Abstract
AP endonuclease 1 (APE1; also known as REF-1) contains a DNA repair domain and a redox regulation domain. APE1 is overexpressed in several human cancers, and disruption of APE1 function has detrimental effects on cancer cell viability. However, the selective contribution of the redox and the DNA repair domains to maintenance of cellular homeostasis in cancer has not been elucidated. In the present study, we used E3330, a small-molecule inhibitor of APE1 redox domain function, to interrogate the functional relevance of sustained redox function in pancreatic cancer. We show that E3330 significantly reduces the growth of human pancreatic cancer cells in vitro. This phenomenon was further confirmed by a small interfering RNA experiment to knockdown APE1 expression in pancreatic cancer cells. Further, the growth-inhibitory effects of E3330 are accentuated by hypoxia, and this is accompanied by striking inhibition in the DNA-binding ability of hypoxia-inducible factor-1α, a hypoxia-induced transcription factor. E3330 exposure promotes endogenous reactive oxygen species formation in pancreatic cancer cells, and the resulting oxidative stress is associated with higher levels of oxidized, and hence inactive, SHP-2, an essential protein tyrosine phosphatase that promotes cancer cell proliferation in its active state. Finally, E3330 treatment inhibits pancreatic cancer cell migration as assessed by in vitro chemokine assays. E3330 shows anticancer properties at multiple functional levels in pancreatic cancer, such as inhibition of cancer cell growth and migration. Inhibition of the APE1 redox function through pharmacologic means has the potential to become a promising therapeutic strategy in this disease.
Introduction
The control of cell proliferation in cancer is deranged (1). The intracellular redox state has been suggested as regulator of growth in cancer cells (2). AP endonuclease 1 (APE1; also known as REF-1) is a dual-function protein, which contains a redox domain and a DNA repair domain (3). APE1 is overexpressed in numerous solid cancers, including prostate and bladder cancers (4, 5), non-small cell lung cancers (6), gliomas (7), medulloblastomas and primitive neurectodermal tumors (8), osteosarcomas (9), and germ cell tumors. In these and other studies, nuclear APE1 overexpression is associated with an adverse prognosis (4, 6, 9) and/or resistance to radiation and chemotherapeutic agents (5, 8). APE1 was recently reported to be overexpressed in pancreatic cancer, and APE1 levels were further up-regulated by exposure to the antimetabolite agent, gemcitabine (10). Conversely, inhibition of APE1 with antisense DNA resulted in profound sensitization of pancreatic cancer cells to gemcitabine (10). APE1 has been implicated in the transactivation of numerous seminal transcription factors involved in cancer initiation and progression, including activator protein-1, nuclear factor-κB (NF-κB), hypoxia-inducible factor-1α (HIF-1α), cyclic AMP response element-binding protein, etc. (reviewed in ref. 3). For example, Gray et al. have recently shown that APE1 plays a critical role in hypoxia-induced HIF-1α-mediated up-regulation of the angiogenic growth factor, vascular endothelial growth factor, in pancreatic cancer (11).
Although the accumulating weight of evidence strongly implicates APE1 as a cancer potentiating molecule with diverse functions, there is scant information at present vis-à-vis the relative importance of the DNA-binding versus redox domains of APE1 in sustaining the growth of cancer cells. Further, it is not known whether selective inhibition of the redox domain by itself can mitigate the transformed phenotype of cancer cells. Previously, APE1 knockdown by RNA interference in human cancer cell lines was shown to block cell proliferation and induce apoptosis (12). Because the deleterious effects on cell viability could be rescued with an unrelated yeast protein that contains AP endonuclease activity but lacks the other functions of human APE1, the authors hypothesized that loss of the AP endonuclease activity, with resultant increase in abasic DNA damage, is sufficient for inducing cell death (12). Nevertheless, the contribution of the APE1 redox domain was not specifically investigated in their report.
In the present study, we investigated whether selective blockade of the APE1 redox function can inhibit pancreatic cancer growth and, if so, what the underlying mechanisms of these phenotypic effects might be. To accomplish this, we used E3330 (IC50, 50 μmol/L), a small-molecule inhibitor of redox activity of APE1 protein that does not impede its DNA repair function (13, 14). The effects of E3330 were compared with that of methoxyamine, a small-molecule inhibitor of the APE1 DNA repair activity (3, 14). Our results show that E3330 inhibits not only the growth but also the migratory abilities of pancreatic cancer cells in vitro. We also elucidate the signaling moieties whose activity is perturbed by APE1 redox inhibition and which likely contributes to the anticancer effects of E3330.
Materials and Methods
Cell Culture
The human cancer cell lines used in this study, PANC1, PK9, BxPC3, CAPAN-1, ASPC-1, and XPA1 cells (purchased from the American Type Culture Collection), were all individually cultured in DMEM supplemented with 10% fetal bovine serum, 100 units/mL penicillin, and 100 units/mL streptomycin (Invitrogen) as described previously (15). hTERT-immortalized nontransformed human pancreatic epithelial cell line HPNE (16) were cultured with medium D, which contains 1 volume of medium M3, 3 volumes of glucose-free DMEM, 5% fetal bovine serum, 5.5 mmol/L glucose, 10 ng/mL epidermal growth factor, and 50 μg/mL gentamycin.
Western Blot Analysis
For APE1 Western blot, refer to our previous report (14). In summary, whole-cell extracts were prepared using nuclear protein extract buffer from pancreatic cancer cells. The cell lysates were separated by SDS-PAGE using a 10% (w/v) polyacrylamide resolving gel and transferred electrophoretically to a nitrocellulose membrane. The blots were blocked with 5% TBS-Tween 20 buffer for 1 h. Immunobloting was done using the APE1 primary polyclonal antibody (Santa Cruz Biotechnology) at a 1:400 dilution at room temperature and the peroxidase-conjugated secondary antibodies (Amersham Pharmacia) overnight at 4°C. All immunoblots were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech). For SHP-2 Western blot, refer to our previous report (17).
