NB |
AE., B. and C. represent the CRF or clades of HIV-1 Env.
Positive results in ELISA with EC50 above the highest tested concentration (100ug/ml) are indicated as >667nM; NB = no detectable binding.
Finally, 2 of 3 of the Env constructs that reacted with the UAs of CH58 and CH59 (AE.A244 gp120Δ11, AE.A244 V1–V2 tags) also reacted with the UAs of the CH01–CH04 V1–V2 bnAb clonal lineage (Table 3, Figure S1E, Figures S5E–H). In contrast, none of the 43 Envs tested reacted with UAs of PG9 (Figures S5I, J) or PG16 (Table 3, Figures S5K, L) and the Env constructs that reacted with the UA of 697D were entirely different than those reacting with the UAs of any of the other mAbs studied (Figures S5M, N).
CH58- and CH59-Like Antibodies in RV144 Vaccinee Plasma
To determine the titers of CH58- and CH59-like antibodies present in RV144 vaccinee plasma, we established assays wherein we tested vaccinee plasma for the ability to block the binding of biotinylated CH58 or CH59 to AE.A244 gp120 Env protein. We found that vaccinee plasma had a mean of 3.2 ug/ml (range 0–13.9 ug/ml) of CH58-like antibody and 2.5 ug/ml (range 0–11.7 ug/ml) of CH59-like antibody (Figure S4B). Thus, 12 of 43 (28%) of RV144 vaccinees studied had >5.0 ug/ml of either CH58- or CH59-like plasma antibody levels, and the ranges of induced V2 antibodies was within the levels required for RV144 V2 mAb-mediated ADCC and tier 1 neutralizing activity.
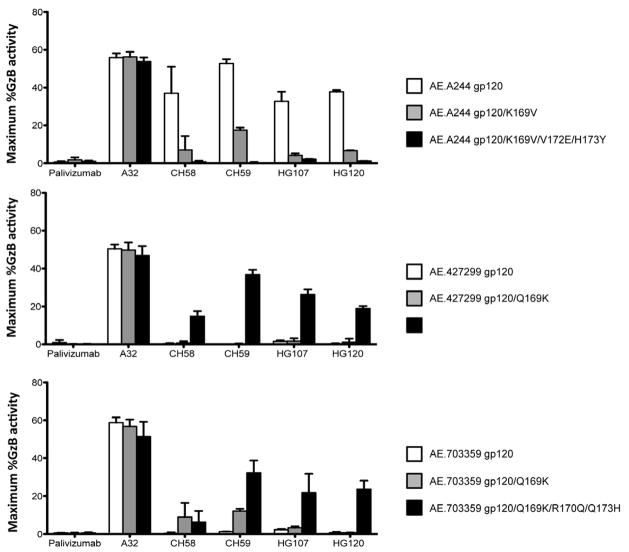
CH58 and CH59 mAbs were derived from the same RV144 vaccinee (347759). To determine the presence of K169 mAbs in other vaccinees, we studied two additional subjects (200184 and 302689) and were able to isolate two additional V2 antibodies (HG107, HG120) with footprints near identical to CH59 (Figure S6, Table S5). Moreover, these two V2 antibodies both neutralized AE.92TH023 HIV-1 (Table S3) and mediated ADCC of tier 2 AE.CM235 infected CD4 CEM.NKRCCR5 T cells (Figure 4). Importantly, when the A244 gp120 and RV144 breakthrough infection 427299 and 703357 Env gp120s were used to coat CD4 CEM.NKRCCR5 T cell targets in ADCC assays, the ability of CH58, CH59, HG107 and HG120 mAbs to mediate ADCC was dependent on CH58, CH58, HG107, HG120 mAb footprint mutations that included 169K (Figure 4).
Figure 4. See also Figures S4 and S6. Effect of V2 mAbs CH58, CH59, HG107, HG120 footprint mutations in HIV-1 vaccine AE.A244 Env and RV144 breakthrough AE.427299 and AE.703357 Envs on ability of V2 mAbs to mediate ADCC.
