Abstract
In the title compound, C36H24O6, the benzoyl groups at the 1- and 8-positions of the naphthalene system are in an anti orientation. Both carbonyl groups form intramolecular O—H⋯O hydrogen bonds with hydroxy groups affording six-membered rings. The benzene rings of the benzoyl groups make dihedral angles of 59.26 (13) and 59.09 (13)° with the naphthalene ring system. Zigzag C—H⋯O chains and ladder C—H⋯O chains between the phenoxybenzoyl groups along the ab diagonals form an undulating checkered sheet. The molecules are further connected into a three-dimensional network by C—H⋯π interactions.
Related literature
For electrophilic aromatic aroylation of the naphthalene core, see: Okamoto & Yonezawa (2009 ▶); Okamoto et al. (2011 ▶, 2013 ▶). For the structures of (2,7-dimethoxynaphthalene-1,8-diyl)bis(4-fluorophenyl)dimethanone and 2,7-dimethoxy-1,8-bis(4-phenoxybenzoyl)naphthalene, see: Watanabe et al. (2010 ▶) andr Hijikata et al. (2010 ▶), respectively.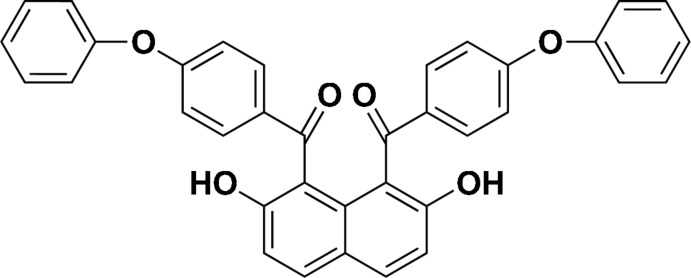
Experimental
Crystal data
C36H24O6
M r = 552.55
Monoclinic,

a = 16.0313 (3) Å
b = 18.4956 (3) Å
c = 12.1238 (2) Å
β = 131.389 (1)°
V = 2696.95 (9) Å3
Z = 4
Cu Kα radiation
μ = 0.75 mm−1
T = 193 K
0.60 × 0.55 × 0.10 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
Absorption correction: numerical (NUMABS; Higashi, 1999 ▶) T min = 0.661, T max = 0.929
22236 measured reflections
4868 independent reflections
4527 reflections with I > 2σ(I)
R int = 0.033
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.096
S = 1.08
4868 reflections
382 parameters
2 restraints
H-atom parameters constrained
Δρmax = 0.20 e Å−3
Δρmin = −0.21 e Å−3
Absolute structure: Flack (1983 ▶), 2389 Friedel pairs
Flack parameter: 0.05 (19)
Data collection: PROCESS-AUTO (Rigaku, 1998 ▶); cell refinement: PROCESS-AUTO; data reduction: PROCESS-AUTO; program(s) used to solve structure: Il Milione (Burla et al., 2007 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEPIII (Burnett & Johnson, 1996 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812052038/rn2112sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812052038/rn2112Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812052038/rn2112Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of the C25—C30 and C31—C36 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O5—H5A⋯O1 | 0.84 | 1.83 | 2.560 (3) | 145 |
| O6—H6A⋯O2 | 0.84 | 1.88 | 2.563 (3) | 138 |
| C26—H26⋯O4i | 0.95 | 2.48 | 3.377 (4) | 157 |
| C27—H27⋯O1i | 0.95 | 2.51 | 3.269 (4) | 137 |
| C32—H32⋯O3ii | 0.95 | 2.49 | 3.382 (4) | 156 |
| C33—H33⋯O2ii | 0.95 | 2.51 | 3.270 (4) | 137 |
| C14—H14⋯Cg1iii | 0.95 | 2.80 | 3.740 (2) | 171 |
| C21—H21⋯Cg2iv | 0.95 | 2.80 | 3.740 (2) | 171 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Acknowledgments
The authors would like to express their gratitude to Professor Keiichi Noguchi, Instrumentation Analysis Center, Tokyo University of Agriculture and Technology, for his technical advice. This work was partially supported by the Ogasawara Foundation for the Promotion of Science & Engineering, Tokyo, Japan.
supplementary crystallographic information
Comment
In the course of our study on electrophilic aromatic aroylation of 2,7-dimethoxynaphthalene, peri-aroylnaphthalene compounds have proven to be formed regioselectively with the aid of suitable acidic mediators (Okamoto & Yonezawa, 2009; Okamoto et al., 2011). As one of the applications of peri-aroylnaphthalene synthetic studies, the authors have integrated the resulting molecular unit to the poly(ether ketone) backbone via nucleophilic aromatic substitution polycondensation (Okamoto et al., 2013). The poly(ether ketone)s composed of 1,8-diaroylenenaphthalene units show unique thermal properties and solubility for organic solvents. These notable properties could arise from the structural features of the 1,8-diaroylene naphthalene units. Under these circumstances, the authors have undertaken the X-ray crystal structural study of several 1,8-diaroylated naphthalene analogues exemplified by (2,7-dimethoxynaphthalene-1,8-diyl)bis(4-fluorophenyl)dimethanone (Watanabe et al., 2010) and 2,7-dimethoxy-1,8-bis(4-phenoxybenzoyl)naphthalene (Hijikata et al., 2010). These molecules have essentially the same non-coplanar features. The two aroyl groups are twisted so they are almost perpendicular to the naphthalene rings.