Preparation of Single-Cell Suspension from Fresh Pancreatic Cancer Tissues and Isolation of Epithelial-Specific Antigen – Positive Primary Pancreatic Cancer Cells
Excess tissue from three freshly resected pancreatic cancer specimens was obtained from the Department of Pathology at the Johns Hopkins Hospital. Before collagenase digestion, the specimens were minced using sterile scalpel blades. To obtain a single-cell suspension, the resultant minced tumor pieces were mixed with ultrapure collagenase IV (Worthington Biochemical) and allowed to incubate at 37°C for 2.5 to 3 h for enzymatic dissociation. The specimen was further mechanically dissociated every 15 to 20 min by pipetting with a 10 mL pipette. Subsequently, cells were filtered through a 40-μm nylon mesh and washed with HBSS/20% fetal bovine serum and then washed twice with HBSS. The cell suspensions were then incubated with anti-human epithelial-specific antigen (ESA; CD326) bead (Miltenyi Biotec). ESA is overexpressed on the surface of pancreatic cancer cells, and ESA-positive primary cancer cells were thus isolated using MACS.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide Assay
E3330 powder (kindly provided by Dr. Honda) was dissolved with pure ethanol. PANC1, XPA1, or HPNE cells were incubated with various concentrations of either E3330 (10-30 μmol/L) or methoxyamine (1 μmol/L-10 mmol/L) for 72 h in 96-well plates (Flow Laboratories). At the culmination of the study, cell viability was evaluated by measuring the mitochondrial-dependent conversion of the tetrazolium salt, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma), to a colored formazan product as described previously (18). A similar analysis was done to assess growth of ESA-positive primary pancreatic cancer cells in the presence of E3330, with the exception that MTT assays were culminated at 48 h.
Trypan Blue Exclusion Assay
Refer to our previous report (19). In summary, 2 × 105 PANC1 cells were placed in one well of a 12-well plate (Costar) and treated with 5 to 30 μmol/L E3330. After a 24, 48, and 72 h of culture, the cells were washed with PBS and stained with trypan blue, and cell viability was examined by counting the live cell numbers.
APE1Small Interfering RNA Experiment
Transient transfection of small interfering RNA (siRNA) was carried out as we reported previously (14). In summary, PANC1 cells were diluted with fresh medium without antibiotics and transferred to 12-well plates at 1 × 105 per well (500 μL/well). Transient transfection of double-stranded RNA was carried out using Oligofectamine 2000 (Life Technologies). Cells were untreated (control) or treated with scrambled APE1 siRNA (25-50 nmol/L) or APE1 siRNA (25-50 nmol/L). After 72 h, the live cell numbers were counted by trypan blue exclusion assay.
Cell Cycle Analysis by Flow Cytometry
Cell cycle analysis was done on PANC1 cells incubated for 48 h with E3330 (10, 20, or 30 μmol/L). The cells were fixed in chilled methanol overnight before staining with propidium iodide (50 Ag/mL) in the presence of 20 μg/mL RNase (Sigma) and 0.1% NP-40 (Sigma). Analysis was done immediately after staining using a FACScan (Becton Dickinson).
Flow Cytometric Detection of Proliferation-Associated Antigen Ki-67
Proliferation-associated antigen Ki-67 was detected as described (20). Briefly, PANC1 cells were fixed for 15 min by gently dropping ice-cold methanol on the cell pellet at −20°C. Fixed cells were washed and incubated for 45 min at room temperature with monoclonal antibodies against Ki-67 antigen (Becton Dickinson Immunocytometry Systems) or control IgG1 (Becton Dickinson Immunocytometry Systems). After washing, green fluorescence intensity was detected by flow cytometry (FACScan; Becton Dickinson). The expression of the proliferation-associated antigens was determined by the mean fluorescence intensity of gated cancer cells and was expressed as the percentage of positively stained cells in relation to the total amount of cells.
Detection of Reactive Oxygen Species by Flow Cytometry
Flow cytometric detection of reactive oxygen species (ROS) was done as described (21). Briefly, PANC1 cells treated with 20 μmol/L E3330 for 2 days and subsequently washed with PBS and resuspended in complete medium followed by incubation with 0.5 μmol/L dihydrorhodamine 123 (Sigma) for 30 min at 37°C. ROS fluorescence intensity was determined by flow cytometry with excitation at 490 nm and emission at 520 nm.
SHP-2 Oxidation Assay by “Positive Labeling” of Oxidized Thiols with PEO-Iodoacetyl biotin
SHP-2 oxidation was measured by “positive labeling” of oxidized thiols as described previously (22, 23). In summary, PANC1 cells were serum starved and subsequently stimulated with E3330 for 2 h. Then, the E3330-treated cells were lysated for 1 h at 25°C in an anaerobic chamber. All free thiols were masked with 10 mmol/L NEM and 10 mmol/L iodoacetamide. Reversibly oxidized thiols were reduced with 4 mmol/L DTT in the anaerobic chamber and then were labeled with 0.5 mmol/L PEO-iodoacetyl biotin (Pierce). Biotin incorporation into immunoprecipitated SHP-2 was detected by Western blotting with horseradish peroxidase–conjugated streptavidin and enhanced chemiluminescence.
HIF-1α DNA-Binding ELISA Assay
HIF-1α DNA binding was assessed by using HIF-1α DNA-binding ELISA assay kit purchased from Active Motif. HIF-1α DNA binding was assessed as described (24). PANC1 or XPA1 cells were treated with E3330 at various doses for 6 h. Then, the cells were harvested and whole-cell proteins were isolated. The procedures of ELISA assay were done according to the manufacturer’s protocol.
NF-κB Reporter Assay
PANC1 or XPA1 cells were plated at 1 × 105 per well in 24-well plates on the day before transfection. The cells were transiently transfected by Fugene 6 Transfection Reagent (Roche Molecular Biochemicals) with 950 ng of a NF-κB reporter plasmid (pNF-κB-Luc; Stratagene) and 50 ng of a construct directing expression of Renilla luciferase under the control of a constitutively active thymidine kinase promoter (pRL-TK; Promega). Twenty-four hours after transfection, the cells were treated by E3330 at various doses for 6 h, and luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
In vitro Migration Assays
Migration of three APE1 expressing pancreatic cancer cells (PANC1, XPA1, and BxPC3) was assayed using 6.5-mm-diameter chambers with 8-μm pore filters (Trans-well, 24-well cell culture; Costar) as described (18). Pancreatic cancer cells were suspended in serum-free medium at a concentration of 2 × 105/mL. Thereafter, 0.2 mL of the cell suspension was added to the upper chamber, and 0.5 mL serum-free medium with 100 ng/mL stromal cell–derived factor-1 was added to the lower chamber. Various concentrations of E3330 were added to the upper chamber. The chambers were incubated for 12 h at 37°C in a humid atmosphere of 5% CO2/95% air. After incubation, the filters were fixed and stained with Diff-Quick reagent (Dade Behring). Cell viability (MTT) assays were done on the upper chamber lysates, to normalize any effects of E3330-induced growth inhibition (reduced cell numbers) on the migration readout. The upper surface of the filters was scraped twice with cotton swabs to remove nonmigrating cells. The experiments were repeated in triplicate wells, and the number of migrating cells in five high-power fields per filter was counted microscopically at ×400 magnification.