Panels show the ability of mAb A32 and RV144 V2 mAbs CH58, CH59, HG107, HG120 to mediate ADCC against gp120-coated CD4 cell (CEMCCR5) target T cells. Data shown is maximum percent granzyme B activity from ADCC. Top panel shows that CH58, CH59, HG107 and HG120 mAbs all mediate high levels of ADCC against WT AE.244 Env coated CD4 T cell targets (white bars), and this killing is mitigated by a single K169V mutation (grey bars), and is abrogated by the full V2 mAb footprint set of mutations (black bars). Middle and lower panels show, in contrast, that none of the CH58, CH59, HG107 and HG120 mAbs mediated ADCC against RV144 breakthrough Env AE.427299 and AE.703357 WT CD4 T cell targets (with V2s that did not match the RV144 vaccine) (white bars), and that ADCC was restored minimally with AE.703357 targets with the Q169K mutation (grey bars), and restored in a pronounced manner in both breakthrough Env targets with the full set of mutations that include Q169K that restored the V2 mAb footprint mutations (black bars). Purified NK cells isolated from a normal donor with Fc-gamma receptor IIIα FF phenotype were used as effector cells. The effector to target ratio was 10:1. Error bars show mean +/− SEM. Each antibody was tested in a wide dose curve starting at 40ug/ml with 4-fold dilutions. See also Figures S6 and Table S5.
Remarkably, like CH59, the light chains of HG107 and HG120 were Vλ3–10 with a common aa glutamate-aspartate (ED) residue motif in LCDR2 that is critical for binding to K169 (Figures S6A–C), in that the D of this motif in CH59 forms a salt bridge with V2 K169 (Figure 3, Figures S6G, H). In CH58, the Ig light chain is Vλ 6–57 and also shares the LCDR2 ED motif (Figure S6C), with the E of the motif binding K169 in the V2-CH58 structure (Figures S6G, H). Overlayed LCDR2 ED motif regions of CH58 and CH59 show striking similarity between the two LC-Env binding regions (Figures S6G, H). Thus, with four V2 antibodies from three different RV144 vaccinees, light chain usage was restricted to Vλ light chains containing an LCDR2 ED motif required for CH58 and CH59 Env V2 K169 binding.
DISCUSSION
Eighty-nine percent of infections in the RV144 efficacy trial were with CRF01_AE HIV-1 strains (Rerks-Ngarm et al., 2009). One correlate of infection risk from the RV144 trial analysis is the genetic analysis of Env sequences in vaccine compared to placebo recipients that demonstrated increased vaccine efficacy against viruses with a V2 K169 residue among vaccines (Rolland et al., 2012). This signature at K169 suggested the hypothesis that RV144 V2 antibodies such as CH58 and CH59 may have mediated, in as yet undefined manners, immune pressure at that site. Based on antibody effector functions shown to be mediated in vitro by mAbs CH58, CH59, HG107 and HG120, the potential mechanisms of antibody-mediated immune pressure include: a) virus neutralization of susceptible CRF01_AE HIV-1 strains, and b) binding HIV-1-infected CD4 T cells and mediation of ADCC, or other as yet undefined effector mechanisms.
A second immune correlate of lowered infection risk is the antibody response to V1–V2 as measured by the clade B gp70 V1–V2 CaseA2 fusion protein (Haynes et al., 2012a). Since gp70 V1–V2 CaseA2 has a V169 and only CH58 binds to this protein, there may be at least two types of RV144 V2 antibodies capable of mediating immune pressure, those that bind to gp70 V1–V2 CaseA2 protein and bind K169 (i.e. CH58-like), and those that do not bind to gp70 V1–V2 CaseA2 protein and bind K169 (i.e. CH59, HG107, HG120-like). Critical studies going forward will be to perform new efficacy trials in humans and perform passive protection trials in rhesus macaques with RV144 V2 antibodies with R5 SHIVs derived from RV144 trial breakthrough infections to directly explore the protective effect of these two types of V2 mAbs. Nonetheless, the studies in the present report describe two types of V2 antibodies induced by the RV144 vaccine that recognize K169, define their structures and effector function capabilities, and demonstrate light chain conserved usage for binding to the Env V2 K169 site of immune pressure.
