Abstract
The chiral title compound, C19H32O2, contains a [4.3.0]-bicyclic moiety in which the shared C—C bond presents a trans configuration and a side chain in which the C=C double bond shows an E conformation. The conformations of five- and six-membered rings are envelope (with the bridgehead atom bearing the methyl substituent as the flap) and chair, respectively, with a dihedral angle of 4.08 (17)° between the idealized planes of the rings. In the crystal, the molecules are self-assembled via classical O—H⋯O hydrogen bonds, forming chains along [112]; these chains are linked by weak non-classical C—H⋯O hydrogen bonds, giving a two-dimensional supramolecular structure parallel to (010). The absolute configuration was established according to the configuration of the starting material.
Related literature
The title compound is a precursor of the hormonally active form of vitamin D3. For general background to vitamin D3, see: Heaney (2008 ▶); Henry (2011 ▶). For related structures, see: Maehr & Uskokovic (2004 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶).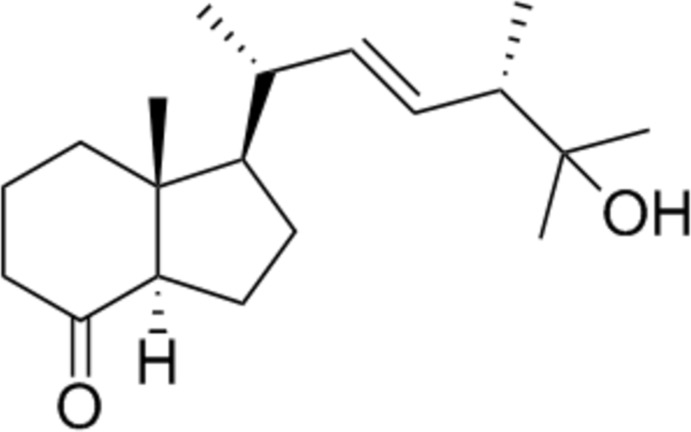
Experimental
Crystal data
C19H32O2
M r = 292.45
Monoclinic,

a = 20.057 (4) Å
b = 7.3816 (15) Å
c = 13.700 (3) Å
β = 112.324 (4)°
V = 1876.3 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.07 mm−1
T = 293 K
0.45 × 0.36 × 0.18 mm
Data collection
Bruker SMART 1000 CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.602, T max = 0.745
4958 measured reflections
3254 independent reflections
2389 reflections with I > 2σ(I)
R int = 0.018
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.152
S = 1.02
3254 reflections
196 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.14 e Å−3
Δρmin = −0.11 e Å−3
Data collection: SMART (Bruker, 1998 ▶); cell refinement: SAINT (Bruker, 1998 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶) and Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: SHELXTL (Sheldrick, 2008 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812051343/pv2614sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812051343/pv2614Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812051343/pv2614Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O7′—H7′⋯O4i | 0.82 | 2.08 | 2.876 (3) | 164 |
| C3A—H3A1⋯O7′ii | 0.98 | 2.56 | 3.523 (3) | 166 |
Symmetry codes: (i) 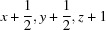 ; (ii)
; (ii) 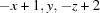 .
.
Acknowledgments
This work was supported financially by the Spanish Ministry of Foreign Affairs and Cooperation (PCIA/030052/10) and the Xunta de Galicia (INCITE845B, INCITE08PXIB314255PR).