The molecular structure of the title compound is displayed in Fig. 1. Two benzoyl groups are on the 1,8-positions of the naphthalene ring and are in an anti orientation relative to one another. The benzene rings of the benzoyl groups make dihedral angles with the naphthalene ring of 59.26 (13) and 59.09 (13)°, respectively. The dihedral angles between the benzene rings of the benzoyl groups and those of the phenoxy groups are 69.05 (13) and 69.02 (13)°. Both carbonyl groups form intramolecular O—H···O hydrogen bonds with hydroxy groups affording six-membered rings. (Fig. 1, Table 1).
In the crystal structure, the molecular packing of the title compound is stabilized mainly by C—H···O and C—H···π interactions. The aromatic hydrogen atoms of the phenoxy groups form two types of intermolecular C—H···O interactions with the ethereal oxygen atom of the phenoxy groups(C26—H26···O4i= 2.48 Å, C32—H32···O3ii= 2.49 Å; Fig. 2 and Table 1) and the carbonyl oxygen atom (C27—H27···O1i= 2.51 Å, C33—H33···O2ii= 2.51 Å; Fig. 2 and Table 1). Intermolecular C—H···π interactions between the aromatic hydrogen atom of the benzoyl group and the centroid of the benzene ring of the phenoxy group (C14—H14···Cg1iii= 2.80 Å, C21—H21···Cg2iv= 2.80 Å; Fig. 3 and Table 1) are observed.
Experimental
To a stirring solution of 1,8-bis(4-phenoxybenzoyl)-2,7-dimethoxynaphthalene (1.0 mmol, 580 mg) in dichloromethane (1.0 ml) at 0°C was added 1.0 M boron tribromide solution in dichloromethane (4.4 ml) slowly, and the reaction mixture was allowed to reach the room temperature. After the reaction mixture had been stirred at room temperature for 48 h, the reaction mixture was cooled to 0 oC and very slowly quenched with water and extracted with CHCl3. The organic layer thus obtained was dried over anhydrous MgSO4. The solvent was removed under reduced pressure to give a cake. The crude product was purified by column chromatography (silica gel, CHCl3) to give the title compound (isolated yield 88%). Single crystals suitable for X-ray diffraction were obtained by crystallization from Et2O-hexane (v/v = 1:2).
1H NMR δ (300 MHz, CDCl3): 6.82–6.84 (4H, m), 7.08–7.26 (10H, m), 7.40 (4H, t, J=7.9 Hz) 7.86 (2H, d, J=8.9 Hz), 11.29 (2H, s) p.p.m.
13C NMR δ (75 MHz, CDCl3): 115.13, 117.03, 117.28, 120.02, 122.02, 124.46, 130.00, 130.68, 133.79, 136.09, 155.58, 161.74, 195.80 p.p.m.
IR (KBr): 3396(O—H), 1620 (C=O), 1608, 1583, 1487 (Ar, naphthalene) cm-1.
HRMS (m/z): [M + H]+ calcd for C36H25O6, 553.1651 found, 553.1637.
m.p. 464.6–465.9 K.
Refinement
All the H atoms could be located in difference Fourier maps. All the H atoms were subsequently refined as riding atoms, with O5—H5A = 0.84, O6—H6A = 0.84, C—H = 0.95 (aromatic) Å, Uiso(H) = 1.2Ueq(O) and Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.

The molecular structure of title compound, showing 30% probability displacement ellipsoids. The intramolecular O—H···O hydrogen bond is shown as a dashed line.
Fig. 2.
A partial crystal packing diagram of title compound. The intermolecular C—H···O interactions are shown as dashed lines.
Fig. 3.

A partial crystal packing diagram of title compound. The intermolecular C—H···π interactions are shown as dashed lines.