A key task for the HIV-1 vaccine development field is to improve the degree of vaccine efficacy seen in the RV144 clinical trial with subsequent vaccine designs. Vaccine designers generally focus on regions of conservation. For RNA viruses such as influenza and HIV-1, which are highly divergent and capable of rapid genetic alteration, conserved regions on Env are generally well-protected from humoral recognition, and it is the divergent regions that may be more susceptible to antibody-mediated neutralization. Indeed, antibodies directed against the variable head region of influenza hemagglutinin are the source of the vaccine protection elicited by the seasonal influenza vaccines (Karlsson Hedestam et al., 2008). With the RV144 trial, it also seems a variable region – in this case, around residue 169 of V2 – is the site of successful vaccine-induced immune pressure. Virologically, it makes sense that selection and/or immune pressure could be identified by variation. Our results with RV144 trial antibodies CH58, CH59, HG107 and HG120 mAbs indicate that this variation may include not only sequence diversity, but also conformational changes in the structure of the same aa sequence. Despite extraordinary variation in both sequence and structure, the humoral immune system appears capable of recognizing V1–V2 in the setting of vaccination with a restricted Ig light chain LCDR2 motif– and Env immunogens that focus the elicited response to this V2 region should be explored.
EXPERIMENTAL PROCEDURES
Production of Recombinant Antibodies
Three RV144 vaccine-recipients 347759, 200134, and 302689 were studied for isolation of HIV-1 antibodies (Figure S3C). MAbs CH58 and CH59 from RV144 vaccine-recipients 347759 and mAb HG107 from RV144 vaccine-recipients 200134 were isolated by screening clonal cultures of memory B cells with either AE.A244 gp120 (for CH58 and CH59) or A244 V1–V2 tags (for CH107) followed by isolation of VH and VL genes as described (Bonsignori et al., 2011; Liao et al., 2011; Liao et al., 2009). MAb HG120 was isolated from RV144 vaccine-recipients 302689 by flow sorting HIV-1 V2-specific single memory B cells cultured overnight (Bonsignori et al., 2011) using dual color fluorochrome-labeled AE.A244 V1–V2 tags followed by isolation of VH and VL genes as described (Bonsignori et al., 2011; Moody et al., 2012). In the memory B cell culture system used to isolate CH58 and CH59 (Bonsignori et al., 2011), of 5,232 IgG+ memory B cell cultures, 5 cultures were scored as positive for V2 reactivity in the initial culture screen. This yielded a frequency of V2-reactive memory B cells of 0.096%, a frequency comparable to the frequency of immunodominant V3 antibody producing B cells (0.13%) induced in the VAX004 HIV vaccine clinical trial (Bonsignori et al., 2009).
VH and VL genes of 697D (Gorny et al., 1994) were obtained from M. Gorny, and S. Zolla-Pazner (New York University, NY). V1–V2 bnAbs PG9 and PG16 (Walker et al., 2009) were provided by D. Burton (Scripps Institute, La Jolla, CA). MAbs 7B2 and A32 were provided by James Robinson, Tulane University, LA), and Synagis (MedImmune, LLC; Gaithersburg, MD), a human anti-respiratory syncytial virus mAb, was used as negative controls.
The reverted unmutated ancestor antibodies of CH58, CH59, CH01, 697D, PG9 and PG16 were inferred and produced as described (Bonsignori et al., 2011; Haynes et al., 2012b; Liao et al., 2011; Ma et al., 2011).
Production of Recombinant HIV-1 Proteins
Sequences of all HIV-1 Env proteins used in the study were summarized in (Table S4). RV144 vaccine proteins AE.A244 gp120, and B.MN gp120 were supplied by GSID (Global Solutions for Infectious Diseases, South San Francisco, CA) or produced as described (Alam et al., 2012; Liao et al., 2006), HIV-1 Env A.92RW020, B.HXB/BAL and C.97ZA012 were scaffolded on J08 protein or murine leukemia virus (MLV) gp70 protein and produced as fusion proteins as described (McLellan et al., 2011; Pinter et al., 1998). MLV gp70 carrier protein without V1–V2 sequence was similarly produced and used as negative control. AE.703357 and AE.427299 Env gp160 sequences were obtained from RV144 subjects (placebo arm) and represented breakthrough infection Envs with V2 sequences that do not match the RV144 AE vaccine strains A244 and 92TH023 and used to produce recombinant gp120 proteins and gp120 mutants to match the V2 sequences of RV144 vaccine AE strains, AE.A244 and AE.92TH023. HIV-1 Env V1–V2 tags proteins were designed with Ig leader (METDTLLLWVLLLWVPGSTGD) serving as a mature protein cleavage and secretion signal at the N-terminus and with the C-terminal avi-tag followed by His6-tag for purification, produced in 293F cells by transfection and purified by nickel columns. Other recombinant HIV-1 Envs and Env mutants used in the study were produced as described (Liao et al., 2006; Ma et al., 2011).