supplementary crystallographic information
Comment
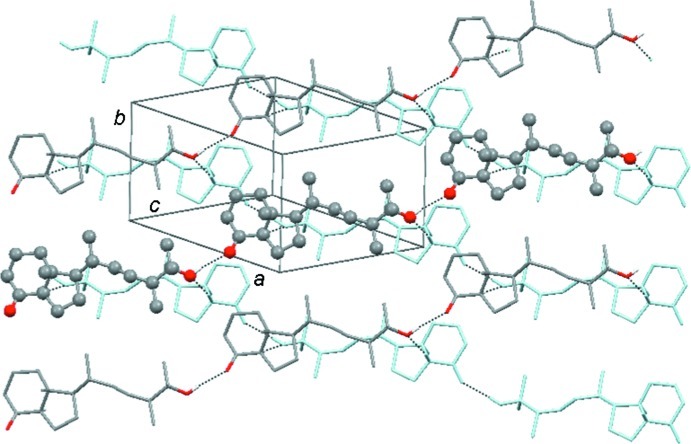
The title compound is a precursor of 1α,25-dihydroxyvitamin D3 (calcitriol) analogue which is the hormonally active form of vitamin D3. Besides regulating calcium homeostasis, this form is also involved in other cellular processes such as cell differentiation; immune system regulation and gene transcription (Henry, 2011). Nevertheless, the clinical utility of this hormone for treatment of cancers and skin disorders is limited by its hypercalcemic effects (Heaney, 2008), for this purpose the design and synthesis of more selective biological-effect analogues is of paramount importance. In the title compound (Figure 1), the C3A—C7A shared bond of the bicyclic moiety presents a trans configuration. Besides, the 5-membered ring adopts an envelope conformation with puckering parameters Q = 0.462 (3) Å and φ = 136.5 (4)° and with the bridgehead C7A atom bearing the methyl substituent as the flap (Cremer & Pople, 1975) and the 6-membered ring presents a chair conformation with puckering parameters Q = 0.556 (3) Å, θ = 169.5 (3)° and φ = 133.4 (18)° (Cremer & Pople, 1975). The value for the dihedral angle between the idealized planes of the rings is 4.08 (17)°. All bond lengths and bond angles are normal comparable to those observed in similar crystal structures (Maehr & Uskokovic, 2004). In the crystal structure, the molecules are self-assembled via classical O—H···O hydrogen bonds to form a chain along [112], the resulting chains are connected by weak non-classical C—H···O hydrogen bonds to create a two-dimensional supramolecular structure (Table 1, Figure 2).
Experimental
Over a stirring solution of inhoffen-lythgoe diol (2.1 g; 7.2 mmol) in CH2Cl2 (10 ml), PDC S-methyl-3-hydroxy-2-methylpropionate (5.4 g; 14.4 mmol) was added. The mixture was stirred at room temperature for 16 h then it was quenched with ethylic ether (20 ml) and stirred one more hour. The solid precipitated was filtered over celite, the organic layer was concentrated and the residue was purified by flash column chromatography on silica gel (10% ethyl acetate/hexane) to afford the title compound (1.8 g; 80%). The crystals were obtained by slow evaporation in a closed camera of a solution of the compound in a mixture of ethyl acetate/hexane (7:3).
Refinement
All H-atoms were positioned and refined using a riding model with O–H = 0.82 Å and C–H = 0.98, 0.97, 0.96 and 0.93 Å for methine, methylelne, methyl and vinyl H-atoms, respectively. The H-atoms were allowed Uiso = 1.5Ueq(O/C-methyl) or 1.2Ueq(the rest of the C atoms). Due to insufficient anamolous dispersion effects, an absolute structure was not established in this analysis and 1457 Friedel pairs were not merged. However, the absolute configuration of the title compound was established according to the configuration of starting material.
Figures
Fig. 1.
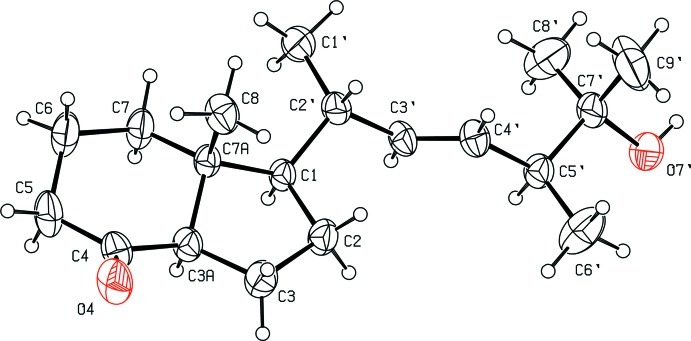
The molecular structure of the title compound. Non-H atoms are present as displacement ellipsoids at the 30% probability level.
Fig. 2.
View of two-dimensional supramolecular organization in the crystal structure of the title compound. H atoms not involved in hydrogen bonds (dashed lines) have been omitted for clarify.