Flow Cytometric Detection of Surface CD44 Expression in PANC1 Cells
PANC1 cells were stained with PE-labeled rat IgG2b (isotype control) antibody or PE-labeled rat anti-human CD44 (eBioscience) at concentration of 0.125 μg antibody per 1 million of cells as suggested by the antibody provider. After incubation for 30 min at 4°C, the cells were washed with PBS and analyzed by flow cytometry for CD44 expression.
Results
Pancreatic Cancer Cells Overexpress APE1
To examine the expression of APE1 protein in pancreatic cancer cells, we did Western blot analysis of six human pancreatic cancer cell lines: PANC1, BxPC3, CAPAN-1, ASPC-1, XPA1, and PK9. As shown in Fig. 1A, all tested pancreatic cancer cell lines expressed APE1 protein at a considerably higher level than HPNE and HPDE, the nonneoplastic human pancreatic cell lines.
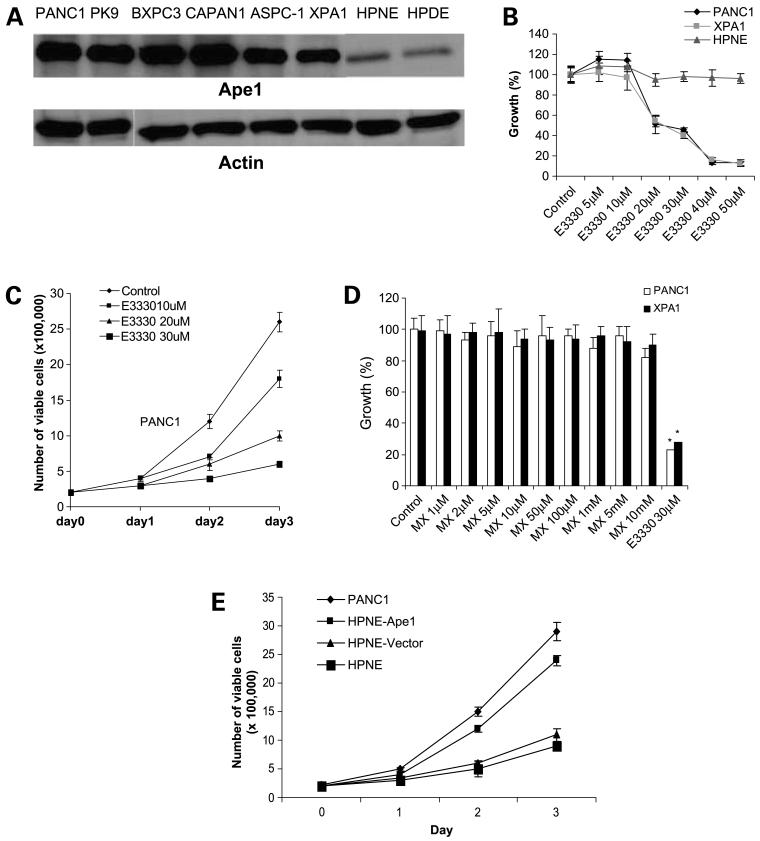
Figure 1.
A, APE1 expression in pancreatic cancer cells (Western blot). The total cell lysates are analyzed by Western blot. All six tested pancreatic cancer cell lines PANC1, PK9, BxPC3, CAPAN-1, ASPC-1, and XPA1 express a dominant level of APE1. Non-tumorigenic human pancreatic cell line HPNE and HPDE express lower level of APE1 compared with these malignant cell lines. B, dose-dependent inhibition of growth of pancreatic cancer cells (PANC1 and XPA1) but not the nonneoplastic human pancreatic epithelial cells HPNE by E3330. PANC1, XPA1, or HPNE cells were treated with various dose of E3330 for 72 h. Then, MTT assays were done to analyze cell viability. C, trypan blue staining and cell count for E3330-treated PANC1 cells. D, methoxyamine, the APE1 DNA repair domain inhibitor, has no growth-inhibitory effects on pancreatic cancer cells. Both PANC1 and XPA1 cells were treated with methoxyamine for 72 h followed by a MTT assay. E, difference of spontaneous growth rate between low APE1-expressing pancreatic cell (HPNE) and high APE1-expressing pancreatic cells (PANC1). Transduction of exogenous wild-type APE1 into HPNE cells enhances their growth rate.
Inhibition of APE1Redox Function with E3330 Inhibits the Growth of Human Pancreatic Cancer Cell Lines but Not That of Nonneoplastic Human Pancreatic Epithelial Cells
We determined the in vitro growth of PANC1 and XPA1 cells exposed to increasing dosages of E3330. In normoxic conditions, at dosages of E3330 <10 μmol/L, there was no affect on the growth of PANC1 and XPA1 cells as examined by MTT assay. However, at dosages of E3330 ≥20 μmol/L, there was significant inhibition of in vitro growth of PANC1 and XPA1 cells. In contrast, this compound had no suppressive effects on in vitro growth of HPNE cells, which have minimal APE1 expression, even at 50 μmol/L E3330 exposure (Fig. 1B). Further studies show that E3330 also inhibits growth of other pancreatic cancer cell lines, such as MIAPACA, BxPC3, and PK9 (data not shown). Trypan blue exclusion assay also confirmed the inhibitory effect of this compound in PANC1 cell proliferation (Fig. 1C). Our studies also indicated that the DNA repair domain of APE1 was not essential for maintaining the growth of pancreatic cancer cells, because blockade of the function of this domain with the chemical inhibitor methoxyamine did not affect cell viability in PANC1 and XPA1 pancreatic cancer cell lines as measured by MTT assays (Fig. 1D). Further study showed that methoxyamine also had no effect in cell viability of other pancreatic cancer cell lines, such as MIAPACA, BxPC3, and PK9 (data not shown). To address whether there is difference of growth rate between HPNE cells and APE1-overexpressing pancreatic cells, we examined the spontaneous growth rate between HPNE and PANC1 cells. We observed that PANC1 cells have a significant higher growth rate than HPNE cells. To further verify the role of APE1 in regulation of growth of pancreatic cells, we transduced exogenous APE1 into HPNE cells and then observed that HPNE cells transduced with exogenous APE1 exhibit higher growth rate than control cells (Fig. 1E). To further confirm the essential role of APE1 in maintaining pancreatic cancer cell growth, we used siRNA to knockdown APE1 gene expression in PANC1 cells and showed that knockdown of APE1 gene expression affects pancreatic cancer cell growth (Fig. 2A).