Crystal data
| C36H24O6 | F(000) = 1152 |
| Mr = 552.55 | Dx = 1.361 Mg m−3 |
| Monoclinic, Cc | Cu Kα radiation, λ = 1.54187 Å |
| Hall symbol: C -2yc | Cell parameters from 14515 reflections |
| a = 16.0313 (3) Å | θ = 4.4–68.1° |
| b = 18.4956 (3) Å | µ = 0.75 mm−1 |
| c = 12.1238 (2) Å | T = 193 K |
| β = 131.389 (1)° | Block, yellow |
| V = 2696.95 (9) Å3 | 0.60 × 0.55 × 0.10 mm |
| Z = 4 |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 4868 independent reflections |
| Radiation source: rotating anode | 4527 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.033 |
| Detector resolution: 10.000 pixels mm-1 | θmax = 68.1°, θmin = 4.4° |
| ω scans | h = −19→19 |
| Absorption correction: numerical (NUMABS; Higashi, 1999) | k = −22→22 |
| Tmin = 0.661, Tmax = 0.929 | l = −14→14 |
| 22236 measured reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.037 | w = 1/[σ2(Fo2) + (0.0414P)2 + 1.2872P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.096 | (Δ/σ)max < 0.001 |
| S = 1.08 | Δρmax = 0.20 e Å−3 |
| 4868 reflections | Δρmin = −0.21 e Å−3 |
| 382 parameters | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 2 restraints | Extinction coefficient: 0.00222 (11) |
| Primary atom site location: structure-invariant direct methods | Absolute structure: Flack (1983), 2389 Friedel pairs |
| Secondary atom site location: difference Fourier map | Flack parameter: 0.05 (19) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.46616 (15) | 0.24827 (8) | 0.4776 (2) | 0.0586 (5) | |
| O2 | 0.36810 (15) | 0.00189 (8) | 0.4778 (2) | 0.0589 (5) | |
| O3 | 0.78626 (13) | −0.00972 (9) | 0.69269 (17) | 0.0540 (4) | |
| O4 | 0.26329 (13) | 0.25987 (9) | 0.69275 (17) | 0.0540 (4) | |
| O5 | 0.3598 (2) | 0.31569 (12) | 0.2334 (3) | 0.0889 (7) | |
| H5A | 0.4028 | 0.3117 | 0.3252 | 0.107* | |
| O6 | 0.2301 (2) | −0.06560 (12) | 0.2333 (3) | 0.0887 (7) | |
| H6A | 0.2881 | −0.0652 | 0.3226 | 0.106* | |
| C1 | 0.33277 (19) | 0.18893 (13) | 0.2506 (2) | 0.0461 (5) | |
| C2 | 0.3049 (3) | 0.25255 (17) | 0.1699 (3) | 0.0664 (8) | |
| C3 | 0.2213 (3) | 0.2516 (3) | 0.0165 (4) | 0.0985 (15) | |
| H3 | 0.2020 | 0.2950 | −0.0374 | 0.118* | |
| C4 | 0.1684 (3) | 0.1906 (3) | −0.0546 (3) | 0.1016 (16) | |
| H4 | 0.1154 | 0.1909 | −0.1591 | 0.122* | |
| C5 | 0.1337 (2) | 0.0591 (3) | −0.0547 (3) | 0.1027 (16) | |
| H5 | 0.0822 | 0.0587 | −0.1592 | 0.123* | |
| C6 | 0.1529 (3) | −0.0033 (3) | 0.0189 (4) | 0.1012 (15) | |
| H6 | 0.1190 | −0.0471 | −0.0339 | 0.121* | |
| C7 | 0.2216 (2) | −0.00274 (17) | 0.1702 (3) | 0.0661 (8) | |
| C8 | 0.27453 (17) | 0.06111 (13) | 0.2507 (2) | 0.0460 (5) | |
| C9 | 0.26698 (17) | 0.12502 (15) | 0.1775 (2) | 0.0487 (5) | |
| C10 | 0.1889 (2) | 0.1250 (2) | 0.0211 (3) | 0.0757 (9) | |
| C11 | 0.43775 (19) | 0.19208 (11) | 0.4067 (2) | 0.0426 (5) | |
| C12 | 0.51884 (16) | 0.13161 (11) | 0.4748 (2) | 0.0369 (4) | |
| C13 | 0.59891 (17) | 0.12824 (12) | 0.6275 (2) | 0.0422 (5) | |
| H13 | 0.5936 | 0.1599 | 0.6843 | 0.051* | |