HIV-1 Env V2 Peptides
HIV-1 AE.A244 gp120 and AE.A244 V2-171 peptide (LRDKKQKVHALFYKLDIVPIED) spanning from L165 to D186 and its 22 derivative alanine-scanning peptides (Table S4) were produced (CPC Scientific Inc., San Jose, CA) for epitope mapping of V2 mAbs.
SPR Kinetics Measurements
Binding Kd and rate constant measurements of mAbs to AE.A244 gp120 and AE.A244 V1–V2 tags were carried out on BIAcore 3000 instruments as described (Alam et al., 2011; Alam et al., 2007; Alam et al., 2008). All data analysis was performed using the BIAevaluation 4.1 analysis software (GE Healthcare).
Determination of CH58- and CH59-Like Antibodies in Plasma
The levels of CH58 and CH59-like antibodies in RV144 vaccinee plasma (n = 43) were determined by blocking of the biotinylated mAbs CH58 and CH59 to recombinant AE.A244 gp120 protein by serial dilutions of plasma of the RV144 vaccinees (Alam et al., 2008).
Analysis of Binding of mAbs to HIV-1-Infected Cells
Analysis PB CD4+ T cells infected with replication-competent IMC (Edmonds et al., 2010) was performed as described (Ferrari et al., 2011; Pollara et al., 2011) except with a 2 hr. incubation with primary antibody.
ADCC assays
Assays for determining ADCC activity mediated by RV144 mAbs were carried by luciferase ADCC assay and ADCC-GTL assay (Bonsignori et al., 2012; Pollara et al., 2011; Trkola et al., 1999) using the methods as reported and described in detail in Supplemental Experimental Procedure.
HIV-1 Virion Capture Assays
The ability of mAbs CH58 and CH59 to capture HIV-1 virions was tested by HIV-1 p24 assay using the methods as described (Burrer et al., 2005) and to capture infectious virions of HIV-1 92TH023 or CM244 was performed as described (Leaman et al., 2010; Liu et al., 2011). Briefly, the virus particles or infectious virions in the uncaptured and captured fraction were measured by viral RNA with HIV-1 gag real time RT-PCR or TZM-bl infectious virus capture readout, respectively. The captured RNA represents the viral RNA (rVirion) after subtracting the background value (virus only). The percentage of captured infectious virion (iVirion) was calculated as 100− the uncaptured virus infectivity/virus only infectivity × 100%.
Neutralization Assays
Neutralizing activity of mAbs were assayed in TZM-bl cells-based (Seaman et al., 2010) (Montefiori, 2005) and/or A33R cells-based neutralization assays (Montefiori et al., 2012).
Crystallographic analysis of CH58 and CH59
CH58 and CH59 Fab preparation, crystallization and data collection were described in Supplemental Experimental Procedures. All diffraction data were processed with the HKL2000 suite (Otwinowski and Minor, 1997), model building and refinement were performed in COOT (Emsley and Cowtan, 2004) and PHENIX (Adams et al., 2002), respectively. Automatic model building was performed by Arp/wArp (Langer et al., 2008). Structure determination, model building and refinement were described in Supplemental Experimental Procedures. Coordinates and structure factors for unbound CH58 Fab, as well as CH58 and CH59 Fabs in complex with a V2 peptide have been deposited with the Protein Data Bank under accession codes 4HQQ, 4HPO and 4HPY, respectively.
Molecular Modeling
Homology models of the light chains for HG107 and HG120 were constructed using ROSETTA 3.3’s threading protocol (Leaver-Fay et al., 2011) with the light chain of CH59 from the crystal structure of the V2-CH59 complex as the template. The model with the best energy score was selected from 100 models generated for each light chain.
Supplementary Material
HIGHLIGHTS.
A comprehensive analysis of RV144 HIV-1 vaccine-induced V2 antibodies.
Light chain signature of antibodies that bind at vaccine-induced immune pressure site.
V2 antibodies bind to field HIV-1 isolate Envs expressed on CD4+ infected T cells
Structure of RV144 HIV-1 vaccine-induced V2 antibodies.