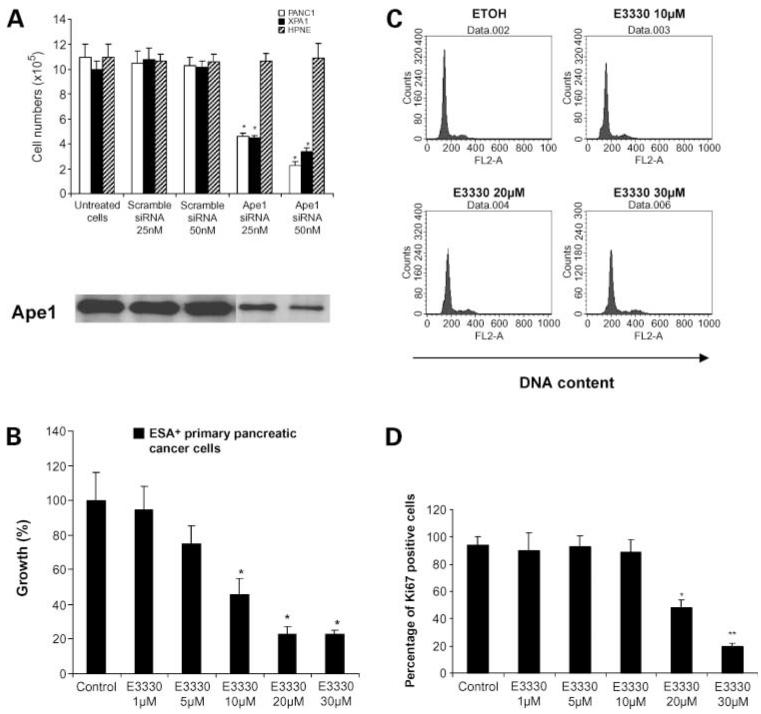
Figure 2.
APE1 is essential for pancreatic cancer cell growth in vitro. A, knockdown of APE1 expression affects cell growth in trypan blue exclusion assay. PANC1, XPA1, or HPNE cells were transfected with scramble (control) siRNA 25 or 50 nmol/L or APE1 siRNA 25 or 50 nmol/L. After transfection, the cells were cultivated for 3 d. The live cell numbers were counted after trypan blue staining. The MTT assays were carried out to examine the cell viability. Mean of three independent experiments; bars, SD. B, APE1 redox domain inhibitor E3330 inhibits the in vitro growth of primary pancreatic cancer cells. ESA-positive primary pancreatic cancer cells were isolated from three independent freshly resected pancreatic cancer tissue specimens using ESA-positive MACS bead. The isolated pancreatic cancer cells were then treated with various doses of E3330. After 48-h incubation, the MTT assay was done to examine cell viability. Columns, mean of three independent experiments; bars, SD. C, DNA contents of PANC1 cells treated with ethanol (as control cells) or with E3330 at various doses (10, 20, and 30 μmol/L) for 48 h. Representative result from three independent experiments. D, percentage of Ki-67-positive cells in E3330-treated PANC1 cells in flow cytometry assay. PANC1 cells were treated with either with ethanol (as control cells) or with various doses of E3330 (1, 5, 10, 20, or 30 μmol/L) for 48 h. The cells were then stained with FITC-conjugated anti-Ki-67 antibody and read on flow cytometry. Columns, mean of three independent experiments; bars, SD. *, P < 0.05; **, P < 0.01.
E3330 Inhibits the In vitro Growth of Primary Pancreatic Cancer Cells
In light of the growth-inhibitory effects of E3330 on pancreatic cancer cell lines, we examined the behavior of ESA-positive primary pancreatic cancer cells to E3330. We isolated ESA-positive cells from a fresh pancreatic cancer resection and plated the viable cells obtained in the presence of E3330. We found a dose-dependent reduction of in vitro growth with E3330 exposure (Fig. 2B), consistent with our observations in pancreatic cancer cell lines.
E3330 Does Not Induce G1 or G2 Arrest but Rather Promotes Exit of Cell Cycle of PANC1 Cells
Because E3330 inhibits PANC1 growth (Fig. 1), we examined whether E3330 affected the cell cycle kinetics of these cancer cells. Our previous study in mouse embryonic stem cells and embryonic stem cell-derived embryoid body cells revealed that knockdown of APE1 expression induced G1 arrest in cell cycle (14). In contrast, we find that E3330 treatment of PANC1 cells induced neither G1 nor G2 arrest (Fig. 2C). E3330-treated cells and control cells (treated by ethanol, a vehicle of E3330) have similar DNA contents. Ki-67 is a nuclear antigen expressed in all phases of the cell cycle, except in G0 cells. The frequency of Ki-67-expressing PANC1 cells is significantly reduced compared with control cells on E3330 exposure (Fig. 2D), indicating that E3330 promotes exit of cell cycle in PANC1 cells rather than inducing G1 or G2 arrest.
Hypoxia Potentiates E3330-Mediated Inhibition of Pancreatic Cancer Cell Growth
Hypoxia is a hallmark of the tumor microenvironment and hypoxic tumor cells are relatively resistant to radiation and chemotherapy (25). We examined the regulatory role of E3330 in pancreatic cancer growth under hypoxic conditions. The cells were treated with 100 μmol/L CoCl2, which is an established system to model hypoxia in culture settings (26); the cells were then treated with E3330. Our data show that, in the setting of hypoxia, E3330 is more effective in inhibiting PANC1 and XPA1 cell growth compared with normoxic conditions, suggesting an enhanced requirement for APE1 redox function in hypoxia (Fig. 3A and B).
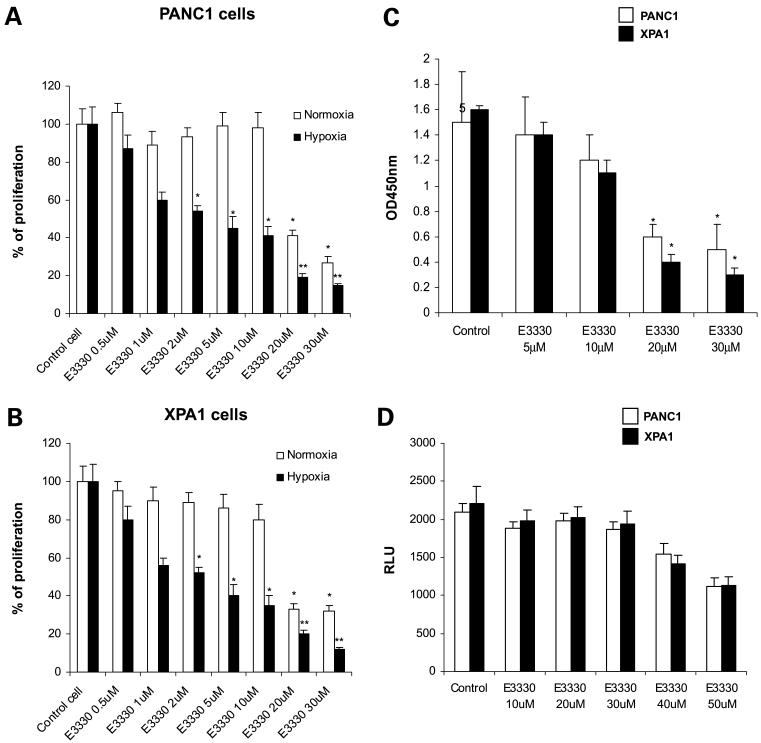
Figure 3.