| C14 | 0.68556 (17) | 0.07991 (13) | 0.6977 (2) | 0.0452 (5) | |
| H14 | 0.7375 | 0.0764 | 0.8019 | 0.054* | |
| C15 | 0.69610 (16) | 0.03646 (11) | 0.6145 (2) | 0.0399 (4) | |
| C16 | 0.61693 (17) | 0.03743 (12) | 0.4626 (2) | 0.0413 (5) | |
| H16 | 0.6240 | 0.0065 | 0.4066 | 0.050* | |
| C17 | 0.52707 (17) | 0.08406 (12) | 0.3929 (2) | 0.0405 (5) | |
| H17 | 0.4707 | 0.0836 | 0.2888 | 0.049* | |
| C18 | 0.32565 (17) | 0.05816 (11) | 0.4067 (2) | 0.0427 (5) | |
| C19 | 0.31261 (16) | 0.11833 (11) | 0.4748 (2) | 0.0368 (4) | |
| C20 | 0.38516 (17) | 0.12173 (12) | 0.6276 (2) | 0.0421 (5) | |
| H20 | 0.4471 | 0.0899 | 0.6844 | 0.050* | |
| C21 | 0.36894 (18) | 0.17017 (13) | 0.6976 (2) | 0.0458 (5) | |
| H21 | 0.4215 | 0.1738 | 0.8018 | 0.055* | |
| C22 | 0.27547 (17) | 0.21371 (11) | 0.6151 (2) | 0.0399 (4) | |
| C23 | 0.20247 (17) | 0.21294 (12) | 0.4623 (2) | 0.0412 (5) | |
| H23 | 0.1397 | 0.2441 | 0.4063 | 0.049* | |
| C24 | 0.22251 (17) | 0.16599 (12) | 0.3928 (2) | 0.0402 (5) | |
| H24 | 0.1746 | 0.1662 | 0.2886 | 0.048* | |
| C25 | 0.83934 (16) | −0.02601 (13) | 0.6401 (2) | 0.0449 (5) | |
| C26 | 0.87387 (19) | −0.09629 (14) | 0.6575 (3) | 0.0530 (6) | |
| H26 | 0.8565 | −0.1317 | 0.6964 | 0.064* | |
| C27 | 0.9345 (2) | −0.11477 (15) | 0.6174 (3) | 0.0567 (6) | |
| H27 | 0.9577 | −0.1634 | 0.6274 | 0.068* | |
| C28 | 0.9614 (2) | −0.06375 (15) | 0.5636 (3) | 0.0553 (6) | |
| H28 | 1.0036 | −0.0769 | 0.5372 | 0.066* | |
| C29 | 0.92693 (19) | 0.00719 (15) | 0.5477 (3) | 0.0532 (6) | |
| H29 | 0.9457 | 0.0428 | 0.5108 | 0.064* | |
| C30 | 0.86529 (18) | 0.02621 (13) | 0.5855 (2) | 0.0480 (5) | |
| H30 | 0.8410 | 0.0747 | 0.5740 | 0.058* | |
| C31 | 0.15740 (18) | 0.27605 (13) | 0.6400 (2) | 0.0447 (5) | |
| C32 | 0.1402 (2) | 0.34624 (14) | 0.6574 (3) | 0.0536 (6) | |
| H32 | 0.1966 | 0.3815 | 0.6966 | 0.064* | |
| C33 | 0.0399 (2) | 0.36505 (15) | 0.6174 (3) | 0.0565 (6) | |
| H33 | 0.0268 | 0.4137 | 0.6274 | 0.068* | |
| C34 | −0.0414 (2) | 0.31361 (15) | 0.5632 (3) | 0.0553 (6) | |
| H34 | −0.1098 | 0.3267 | 0.5370 | 0.066* | |
| C35 | −0.0229 (2) | 0.24338 (15) | 0.5473 (3) | 0.0536 (6) | |
| H35 | −0.0788 | 0.2079 | 0.5097 | 0.064* | |
| C36 | 0.0773 (2) | 0.22397 (13) | 0.5859 (2) | 0.0480 (5) | |
| H36 | 0.0902 | 0.1754 | 0.5751 | 0.058* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0674 (11) | 0.0398 (8) | 0.0723 (11) | 0.0032 (8) | 0.0477 (10) | −0.0045 (8) |
| O2 | 0.0679 (12) | 0.0406 (9) | 0.0727 (11) | 0.0090 (8) | 0.0484 (10) | 0.0052 (8) |
| O3 | 0.0489 (9) | 0.0693 (11) | 0.0468 (8) | 0.0229 (8) | 0.0329 (8) | 0.0118 (7) |
| O4 | 0.0441 (9) | 0.0691 (11) | 0.0456 (8) | 0.0075 (8) | 0.0284 (7) | −0.0114 (7) |
| O5 | 0.1121 (19) | 0.0656 (13) | 0.1298 (19) | 0.0416 (13) | 0.0974 (18) | 0.0511 (13) |
| O6 | 0.0801 (15) | 0.0650 (13) | 0.1276 (19) | −0.0250 (11) | 0.0715 (14) | −0.0503 (13) |
| C1 | 0.0473 (12) | 0.0584 (14) | 0.0462 (12) | 0.0211 (10) | 0.0368 (11) | 0.0146 (10) |
| C2 | 0.0748 (18) | 0.0779 (19) | 0.0782 (18) | 0.0407 (15) | 0.0640 (17) | 0.0389 (15) |