Crystal data
| C19H32O2 | F(000) = 648 |
| Mr = 292.45 | Dx = 1.035 Mg m−3 |
| Monoclinic, C2 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: C 2y | Cell parameters from 1729 reflections |
| a = 20.057 (4) Å | θ = 2.2–23.0° |
| b = 7.3816 (15) Å | µ = 0.07 mm−1 |
| c = 13.700 (3) Å | T = 293 K |
| β = 112.324 (4)° | Prism, colourless |
| V = 1876.3 (6) Å3 | 0.45 × 0.36 × 0.18 mm |
| Z = 4 |
Data collection
| Bruker SMART 1000 CCD diffractometer | 3254 independent reflections |
| Radiation source: fine-focus sealed tube | 2389 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.018 |
| φ and ω scans | θmax = 25.0°, θmin = 1.6° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −23→14 |
| Tmin = 0.602, Tmax = 0.745 | k = −8→8 |
| 4958 measured reflections | l = −13→16 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.048 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.152 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0934P)2 + 0.0697P] where P = (Fo2 + 2Fc2)/3 |
| 3254 reflections | (Δ/σ)max < 0.001 |
| 196 parameters | Δρmax = 0.14 e Å−3 |
| 1 restraint | Δρmin = −0.11 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.31315 (12) | −0.0108 (3) | 0.83385 (16) | 0.0467 (6) | |
| H1 | 0.3524 | 0.0312 | 0.8135 | 0.056* | |
| C2 | 0.32284 (17) | −0.2166 (4) | 0.8512 (2) | 0.0663 (8) | |
| H2A | 0.3046 | −0.2556 | 0.9039 | 0.080* | |
| H2B | 0.3735 | −0.2484 | 0.8752 | 0.080* | |
| C3 | 0.28017 (17) | −0.3084 (4) | 0.7442 (2) | 0.0737 (8) | |
| H3A | 0.3107 | −0.3896 | 0.7239 | 0.088* | |
| H3B | 0.2394 | −0.3759 | 0.7469 | 0.088* | |
| C3A | 0.25554 (14) | −0.1495 (3) | 0.66904 (18) | 0.0554 (7) | |
| H3A1 | 0.2968 | −0.1140 | 0.6516 | 0.066* | |
| C4 | 0.19282 (15) | −0.1736 (4) | 0.5661 (2) | 0.0619 (7) | |
| O4 | 0.16051 (12) | −0.3154 (3) | 0.53886 (15) | 0.0849 (7) | |
| C5 | 0.17509 (18) | −0.0054 (5) | 0.4997 (2) | 0.0807 (9) | |
| H5A | 0.1294 | −0.0224 | 0.4411 | 0.097* | |
| H5B | 0.2118 | 0.0139 | 0.4708 | 0.097* | |
| C6 | 0.17031 (19) | 0.1620 (5) | 0.5617 (2) | 0.0827 (10) | |
| H6A | 0.1253 | 0.1584 | 0.5727 | 0.099* | |
| H6B | 0.1695 | 0.2690 | 0.5202 | 0.099* | |
| C7 | 0.23275 (15) | 0.1787 (4) | 0.6691 (2) | 0.0673 (8) | |
| H7A | 0.2769 | 0.2040 | 0.6582 | 0.081* | |
| H7B | 0.2236 | 0.2798 | 0.7075 | 0.081* | |
| C7A | 0.24264 (12) | 0.0069 (3) | 0.73468 (16) | 0.0472 (6) | |