Hypoxia enhances E3330-mediated growth inhibition in pancreatic cancer cells. A, PANC1 cells. B, XPA1 cells. In normoxia or hypoxia conditions (cells were treated with 100 μmol/L CoCl2), the cells were treated with E3330 at various dose for 48 h. The MTT assay was done. Columns, mean of three independent experiments; bars, SD. *, P < 0.05; **, P < 0.01. C, E3330 inhibits HIF-1α activity. HIF-1α DNA-binding activity was analyzed by ELISA. Columns, mean of three independent experiments; bars, SD. *, P < 0.05. D, NF-κB activity assay in pancreatic cancer cells treated with E3330. Pancreatic cancer cells were transfected with NF-κB reporter plasmid pNF-κB-Luc. The cells were treated with E3330 at various doses for 6 h, and the luciferase activities were measured using a Dual-Luciferase Reporter Assay System.
E3330 Inhibits the DNA-Binding Activity of HIF-1α
Given the differences in effects of E3330 between hypoxia and normoxic conditions, we explored whether blockade of APE1 redox function has deleterious effects on survival signals activated in hypoxia. HIF-1α, a hypoxia-induced transcription factor, is expressed by many cancers, including pancreatic cancer (27, 28). HIF-1α transcriptionally activates a diverse repertoire of genes, including angiogenic growth factors, and facilitates cancer cell survival in the hypoxic milieu of tumors (29, 30). In our study, exposure of PANC1 cells to E3330 at dosages >20 μmol/L dramatically inhibited nuclear HIF-1α protein DNA-binding activity (Fig. 3C), suggesting the requirement of APE1 redox function domain for the transcriptional activity of HIF-1α. However, E3330 at ≤30 μmol/L had no significant inhibitory effect in NF-κB activity, although this inhibitory effect is obvious when the E3330 dose is ≥40 μmol/L (Fig. 3D).
E3330 Exposure Increase Endogenous ROS Levels in PANC1 Cells
It has been suggested that APE1 suppresses intracellular ROS production induced by exogenous stimuli such as TNF cytokines (31, 32). Elevated intracellular ROS levels can rapidly overwhelm endogenous antioxidant mechanisms and induce oxidative stress-related damage. We hypothesized that the APE1 redox domain controls endogenous ROS level; hence, inhibition of this domain with E3330 would increase ROS production in pancreatic cancer cells. Using dihydrorhodamine 123 as a probe, our flow cytometric analysis revealed that untreated cells have low level of endogenous ROS, whereas E3330 treatment enhances significantly intracellular ROS level in PANC1 cells (Fig. 4A). To confirm this phenomenon, we also knocked down APE1 expression in PANC1 cells with APE1 siRNA and found that knockdown of APE1 expression resulted in increased level of ROS (Fig. 4B).
Figure 4.
E3330 enhances ROS production in PANC1 cells. A, ROS was detected by flow cytometry using dihydrorhodamine 123 as a probe. The cells were treated with ethanol (as control cells) or with E3330 at the indicated dose for 2 d. The cells were then harvested, stained with 0.5 μmol/L dihydrorhodamine 123 for 30 min, and analyzed on flow cytometry. Representative results from three independent experiments. B, knockdown of APE1 expression enhance endogenous ROS level in PANC1 cells. PANC1 cells were transfected with control siRNA or APE1 siRNA. After 72 h, the intracellular ROS level was analyzed by labeling cells with dihydrorhodamine 123 and assessed by flow cytometry. MFI, mean fluorescence intensity. C, SHP-2 expression in mitochondria of PANC1 cells in Western blot assay: 50 μg whole-cell lysate of PANC1 cells or 50 μg purified mitochondria of PANC1 cells were assayed in their SHP-2 protein level. D, inhibition of APE1 redox function with E3330 increases SHP-2 oxidation and inactivation in pancreatic carcinoma. PANC1 cells were treated with ethanol (as control cells) or indicated dose of E3330 for 48 h. The cells were lysated in an anaerobic chamber with 1 mL O2 free lysis buffer. The level of oxidation of SHP-2 is examined by immunoprecipitation and blot assay. Representative results from three independent experiments.
E3330 Treatment of PANC1 Cell Increases SHP-2 Oxidation, Thus Inactivating This Oncogenic Tyrosine Phosphatase
SHP-2 is a cytoplasmic Src homology domain containing protein tyrosine phosphatase, which is ubiquitously expressed in most somatic cells, and plays a critical role during development and hematopoiesis (33). Aberrant activity of this phosphatase has been implicated in a growing list of human cancers (34-37), and gain-of-function somatic mutations of PTPN11; the gene encoding SHP-2 have been reported in hematologic malignancies (38, 39). Oxidation of SHP-2 reduces its activity (40). In light of increased intracellular ROS levels on E3330 exposure, we hypothesized that blockade of APE1 redox domain would enhance SHP-2 oxidation in PANC1 cells, leading to inactivation of this phosphatase. We examined the expression of SHP-2 in mitochondria and our result show that SHP-2 protein is localized in mitochondria assayed in Western blot assay with fraction of mitochondria (Fig. 4C). Indeed, as shown in Fig. 4D, E3330-untreated PANC1 cells express the active, reduced form of SHP-2, whereas E3330 treatment induces oxidation and hence inactivation of SHP-2. To the best of our knowledge, this is the first demonstration for the requirement of APE1 redox function in sustaining SHP-2 activation in cancer cells.
E3330 Inhibits Migration of Pancreatic Cancer Cells
The cellular redox state can have profound effects on not only viability but also migratory abilities of cancer cells. Currently, the role of APE1 in regulation of cell migration is not defined. In the present study, we determined whether inhibition of APE1 redox function had an effect on migration of three APE1-overexpressing pancreatic cancer cell lines. The well-established pancreatic cancer chemokine stromal cell–derived factor-1, also known as CXCL12, was used for directing migration in culture chambers. Addition of E3330 in the culture significantly affected the in vitro migration of all three lines independent of any effects on growth inhibition (Fig. 5A).