| C3 | 0.081 (2) | 0.161 (4) | 0.080 (2) | 0.077 (3) | 0.064 (2) | 0.080 (3) |
| C4 | 0.0517 (18) | 0.217 (5) | 0.0412 (15) | 0.054 (3) | 0.0328 (14) | 0.038 (2) |
| C5 | 0.0345 (14) | 0.219 (5) | 0.0422 (15) | 0.004 (2) | 0.0201 (13) | −0.042 (2) |
| C6 | 0.0496 (17) | 0.167 (4) | 0.082 (2) | −0.031 (2) | 0.0411 (18) | −0.083 (3) |
| C7 | 0.0424 (13) | 0.080 (2) | 0.0774 (18) | −0.0113 (13) | 0.0403 (14) | −0.0387 (15) |
| C8 | 0.0302 (10) | 0.0581 (14) | 0.0470 (12) | 0.0028 (9) | 0.0244 (9) | −0.0147 (10) |
| C9 | 0.0329 (11) | 0.0798 (15) | 0.0347 (10) | 0.0158 (11) | 0.0230 (9) | −0.0004 (11) |
| C10 | 0.0336 (12) | 0.159 (3) | 0.0319 (11) | 0.0234 (16) | 0.0206 (10) | −0.0003 (16) |
| C11 | 0.0501 (12) | 0.0401 (11) | 0.0516 (12) | 0.0021 (9) | 0.0396 (11) | 0.0009 (9) |
| C12 | 0.0329 (10) | 0.0386 (10) | 0.0411 (10) | −0.0010 (8) | 0.0252 (9) | −0.0009 (8) |
| C13 | 0.0372 (11) | 0.0491 (11) | 0.0418 (10) | −0.0025 (9) | 0.0269 (10) | −0.0094 (9) |
| C14 | 0.0351 (11) | 0.0609 (13) | 0.0352 (10) | 0.0035 (9) | 0.0213 (9) | −0.0013 (9) |
| C15 | 0.0341 (10) | 0.0437 (11) | 0.0426 (10) | 0.0057 (9) | 0.0257 (9) | 0.0040 (9) |
| C16 | 0.0405 (11) | 0.0445 (11) | 0.0421 (10) | 0.0030 (9) | 0.0287 (9) | −0.0050 (8) |
| C17 | 0.0346 (10) | 0.0504 (12) | 0.0362 (10) | 0.0050 (9) | 0.0232 (9) | 0.0013 (8) |
| C18 | 0.0355 (10) | 0.0386 (11) | 0.0523 (12) | 0.0013 (9) | 0.0283 (10) | −0.0003 (9) |
| C19 | 0.0377 (10) | 0.0384 (10) | 0.0410 (10) | 0.0008 (8) | 0.0289 (9) | 0.0017 (8) |
| C20 | 0.0385 (11) | 0.0489 (11) | 0.0418 (10) | 0.0101 (9) | 0.0278 (9) | 0.0092 (9) |
| C21 | 0.0400 (11) | 0.0627 (13) | 0.0346 (10) | 0.0052 (10) | 0.0247 (9) | 0.0016 (9) |
| C22 | 0.0392 (11) | 0.0460 (11) | 0.0404 (10) | 0.0016 (9) | 0.0289 (9) | −0.0025 (9) |
| C23 | 0.0379 (10) | 0.0450 (11) | 0.0405 (10) | 0.0104 (9) | 0.0259 (9) | 0.0054 (8) |
| C24 | 0.0350 (11) | 0.0509 (12) | 0.0360 (10) | 0.0034 (9) | 0.0241 (9) | −0.0008 (8) |
| C25 | 0.0310 (10) | 0.0608 (14) | 0.0350 (10) | 0.0065 (9) | 0.0184 (9) | −0.0036 (9) |
| C26 | 0.0390 (12) | 0.0598 (14) | 0.0540 (13) | 0.0056 (10) | 0.0281 (11) | −0.0001 (11) |
| C27 | 0.0425 (12) | 0.0593 (15) | 0.0600 (14) | 0.0066 (11) | 0.0303 (11) | −0.0098 (12) |
| C28 | 0.0370 (11) | 0.0753 (16) | 0.0529 (12) | −0.0033 (11) | 0.0294 (11) | −0.0173 (12) |
| C29 | 0.0395 (12) | 0.0713 (16) | 0.0431 (12) | −0.0088 (11) | 0.0248 (10) | −0.0100 (11) |
| C30 | 0.0368 (11) | 0.0516 (13) | 0.0411 (11) | 0.0009 (9) | 0.0196 (10) | −0.0073 (9) |
| C31 | 0.0428 (11) | 0.0600 (14) | 0.0358 (10) | 0.0109 (10) | 0.0280 (10) | 0.0043 (9) |
| C32 | 0.0589 (15) | 0.0611 (14) | 0.0552 (13) | 0.0054 (11) | 0.0439 (13) | 0.0000 (11) |
| C33 | 0.0672 (16) | 0.0597 (15) | 0.0581 (14) | 0.0193 (13) | 0.0481 (13) | 0.0102 (11) |
| C34 | 0.0507 (13) | 0.0771 (17) | 0.0521 (12) | 0.0203 (13) | 0.0400 (11) | 0.0176 (12) |
| C35 | 0.0514 (14) | 0.0697 (16) | 0.0444 (12) | 0.0038 (12) | 0.0336 (11) | 0.0090 (11) |