| C8 | 0.17703 (13) | −0.0288 (5) | 0.7620 (2) | 0.0706 (8) | |
| H8A | 0.1349 | −0.0386 | 0.6982 | 0.106* | |
| H8B | 0.1710 | 0.0694 | 0.8038 | 0.106* | |
| H8C | 0.1838 | −0.1397 | 0.8012 | 0.106* | |
| C1' | 0.31086 (19) | 0.2943 (4) | 0.9234 (2) | 0.0799 (10) | |
| H1'1 | 0.3417 | 0.3421 | 0.8905 | 0.120* | |
| H1'2 | 0.3227 | 0.3496 | 0.9914 | 0.120* | |
| H1'3 | 0.2615 | 0.3200 | 0.8801 | 0.120* | |
| C2' | 0.32153 (14) | 0.0889 (4) | 0.93674 (19) | 0.0552 (7) | |
| H2' | 0.2857 | 0.0405 | 0.9624 | 0.066* | |
| C3' | 0.39471 (14) | 0.0496 (4) | 1.01717 (17) | 0.0554 (7) | |
| H3' | 0.4331 | 0.0888 | 1.0004 | 0.067* | |
| C4' | 0.41212 (13) | −0.0327 (4) | 1.10809 (18) | 0.0578 (7) | |
| H4' | 0.3744 | −0.0706 | 1.1268 | 0.069* | |
| C5' | 0.48679 (13) | −0.0715 (4) | 1.18480 (19) | 0.0564 (7) | |
| H5' | 0.5196 | −0.0367 | 1.1503 | 0.068* | |
| C6' | 0.4971 (2) | −0.2739 (5) | 1.2070 (4) | 0.1075 (14) | |
| H6'1 | 0.4895 | −0.3375 | 1.1425 | 0.161* | |
| H6'2 | 0.4631 | −0.3156 | 1.2359 | 0.161* | |
| H6'3 | 0.5452 | −0.2962 | 1.2566 | 0.161* | |
| C7' | 0.50827 (14) | 0.0412 (5) | 1.2868 (2) | 0.0674 (8) | |
| O7' | 0.58126 (10) | −0.0077 (3) | 1.34707 (14) | 0.0824 (7) | |
| H7' | 0.5959 | 0.0494 | 1.4026 | 0.124* | |
| C8' | 0.5057 (2) | 0.2410 (5) | 1.2604 (3) | 0.1151 (17) | |
| H8'1 | 0.5409 | 0.2673 | 1.2307 | 0.173* | |
| H8'2 | 0.5159 | 0.3110 | 1.3235 | 0.173* | |
| H8'3 | 0.4585 | 0.2715 | 1.2104 | 0.173* | |
| C9' | 0.4620 (2) | 0.0049 (11) | 1.3478 (3) | 0.145 (2) | |
| H9'1 | 0.4765 | 0.0822 | 1.4086 | 0.218* | |
| H9'2 | 0.4672 | −0.1194 | 1.3699 | 0.218* | |
| H9'3 | 0.4126 | 0.0287 | 1.3040 | 0.218* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0422 (12) | 0.0519 (14) | 0.0368 (10) | −0.0019 (12) | 0.0048 (9) | −0.0014 (11) |
| C2 | 0.0731 (18) | 0.0573 (17) | 0.0500 (13) | 0.0055 (14) | 0.0027 (13) | −0.0010 (13) |
| C3 | 0.088 (2) | 0.0542 (16) | 0.0582 (15) | 0.0030 (15) | 0.0040 (14) | −0.0063 (14) |
| C3A | 0.0536 (14) | 0.0559 (16) | 0.0444 (13) | −0.0055 (12) | 0.0048 (11) | −0.0064 (11) |
| C4 | 0.0613 (16) | 0.0701 (19) | 0.0438 (13) | −0.0111 (15) | 0.0079 (12) | −0.0117 (13) |
| O4 | 0.0868 (15) | 0.0816 (15) | 0.0583 (11) | −0.0232 (13) | −0.0039 (10) | −0.0162 (11) |
| C5 | 0.086 (2) | 0.090 (2) | 0.0425 (13) | −0.0127 (18) | −0.0011 (13) | 0.0046 (16) |
| C6 | 0.093 (2) | 0.077 (2) | 0.0523 (16) | 0.0033 (18) | −0.0011 (16) | 0.0166 (15) |