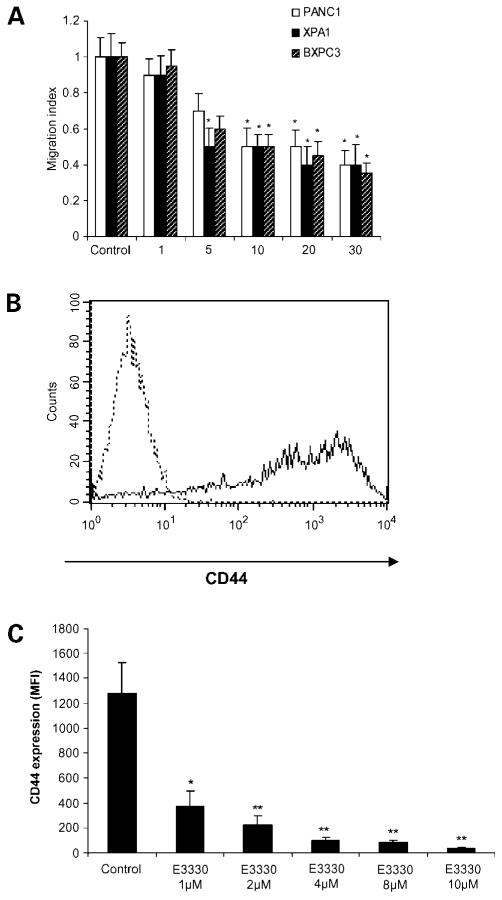
Figure 5.
E3330 inhibits stromal cell – derived factor-1 – mediated migration of pancreatic cancer cells. A, pancreatic cancer cell lines PANC1, XPA1, and BxPC3 were stimulated by stromal cell – derived factor-1 at 100 ng/mL and exposed to varying concentrations of E3330. Stromal cell – derived factor-1 – stimulated migration was inhibited by E3330 at dosages ≥5 μmol/L. Columns, mean of three independent experiments in triplicate wells and normalized for any growth-inhibitory effects of E3330. *, P < 0.05. B, fluorescence-activated cell sorting analysis of CD44 expression in PANC1 cells. Dotted line, cells stained with isotype control antibody; solid line, cells stained with anti-CD44 antibody. C, E3330 inhibits CD44 expression in PANC1 cells. PANC1 cells were treated with E3330 at the indicated dose for 48 h. The cells were then harvested, stained with fluorescence-conjugated anti-CD44 antibody, and analyzed on flow cytometry. Columns, mean of three independent experiments; bars, SD. *, P < 0.05; **, P < 0.01.
E3330 Inhibits CD44 Expression on the Surface of Pancreatic Cancer Cells
In light of the migration inhibition by E3330, we determined if APE1 redox domain had any effects on surface expression of CD44. This molecule is a cell surface glycoprotein overexpressed in many cancers, including pancreatic cancer, and has been implicated in promoting the invasive and migratory capabilities of cancer cells in vitro and in vivo. By flow cytometry, E3330 strikingly inhibited CD44 expression on PANC1 cells, even at the lowest dosages of 1 to 10 μmol/L, where cellular growth itself was unaffected (Fig. 5B and C).
Discussion
Regulation of the intracellular redox environment is critical for cell viability and maintenance of cellular homeostasis. APE1 is a dual-function molecule, containing an AP endonuclease DNA repair domain and a redox function domain (3). Many of the studies to date on the role APE1 in human cancers have ablated total gene function (e.g., by RNA interference or antisense) without selectively dissecting the contribution of the respective functional domains (10, 12). In the present study, we explored the role of APE1 redox regulation in established human pancreatic cancer cell lines. Motivation for this study stemmed from our observation as well as those of other groups (10, 11), showing APE1 overexpression in human pancreatic cancer cells. Our studies were facilitated by the availability of E3330, a small-molecule inhibitor of APE1 redox function (14). An APE1−/− human cell line cannot be generated because APE1 is essential for early embryonic development (41); thus, identification and development of a specific inhibitor of this protein will significantly help as a tool in APE1 functional study and therapeutic potential. Within APE1 protein, there are three E3330-binding sites, and all of these sites are located within the redox domain, none are located in the DNA repair domain of APE1 protein (13), and it affects redox activity of several downstream targets of APE1. These data indicate that E3330 is a specific inhibitor of APE1 redox function without interfering with its DNA repair role (3, 42). It is suggested that methoxyamine is a specific inhibitor of APE1 DNA repair function (3). Interestingly, although DNA repair function of APE1 protein has been suggested to play a key role in sustaining cell viability and proliferation in colon cancer cells when APE1 gene expression was knocked down by siRNA (12), the recent studies show that methoxyamine, the DNA repair domain inhibitor, actually has no cytotoxicity and no influence in cell proliferation (43, 44). Our data presented in this study are consistent with their demonstration. We suggest that novel APE1-independent DNA base excision repair, such as NEIL1/2/PNK-dependent base excision repair, which can bypass APE1 (45), or APE2-dependent base excision repair, may function at this stage to replace the base excision repair function of APE1 when it is inhibited by methoxyamine. So, the redox function may be more important than its DNA repair function to sustain cell survival and proliferation in pancreatic cancer.
APE1 is overexpressed in numerous solid cancers, including prostate and bladder cancers (4, 5), non-small cell lung cancers (6), gliomas (7), medulloblastomas and primitive neurectodermal tumors (8), osteosarcomas (9), and germ cell tumors (46). In this study, we confirmed increased expression of APE1 protein in pancreatic cancer cell lines compared with the nonneoplastic HPNE and HPDE cell line. Thus, aberrant expression of APE1 may be a general phenomenon in human cancer and suggests an increased requirement for one or both functional domains of this critical homeostatic protein in cancer cells. Further, E3330-induced inhibition of growth in pancreatic cancer cell lines, but not in the HPNE cells, implies that neoplastic cells have a greater dependence on sustained APE1 redox function for their viability than nonneoplastic cells. This important observation provides a therapeutic window in terms of therapeutic targeting of the APE1 redox domain using E3330 or more potent small-molecule inhibitors.