| C36 | 0.0547 (13) | 0.0530 (13) | 0.0417 (11) | 0.0106 (10) | 0.0342 (11) | 0.0075 (9) |
Geometric parameters (Å, º)
| O1—C11 | 1.228 (3) | C16—H16 | 0.9500 |
| O2—C18 | 1.230 (3) | C17—H17 | 0.9500 |
| O3—C15 | 1.380 (2) | C18—C19 | 1.481 (3) |
| O3—C25 | 1.392 (3) | C19—C20 | 1.393 (3) |
| O4—C22 | 1.377 (2) | C19—C24 | 1.397 (3) |
| O4—C31 | 1.395 (3) | C20—C21 | 1.372 (3) |
| O5—C2 | 1.355 (4) | C20—H20 | 0.9500 |
| O5—H5A | 0.8400 | C21—C22 | 1.383 (3) |
| O6—C7 | 1.348 (4) | C21—H21 | 0.9500 |
| O6—H6A | 0.8400 | C22—C23 | 1.391 (3) |
| C1—C2 | 1.401 (3) | C23—C24 | 1.387 (3) |
| C1—C9 | 1.435 (4) | C23—H23 | 0.9500 |
| C1—C11 | 1.485 (3) | C24—H24 | 0.9500 |
| C2—C3 | 1.399 (5) | C25—C26 | 1.373 (3) |
| C3—C4 | 1.329 (6) | C25—C30 | 1.382 (3) |
| C3—H3 | 0.9500 | C26—C27 | 1.387 (4) |
| C4—C10 | 1.423 (6) | C26—H26 | 0.9500 |
| C4—H4 | 0.9500 | C27—C28 | 1.370 (4) |
| C5—C6 | 1.364 (6) | C27—H27 | 0.9500 |
| C5—C10 | 1.428 (6) | C28—C29 | 1.387 (4) |
| C5—H5 | 0.9500 | C28—H28 | 0.9500 |
| C6—C7 | 1.381 (5) | C29—C30 | 1.381 (3) |
| C6—H6 | 0.9500 | C29—H29 | 0.9500 |
| C7—C8 | 1.404 (3) | C30—H30 | 0.9500 |
| C8—C9 | 1.435 (4) | C31—C32 | 1.372 (3) |
| C8—C18 | 1.484 (3) | C31—C36 | 1.376 (3) |
| C9—C10 | 1.422 (3) | C32—C33 | 1.383 (4) |
| C11—C12 | 1.484 (3) | C32—H32 | 0.9500 |
| C12—C13 | 1.392 (3) | C33—C34 | 1.379 (4) |
| C12—C17 | 1.396 (3) | C33—H33 | 0.9500 |
| C13—C14 | 1.375 (3) | C34—C35 | 1.374 (4) |
| C13—H13 | 0.9500 | C34—H34 | 0.9500 |
| C14—C15 | 1.385 (3) | C35—C36 | 1.390 (3) |
| C14—H14 | 0.9500 | C35—H35 | 0.9500 |
| C15—C16 | 1.383 (3) | C36—H36 | 0.9500 |
| C16—C17 | 1.387 (3) | ||
| C15—O3—C25 | 119.93 (16) | O2—C18—C8 | 120.5 (2) |
| C22—O4—C31 | 119.97 (16) | C19—C18—C8 | 121.35 (18) |
| C2—O5—H5A | 109.5 | C20—C19—C24 | 118.61 (18) |
| C7—O6—H6A | 109.5 | C20—C19—C18 | 118.29 (18) |
| C2—C1—C9 | 119.7 (2) | C24—C19—C18 | 122.58 (18) |
| C2—C1—C11 | 115.2 (2) | C21—C20—C19 | 121.27 (19) |
| C9—C1—C11 | 124.8 (2) | C21—C20—H20 | 119.4 |
| O5—C2—C3 | 117.3 (3) | C19—C20—H20 | 119.4 |
| O5—C2—C1 | 122.7 (3) | C20—C21—C22 | 119.41 (19) |
| C3—C2—C1 | 120.0 (3) | C20—C21—H21 | 120.3 |
| C4—C3—C2 | 120.9 (3) | C22—C21—H21 | 120.3 |
| C4—C3—H3 | 119.6 | O4—C22—C21 | 116.27 (18) |
| C2—C3—H3 | 119.6 | O4—C22—C23 | 122.82 (18) |
| C3—C4—C10 | 121.9 (3) | C21—C22—C23 | 120.80 (18) |
| C3—C4—H4 | 119.1 | C24—C23—C22 | 119.10 (19) |
| C10—C4—H4 | 119.1 | C24—C23—H23 | 120.4 |
| C6—C5—C10 | 121.7 (3) | C22—C23—H23 | 120.4 |
| C6—C5—H5 | 119.2 | C23—C24—C19 | 120.58 (18) |
| C10—C5—H5 | 119.2 | C23—C24—H24 | 119.7 |
| C5—C6—C7 | 120.0 (3) | C19—C24—H24 | 119.7 |
| C5—C6—H6 | 120.0 | C26—C25—C30 | 121.1 (2) |
| C7—C6—H6 | 120.0 | C26—C25—O3 | 116.2 (2) |
| O6—C7—C6 | 116.1 (3) | C30—C25—O3 | 122.5 (2) |
| O6—C7—C8 | 122.9 (3) | C25—C26—C27 | 119.0 (2) |
| C6—C7—C8 | 121.0 (4) | C25—C26—H26 | 120.5 |
| C7—C8—C9 | 119.8 (2) | C27—C26—H26 | 120.5 |
| C7—C8—C18 | 115.2 (2) | C28—C27—C26 | 120.8 (2) |