Acknowledgments
This study was supported by Collaboration for AIDS Vaccine Discovery grants from Bill & Melinda Gates Foundation, by grants from NIH/NIAID (AI067854, the Center for HIV/AIDS Vaccine Immunology, and AI100645, the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery), by research funds from the Department of Veterans Affairs, by an Interagency Agreement Y1-AI-2642-12 between U.S. Army Medical Research and Material Command (USAMRMC), by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. and the U.S. Department of Defense (DOD), and by intramural NIH support for the NIAID Vaccine Research Center. The authors thank Andrew Foulger, Haiyan Chen, Melissa Cooper, Sean Crawford, Thomas Jeffries, James Holland, Radha De, Richard Scearce, Lee Jeffries Jr., Annie Hogan, Jamie Pritchett, Daria Pause, Erika Solomon, Laura Sutherland, Julie Blinn and Krissey Lloyd for expert technical assistance in antibody and HIV-1 Env protein production; Dawn Marshall and John Whitesides for expert technical assistance in flow cytometry; and Kelly Soderberg, Jennifer Kircherr, and Charla Andrews for project management. Flow cytometry supported by the NIH grants S10RR019145, UC6 AI058607, AI64518 and P30 AI051445. The opinions herein are those of the authors and should not be construed as official or representing the views of the U.S. Department of Health and Human Services, National Institute for Allergy and Infectious Diseases, the Department of Defense, the Department of the Army, or the Department of Veteran Affairs. Materials from the RV144 clinical trial were provided by the Ministry of Public Health, Thailand, the Thai AIDS Vaccine Evaluation Group, and the USMHRP through the Henry M. Jackson Foundation for the Advancement of Military Medicine.
Footnotes
Supplemental Information includes six figure Legends, five tables and additional experimental procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Alam SM, Liao HX, Dennison SM, Jaeger F, Parks R, Anasti K, Foulger A, Donathan M, Lucas J, Verkoczy L, et al. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. Journal of virology. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Liao HX, Tomaras GD, Bonsignori M, Tsao CY, Hwang KK, Chen H, Lloyd KE, Bowman C, Sutherland L, et al. Antigenicity and Immunogenicity of RV144 Vaccine AIDSVAX Clade E Envelope Immunogen is Enhanced by a gp120 N-terminal Deletion. Journal of virology. 2012 doi: 10.1128/JVI.00718-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, Haynes BF. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Scearce RM, Parks RJ, Plonk K, Plonk SG, Sutherland LL, Gorny MK, Zolla-Pazner S, Vanleeuwen S, Moody MA, et al. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. Journal of virology. 2008;82:115–125. doi: 10.1128/JVI.00927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. Analysis of a Clonal Lineage of HIV-1 Envelope V2/V3 Conformational Epitope-Specific Broadly Neutralizing Antibodies and Their Inferred Unmutated Common Ancestors. Journal of virology. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Moody MA, Parks RJ, Holl TM, Kelsoe G, Hicks CB, Vandergrift N, Tomaras GD, Haynes BF. HIV-1 envelope induces memory B cell responses that correlate with plasma antibody levels after envelope gp120 protein vaccination or HIV-1 infection. J Immunol. 2009;183:2708–2717. doi: 10.4049/jimmunol.0901068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Pollara J, Moody MA, Alpert MD, Chen X, Hwang KK, Gilbert PB, Huang Y, Gurley TC, Kozink DM, et al. ADCC-Mediating Antibodies from an HIV-1 Vaccine Efficacy Trial Target Multiple Epitopes and Preferentially Use the VH1 Gene Family. Journal of virology. 2012 doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrer R, Haessig-Einius S, Aubertin AM, Moog C. Neutralizing as well as non-neutralizing polyclonal immunoglobulin (Ig)G from infected patients capture HIV-1 via antibodies directed against the principal immunodominant domain of gp41. Virology. 2005;333:102–113. doi: 10.1016/j.virol.2004.12.034. [DOI] [PubMed] [Google Scholar]
- Doria-Rose NA, Georgiev I, O’Dell S, Chuang GY, Staupe RP, McLellan JS, Gorman J, Pancera M, Bonsignori M, Haynes BF, et al. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. Journal of virology. 2012;86:8319–8323. doi: 10.1128/JVI.00696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds TG, Ding H, Yuan X, Wei Q, Smith KS, Conway JA, Wieczorek L, Brown B, Polonis V, West JT, et al. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology. 2010;408:1–13. doi: 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, et al. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. Journal of virology. 2011;85:7029–7036. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Moore JP, Conley AJ, Karwowska S, Sodroski J, Williams C, Burda S, Boots LJ, Zolla-Pazner S. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. Journal of virology. 