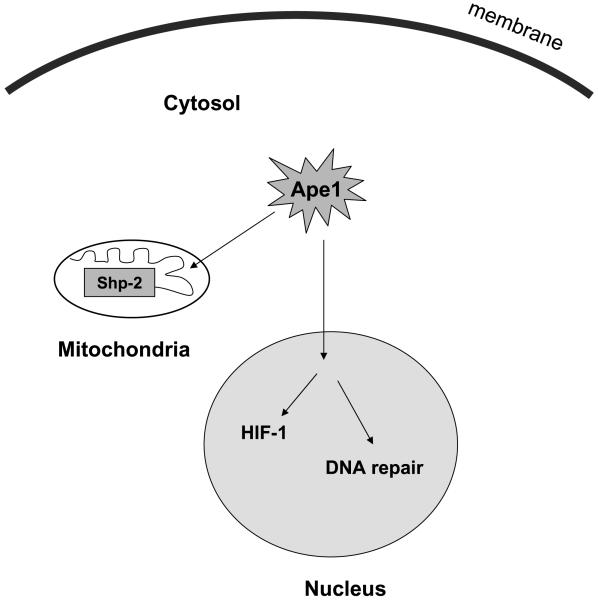
Although APE1/REF-1 subcellular localization is mainly nucleus (32), recently, there is emerging evidence showing that APE1 protein is localized in both nucleus and cytoplasm (31, 47). Furthermore, in liver cancer tissue, cytoplasmic expression of APE1 is higher in cancer cells than in nonmalignant liver cells within the tumor (47). Cytoplasmic APE1/REF-1 is associated with mitochondria (32), similar to its homologous gene Apn1 in yeast (48). This encouraged us to examine the potential extranuclear function of APE1 protein localized within the cytosol of pancreatic cancer cells. We were particularly interested to examine the relevance between cytoplasmic protein tyrosine phosphatase, such as SHP-2, and APE1-mediated redox regulation, because recent observation shows SHP-2 can be localized within mitochondria. SHP-2 is essential for cellular growth and progenitor cell differentiation (17), and recent studies have established a direct role for SHP-2 in cell survival related to oxidative stress-related cellular signaling. SHP-2 is activated in diverse human cancers, and this phosphatase has been implicated as a critical effector for the transforming ability of several credentialed oncogenes, like Ras, Bcr-Abl, and mutant epidermal growth factor receptor (35, 37). The activity of SHP-2 can be negatively regulated by oxidation (40). Previous studies have shown that ROS can induce transient inactivation of SHP-2 in T cells (23). Endogenous APE1 may function as a regulator to maintain ROS balance because knockdown of APE1 expression by siRNA for 48 h significantly increases ROS level (49). In the present study, we confirmed the expression of active (nonoxidized) SHP-2 phosphatase in untreated PANC1 cells. We hypothesized that E3330-induced up-regulation of endogenous ROS levels in PANC1 will suppress SHP-2 activity via oxidation. We confirmed this hypothesis to be true, and our data provide the first evidence linking inhibition of APE1 redox domain with inactivation of SHP-2 by oxidation. We believe that inactivation of SHP-2 may be one mechanism by which E3330 exerts its antiproliferative effects in pancreatic cancer. Further, we propose APE1 may activate SHP-2 through a redox mechanism within mitochondria, a novel mechanism regarding its role in cancer cell regulation (Fig. 6).
Figure 6.
Proposed model about the role of APE1 regulating SHP-2 activity in mitochondria and HIF-1α activity in nucleus through redox mechanism in pancreatic cancer cells.
In conclusion, we show that E3330, a small-molecule inhibitor of APE1 redox domain function, directly inhibits growth and migration of human pancreatic cancer cells. Further, we establish that blockade of APE1 redox domain can inhibit the transcription activating function of HIF-1α, inactivate SHP-2, and down-regulate CD44 expression on the cell surface, all of which have likely mitigating effects on the transformed phenotype of pancreatic cancer cells. Our results suggest that small-molecule inhibition of the APE1 redox domain with E3330, or other more potent chemical inhibitors, is a potential therapeutic strategy in pancreatic cancer. However, we showed previously that E3330 also inhibits hematopoiesis, including reduction of colony-forming units-granulocyte-macrophage colony formation (14). This might limit its potential use as an anticancer agent in pancreatic cancer because of its potential side effect in hematopoiesis suppression. To solve this problem, it is recommended to combine E3330 and hematopoietic stimulatory factor for the future in vivo trial in cancer therapy, such as granulocyte-monocyte-colony-stimulating factor, stem cell factor, etc., which may overcome the potential hematopoietic suppression effect of E3330.
Acknowledgments
We thank Dr. Shibin Zhou for the use of the anaerobic chamber and Dr. Honda (Tokyo Institute of Technology) for kindly providing E3330.
Grant support: Sol Goldman Pancreatic Cancer Research Center and Michael Rolfe Foundation for Pancreatic Cancer Research (A. Maitra).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 3.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med. 2007;28:375–95. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Kelley MR, Cheng L, Foster R, et al. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin Cancer Res. 2001;7:824–30. [PubMed] [Google Scholar]
- 5.Sak SC, Harnden P, Johnston CF, Paul AB, Kiltie AE. APE1 and XRCC1 protein expression levels predict cancer-specific survival following radical radiotherapy in bladder cancer. Clin Cancer Res. 2005;11:6205–11. doi: 10.1158/1078-0432.CCR-05-0045. [DOI] [PubMed] [Google Scholar]
- 6.Puglisi F, Aprile G, Minisini AM, et al. Prognostic significance of Ape1/ref-1 subcellular localization in non-small cell lung carcinomas. Anticancer Res. 2001;21:4041–9. [PubMed] [Google Scholar]
- 7.Bobola MS, Blank A, Berger MS, Stevens BA, Silber JR. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin Cancer Res. 2001;7:3510–8. [PubMed] [Google Scholar]
- 8.McNeill DR, Wilson DM., III A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res. 2007;5:61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–86. [PubMed] [Google Scholar]
- 10.Lau JP, Weatherdon KL, Skalski V, Hedley DW. Effects of gemcitabine on APE/ref-1 endonuclease activity in pancreatic cancer cells, and the therapeutic potential of antisense oligonucleotides. Br J Cancer. 2004;91:1166–73. doi: 10.1038/sj.bjc.6602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray MJ, Zhang J, Ellis LM, et al. HIF-1α, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–20. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- 12.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–70. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu N, Sugimoto K, Tang J, et al. High-performance affinity beads for identifying drug receptors. Nat Biotechnol. 2000;18:877–81. doi: 10.1038/78496. [DOI] [PubMed] [Google Scholar]
- 14.Zou GM, Luo MH, Reed A, Kelley MR, Yoder MC. Ape1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood. 2007;109:1917–22. doi: 10.1182/blood-2006-08-044172. [DOI] [PubMed] [Google Scholar]
- 15.Calhoun ES, Hucl T, Gallmeier E, et al. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 2006;66:7920–8. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 16.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res. 2007;67:2098–106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 17.Zou GM, Chan RJ, Shelley WC, Yoder MC. Reduction of Shp-2 expression by small interfering RNA reduces murine embryonic stem cell-derived in vitro hematopoietic differentiation. Stem Cells. 2006;24:587–94. doi: 10.1634/stemcells.2005-0272. [DOI] [PubMed] [Google Scholar]