| C9—C8—C18 | 124.7 (2) | C28—C27—H27 | 119.6 |
| C10—C9—C8 | 117.7 (3) | C26—C27—H27 | 119.6 |
| C10—C9—C1 | 117.6 (3) | C27—C28—C29 | 119.7 (2) |
| C8—C9—C1 | 124.71 (18) | C27—C28—H28 | 120.2 |
| C9—C10—C4 | 119.0 (3) | C29—C28—H28 | 120.2 |
| C9—C10—C5 | 118.9 (3) | C30—C29—C28 | 120.2 (2) |
| C4—C10—C5 | 122.1 (3) | C30—C29—H29 | 119.9 |
| O1—C11—C12 | 117.7 (2) | C28—C29—H29 | 119.9 |
| O1—C11—C1 | 120.6 (2) | C29—C30—C25 | 119.3 (2) |
| C12—C11—C1 | 121.13 (18) | C29—C30—H30 | 120.4 |
| C13—C12—C17 | 118.70 (18) | C25—C30—H30 | 120.4 |
| C13—C12—C11 | 118.15 (18) | C32—C31—C36 | 121.1 (2) |
| C17—C12—C11 | 122.61 (18) | C32—C31—O4 | 116.1 (2) |
| C14—C13—C12 | 121.21 (19) | C36—C31—O4 | 122.6 (2) |
| C14—C13—H13 | 119.4 | C31—C32—C33 | 119.3 (2) |
| C12—C13—H13 | 119.4 | C31—C32—H32 | 120.3 |
| C13—C14—C15 | 119.11 (19) | C33—C32—H32 | 120.3 |
| C13—C14—H14 | 120.4 | C34—C33—C32 | 120.4 (2) |
| C15—C14—H14 | 120.4 | C34—C33—H33 | 119.8 |
| O3—C15—C16 | 123.03 (18) | C32—C33—H33 | 119.8 |
| O3—C15—C14 | 115.80 (18) | C35—C34—C33 | 119.7 (2) |
| C16—C15—C14 | 121.08 (18) | C35—C34—H34 | 120.1 |
| C15—C16—C17 | 119.23 (19) | C33—C34—H34 | 120.1 |
| C15—C16—H16 | 120.4 | C34—C35—C36 | 120.3 (2) |
| C17—C16—H16 | 120.4 | C34—C35—H35 | 119.8 |
| C16—C17—C12 | 120.47 (18) | C36—C35—H35 | 119.8 |
| C16—C17—H17 | 119.8 | C31—C36—C35 | 119.1 (2) |
| C12—C17—H17 | 119.8 | C31—C36—H36 | 120.5 |
| O2—C18—C19 | 117.5 (2) | C35—C36—H36 | 120.5 |
| C9—C1—C2—O5 | 176.4 (2) | O3—C15—C16—C17 | 178.0 (2) |
| C11—C1—C2—O5 | −9.4 (3) | C14—C15—C16—C17 | 1.6 (3) |
| C9—C1—C2—C3 | −7.1 (3) | C15—C16—C17—C12 | 2.6 (3) |
| C11—C1—C2—C3 | 167.1 (2) | C13—C12—C17—C16 | −3.8 (3) |
| O5—C2—C3—C4 | 175.7 (3) | C11—C12—C17—C16 | 167.6 (2) |
| C1—C2—C3—C4 | −1.0 (4) | C7—C8—C18—O2 | 34.8 (3) |
| C2—C3—C4—C10 | 4.1 (5) | C9—C8—C18—O2 | −151.6 (2) |
| C10—C5—C6—C7 | 4.3 (5) | C7—C8—C18—C19 | −135.9 (2) |
| C5—C6—C7—O6 | 175.6 (3) | C9—C8—C18—C19 | 37.8 (3) |
| C5—C6—C7—C8 | −1.4 (4) | O2—C18—C19—C20 | 26.1 (3) |
| O6—C7—C8—C9 | 176.5 (2) | C8—C18—C19—C20 | −163.0 (2) |
| C6—C7—C8—C9 | −6.7 (3) | O2—C18—C19—C24 | −145.5 (2) |
| O6—C7—C8—C18 | −9.5 (3) | C8—C18—C19—C24 | 25.5 (3) |
| C6—C7—C8—C18 | 167.4 (2) | C24—C19—C20—C21 | 0.5 (3) |
| C7—C8—C9—C10 | 11.6 (3) | C18—C19—C20—C21 | −171.4 (2) |
| C18—C8—C9—C10 | −161.8 (2) | C19—C20—C21—C22 | 3.6 (3) |
| C7—C8—C9—C1 | −168.3 (2) | C31—O4—C22—C21 | −146.0 (2) |
| C18—C8—C9—C1 | 18.3 (3) | C31—O4—C22—C23 | 37.7 (3) |
| C2—C1—C9—C10 | 11.7 (3) | C20—C21—C22—O4 | 178.6 (2) |
| C11—C1—C9—C10 | −161.8 (2) | C20—C21—C22—C23 | −5.0 (3) |
| C2—C1—C9—C8 | −168.4 (2) | O4—C22—C23—C24 | 178.3 (2) |
| C11—C1—C9—C8 | 18.0 (3) | C21—C22—C23—C24 | 2.1 (3) |
| C8—C9—C10—C4 | 171.4 (2) | C22—C23—C24—C19 | 2.1 (3) |
| C1—C9—C10—C4 | −8.7 (3) | C20—C19—C24—C23 | −3.5 (3) |
| C8—C9—C10—C5 | −8.7 (3) | C18—C19—C24—C23 | 168.1 (2) |
| C1—C9—C10—C5 | 171.2 (2) | C15—O3—C25—C26 | −140.9 (2) |
| C3—C4—C10—C9 | 0.9 (4) | C15—O3—C25—C30 | 44.7 (3) |