1994;68:8312–8320. doi: 10.1128/jvi.68.12.8312-8320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Pan R, Williams C, Wang XH, Volsky B, O’Neal T, Spurrier B, Sampson JM, Li L, Seaman MS, et al. Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from HIV-1-infected individuals. Virology. 2012;427:198–207. doi: 10.1016/j.virol.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. The New England journal of medicine. 2012a;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012b;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasavvas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, Koup RA, Madnote S, Arworn D, Shen X, et al. The Thai Phase III HIV Type 1 Vaccine Trial (RV144) Regimen Induces Antibodies That Target Conserved Regions Within the V2 Loop of gp120. AIDS research and human retroviruses. 2012 doi: 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Microbiol. 2008;6:143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman DP, Kinkead H, Zwick MB. In-solution virus capture assay helps deconstruct heterogeneous antibody recognition of human immunodeficiency virus type 1. Journal of virology. 2010;84:3382–3395. doi: 10.1128/JVI.02363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, Whitesides JF, Lu X, Yu JS, Hwang KK, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Overman RG, Yates NL, Alam SM, Vandergrift N, Chen Y, Graw F, Freel SA, Kappes JC, Ochsenbauer C, et al. Dynamic antibody specificities and virion concentrations in circulating immune complexes in acute to chronic HIV-1 infection. Journal of virology. 2011;85:11196–11207. doi: 10.1128/JVI.05601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma BJ, Alam SM, Go EP, Lu X, Desaire H, Tomaras GD, Bowman C, Sutherland LL, Scearce RM, Santra S, et al. Envelope deglycosylation enhances antigenicity of HIV-1 gp41 epitopes for both broad neutralizing antibodies and their unmutated ancestor antibodies. PLoS Pathog. 2011;7:e1002200. doi: 10.1371/journal.ppat.1002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005;Chapter 12(Unit 12):11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, et al. Magnitude and Breadth of the Neutralizing Antibody Response in the RV144 and Vax003 HIV-1 Vaccine Efficacy Trials. J Infect Dis. 2012;206:431–441. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MA, Yates NL, Amos JD, Drinker MS, Eudailey JA, Gurley TC, Marshall DJ, Whitesides JF, Chen X, Foulger A, et al. HIV-1 gp120 vaccine induces affinity maturation in both new and persistent antibody clonal lineages. Journal of virology. 2012;86:7496–7507. doi: 10.1128/JVI.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz F, Cicala C, Van Ryk D, Block KE, Jelicic K, McNally JP, Ogundare O, Pascuccio M, Patel N, Wei D, et al. The genotype of early-transmitting HIV gp120s promotes alpha (4) beta(7)-reactivity, revealing alpha (4) beta(7) +/CD4+ T cells as key targets in mucosal transmission. PLoS Pathog. 2011;7:e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, Kwong PD. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. Journal of virology. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, Kayman SC, Trochev O, Wu Z. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine. 1998;16:1803–1811. doi: 10.1016/s0264-410x(98)00182-0. [DOI] [PubMed] [Google Scholar]
- Plotkin SA, Gilbert PB. Nomenclature for immune correlates of protection after vaccination. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2012;54:1615–1617. doi: 10.1093/cid/cis238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollara J, Hart L, Brewer F, Pickeral J, Packard BZ, Hoxie JA, Komoriya A, Ochsenbauer C, Kappes JC, Roederer M, et al. High-throughput quantitative analysis of HIV-1 and SIV-specific ADCC-mediating antibody responses. Cytometry A. 2011;79:603–612. doi: 10.1002/cyto.a.21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. The New England journal of medicine. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, Sanders-Buell E, Heath L, Magaret CA, Bose M, Bradfield A, et al. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nature medicine. 2011;17:366–371. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. Journal of virology. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Matthews J, Gordon C, Ketas T, Moore JP. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. Journal of virology. 1999;73:8966–8974. doi: 10.1128/jvi.73.11.8966-8974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cardozo T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat Rev Immunol. 2010;10:527–535. doi: 10.1038/nri2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cardozo T, deCamp A, Haynes BF, Kim J, Kong X, Michael N, Rerks-Ngarm S, Williams C. AIDS Vaccine. Bangkok, Thailand: 2011. V2- reactive antibodies in RV144 vaccines’ plasma; p. Abstract No.: OA09.03.p. 77. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.