- 18.Feldmann G, Dhara S, Fendrich V, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–96. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou GM, Reznikoff-Etievant MF, Hirsch F, Milliez J. IFN-γ induces apoptosis in mouse embryonic stem cells, a putative mechanism of its embryotoxicity. Dev Growth Differ. 2000;42:257–64. doi: 10.1046/j.1440-169x.2000.00511.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaaijk P, Kaspers GJ, Van Wering ER, et al. Cell proliferation is related to in vitro drug resistance in childhood acute leukaemia. Br J Cancer. 2003;88:775–81. doi: 10.1038/sj.bjc.6600787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou WC, Chen HY, Yu SL, Cheng L, Yang PC, Dang CV. Arsenic suppresses gene expression in promyelocytic leukemia cells partly through Sp1 oxidation. Blood. 2005;106:304–10. doi: 10.1182/blood-2005-01-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng TC, Fukada T, Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol Cell. 2002;9:387–99. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 23.Kwon J, Qu CK, Maeng JS, Falahati R, Lee C, Williams MS. Receptor-stimulated oxidation of SHP-2 promotes T-cell adhesion through SLP-76-ADAP. EMBO J. 2005;24:2331–41. doi: 10.1038/sj.emboj.7600706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong D, Park EJ, Stephen AG, et al. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 2005;65:9047–55. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 25.Murdoch C, Lewis CE. Macrophage migration and gene expression in response to tumor hypoxia. Int J Cancer. 2005;117:701–8. doi: 10.1002/ijc.21422. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–22. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 27.Semenza GL. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol Med. 2002;8:S62–7. doi: 10.1016/s1471-4914(02)02317-1. [DOI] [PubMed] [Google Scholar]
- 28.Zhong H, De Marzo AM, Laughner E, et al. Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 1999;59:5830–5. [PubMed] [Google Scholar]
- 29.Hirota K, Semenza GL. Regulation of angiogenesis by hypoxia-inducible factor 1. Crit Rev Oncol Hematol. 2006;59:15–26. doi: 10.1016/j.critrevonc.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J, Schmid T, Schnitzer S, Brune B. Tumor hypoxia and cancer progression. Cancer Lett. 2006;237:10–21. doi: 10.1016/j.canlet.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Angkeow P, Deshpande SS, Qi B, et al. Redox factor-1: an extra-nuclear role in the regulation of endothelial oxidative stress and apoptosis. Cell Death Differ. 2002;9:717–25. doi: 10.1038/sj.cdd.4401025. [DOI] [PubMed] [Google Scholar]
- 32.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–84. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 33.Chong ZZ, Maiese K. The Src homology 2 domain tyrosine phosphatases SHP-1 and SHP-2: diversified control of cell growth, inflammation, and injury. Histol Histopathol. 2007;22:1251–67. doi: 10.14670/hh-22.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatakeyama M. The role of Helicobacter pylori CagA in gastric carcinogenesis. Int J Hematol. 2006;84:301–8. doi: 10.1532/IJH97.06166. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Yu WM, Daino H, Broxmeyer HE, Druker BJ, Qu CK. SHP-2 phosphatase is required for hematopoietic cell transformation by Bcr-Abl. Blood. 2007;109:778–85. doi: 10.1182/blood-2006-04-019141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang FM, Liu HQ, Liu SR, Tang SP, Yang L, Feng GS. SHP-2 promoting migration and metastasis of MCF-7 with loss of E-cadherin, dephosphorylation of FAK and secretion of MMP-9 induced by IL-1β in vivo and in vitro. Breast Cancer Res Treat. 2005;89:5–14. doi: 10.1007/s10549-004-1002-z. [DOI] [PubMed] [Google Scholar]
- 37.Zhan Y, O’Rourke DM. SHP-2-dependent mitogen-activated protein kinase activation regulates EGFRvIII but not wild-type epidermal growth factor receptor phosphorylation and glioblastoma cell survival. Cancer Res. 2004;64:8292–8. doi: 10.1158/0008-5472.CAN-03-3143. [DOI] [PubMed] [Google Scholar]
- 38.Tartaglia M, Niemeyer CM, Shannon KM, Loh ML. SHP-2 and myeloid malignancies. Curr Opin Hematol. 2004;11:44–50. doi: 10.1097/00062752-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 39.Loh ML, Vattikuti S, Schubbert S, et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood. 2004;103:2325–31. doi: 10.1182/blood-2003-09-3287. [DOI] [PubMed] [Google Scholar]
- 40.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–70. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 41.Madhusudan S, Middleton MR. The emerging role of DNA repair proteins as predictive, prognostic and therapeutic targets in cancer. Cancer Treat Rev. 2005;31:603–17. doi: 10.1016/j.ctrv.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Kabe Y, Ando K, Hirao S, Yoshida M, Handa H. Redox regulation of NF-κB activation: distinct redox regulation between the cytoplasm and the nucleus. Antioxid Redox Signal. 2005;7:395–403. doi: 10.1089/ars.2005.7.395. [DOI] [PubMed] [Google Scholar]
- 43.Chakravarti D, Badawi AF, Venugopal D, et al. Improved measurement of dibenzo[a,l]pyrene-induced abasic sites by the aldehyde-reactive probe assay. Mutat Res. 2005;588:158–65. doi: 10.1016/j.mrgentox.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 44.She M, Pan I, Sun L, Yeung SC. Enhancement of manumycin A-induced apoptosis by methoxyamine in myeloid leukemia cells. Leukemia. 2005;19:595–602. doi: 10.1038/sj.leu.2403691. [DOI] [PubMed] [Google Scholar]
- 45.Wiederhold L, Leppard JB, Kedar P, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–20. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Thomson BG, Tritt R, Davis M, Perlman EJ, Kelley MR. Apurinic/apyrimidinic endonuclease expression in pediatric yolk sac tumors. Anticancer Res. 2000;20:4153–7. [PubMed] [Google Scholar]
- 47.Di Maso V, Avellini C, Croce LS, et al. Subcellular localization of APE1/Ref-1 in human hepatocellular carcinoma: possible prognostic significance. Mol Med. 2007;13:89–96. doi: 10.2119/2006-00084.DiMaso. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jilani A, Vongsamphanh R, Leduc A, Gros L, Saparbaev M, Ramotar D. Characterization of two independent amino acid substitutions that disrupt the DNA repair functions of the yeast Apn1. Biochemistry. 2003;42:6436–45. doi: 10.1021/bi034163m. [DOI] [PubMed] [Google Scholar]
- 49.Yang S, Misner BJ, Chiu RJ, Meyskens FL., Jr. Redox effector factor-1, combined with reactive oxygen species, plays an important role in the transformation of JB6 cells. Carcinogenesis. 2007;28:2382–90. doi: 10.1093/carcin/bgm128. [DOI] [PubMed] [Google Scholar]