| C3—C4—C10—C5 | −178.9 (3) | C30—C25—C26—C27 | −0.8 (3) |
| C6—C5—C10—C9 | 0.9 (4) | O3—C25—C26—C27 | −175.3 (2) |
| C6—C5—C10—C4 | −179.2 (3) | C25—C26—C27—C28 | 1.1 (4) |
| C2—C1—C11—O1 | 34.9 (3) | C26—C27—C28—C29 | −0.5 (4) |
| C9—C1—C11—O1 | −151.3 (2) | C27—C28—C29—C30 | −0.3 (3) |
| C2—C1—C11—C12 | −136.1 (2) | C28—C29—C30—C25 | 0.5 (3) |
| C9—C1—C11—C12 | 37.7 (3) | C26—C25—C30—C29 | 0.0 (3) |
| O1—C11—C12—C13 | 26.0 (3) | O3—C25—C30—C29 | 174.1 (2) |
| C1—C11—C12—C13 | −162.8 (2) | C22—O4—C31—C32 | −141.1 (2) |
| O1—C11—C12—C17 | −145.5 (2) | C22—O4—C31—C36 | 44.7 (3) |
| C1—C11—C12—C17 | 25.8 (3) | C36—C31—C32—C33 | −1.2 (3) |
| C17—C12—C13—C14 | 0.8 (3) | O4—C31—C32—C33 | −175.38 (19) |
| C11—C12—C13—C14 | −171.0 (2) | C31—C32—C33—C34 | 1.2 (3) |
| C12—C13—C14—C15 | 3.3 (3) | C32—C33—C34—C35 | −0.7 (4) |
| C25—O3—C15—C16 | 37.3 (3) | C33—C34—C35—C36 | 0.2 (3) |
| C25—O3—C15—C14 | −146.1 (2) | C32—C31—C36—C35 | 0.6 (3) |
| C13—C14—C15—O3 | 178.8 (2) | O4—C31—C36—C35 | 174.45 (19) |
| C13—C14—C15—C16 | −4.5 (3) | C34—C35—C36—C31 | −0.1 (3) |
Hydrogen-bond geometry (Å, º)
Cg1 and Cg2 are the centroids of the C25—C30 and C31—C36 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O5—H5A···O1 | 0.84 | 1.83 | 2.560 (3) | 145 |
| O6—H6A···O2 | 0.84 | 1.88 | 2.563 (3) | 138 |
| C26—H26···O4i | 0.95 | 2.48 | 3.377 (4) | 157 |
| C27—H27···O1i | 0.95 | 2.51 | 3.269 (4) | 137 |
| C32—H32···O3ii | 0.95 | 2.49 | 3.382 (4) | 156 |
| C33—H33···O2ii | 0.95 | 2.51 | 3.270 (4) | 137 |
| C14—H14···Cg1iii | 0.95 | 2.80 | 3.740 (2) | 171 |
| C21—H21···Cg2iv | 0.95 | 2.80 | 3.740 (2) | 171 |
Symmetry codes: (i) x+1/2, y−1/2, z; (ii) x−1/2, y+1/2, z; (iii) x, −y, z+1/2; (iv) x+1/2, −y+1/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: RN2112).
References
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., De Caro, L., Giacovazzo, C., Polidori, G., Siliqi, D. & Spagna, R. (2007). J. Appl. Cryst. 40, 609–613.
- Burnett, M. N. & Johnson, C. K. (1996). ORTEPIII Report ORNL-6895. Oak Ridge National Laboratory, Tennessee, USA.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Higashi, T. (1999). NUMABS Rigaku Corporation, Tokyo, Japan.
- Hijikata, D., Takada, T., Nagasawa, A., Okamoto, A. & Yonezawa, N. (2010). Acta Cryst. E66, o2902–o2903. [DOI] [PMC free article] [PubMed]
- Okamoto, A., Hijikata, D., Sakai, N. & Yonezawa, N. (2013). Polym. J. In the press. doi:10.1038/pj.2012.135.
- Okamoto, A., Mitsui, R., Oike, H. & Yonezawa, N. (2011). Chem. Lett. 40, 1283–1284.
- Okamoto, A. & Yonezawa, N. (2009). Chem. Lett. 38, 914–915.
- Rigaku (1998). PROCESS-AUTO Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Watanabe, S., Nagasawa, A., Okamoto, A., Noguchi, K. & Yonezawa, N. (2010). Acta Cryst. E66, o329. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812052038/rn2112sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812052038/rn2112Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812052038/rn2112Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