| C7 | 0.0736 (19) | 0.0615 (18) | 0.0467 (14) | −0.0021 (16) | 0.0002 (13) | 0.0083 (13) |
| C7A | 0.0444 (13) | 0.0503 (14) | 0.0385 (10) | −0.0030 (12) | 0.0061 (9) | 0.0007 (11) |
| C8 | 0.0474 (14) | 0.101 (2) | 0.0565 (14) | −0.0084 (15) | 0.0117 (11) | −0.0040 (16) |
| C1' | 0.095 (2) | 0.072 (2) | 0.0541 (15) | 0.0132 (17) | 0.0068 (15) | −0.0143 (14) |
| C2' | 0.0507 (14) | 0.0651 (18) | 0.0419 (13) | −0.0001 (12) | 0.0086 (11) | −0.0079 (11) |
| C3' | 0.0508 (15) | 0.0722 (19) | 0.0359 (12) | −0.0071 (12) | 0.0081 (11) | −0.0076 (11) |
| C4' | 0.0484 (14) | 0.0715 (19) | 0.0470 (13) | −0.0112 (13) | 0.0108 (11) | −0.0020 (13) |
| C5' | 0.0483 (15) | 0.0643 (18) | 0.0478 (13) | −0.0007 (12) | 0.0081 (11) | −0.0007 (12) |
| C6' | 0.086 (3) | 0.066 (2) | 0.136 (3) | 0.0032 (18) | 0.003 (2) | 0.007 (2) |
| C7' | 0.0483 (15) | 0.096 (3) | 0.0433 (12) | 0.0113 (14) | 0.0012 (11) | −0.0076 (13) |
| O7' | 0.0566 (12) | 0.1072 (18) | 0.0569 (10) | 0.0184 (12) | −0.0083 (9) | −0.0136 (12) |
| C8' | 0.097 (3) | 0.082 (3) | 0.112 (3) | 0.018 (2) | −0.022 (2) | −0.034 (2) |
| C9' | 0.094 (3) | 0.287 (7) | 0.0559 (17) | 0.032 (4) | 0.0299 (19) | 0.005 (3) |
Geometric parameters (Å, º)
| C1—C2 | 1.538 (4) | C8—H8C | 0.9600 |
| C1—C2' | 1.541 (3) | C1'—C2' | 1.533 (4) |
| C1—C7A | 1.550 (3) | C1'—H1'1 | 0.9600 |
| C1—H1 | 0.9800 | C1'—H1'2 | 0.9600 |
| C2—C3 | 1.545 (4) | C1'—H1'3 | 0.9600 |
| C2—H2A | 0.9700 | C2'—C3' | 1.491 (4) |
| C2—H2B | 0.9700 | C2'—H2' | 0.9800 |
| C3—C3A | 1.515 (4) | C3'—C4' | 1.308 (3) |
| C3—H3A | 0.9700 | C3'—H3' | 0.9300 |
| C3—H3B | 0.9700 | C4'—C5' | 1.495 (3) |
| C3A—C4 | 1.501 (3) | C4'—H4' | 0.9300 |
| C3A—C7A | 1.545 (3) | C5'—C6' | 1.523 (5) |
| C3A—H3A1 | 0.9800 | C5'—C7' | 1.540 (4) |
| C4—O4 | 1.213 (3) | C5'—H5' | 0.9800 |
| C4—C5 | 1.500 (4) | C6'—H6'1 | 0.9600 |
| C5—C6 | 1.524 (5) | C6'—H6'2 | 0.9600 |
| C5—H5A | 0.9700 | C6'—H6'3 | 0.9600 |
| C5—H5B | 0.9700 | C7'—O7' | 1.427 (3) |
| C6—C7 | 1.532 (4) | C7'—C9' | 1.490 (5) |
| C6—H6A | 0.9700 | C7'—C8' | 1.515 (6) |
| C6—H6B | 0.9700 | O7'—H7' | 0.8200 |
| C7—C7A | 1.524 (4) | C8'—H8'1 | 0.9600 |
| C7—H7A | 0.9700 | C8'—H8'2 | 0.9600 |
| C7—H7B | 0.9700 | C8'—H8'3 | 0.9600 |
| C7A—C8 | 1.522 (3) | C9'—H9'1 | 0.9600 |
| C8—H8A | 0.9600 | C9'—H9'2 | 0.9600 |
| C8—H8B | 0.9600 | C9'—H9'3 | 0.9600 |
| C2—C1—C2' | 111.5 (2) | H8A—C8—H8B | 109.5 |
| C2—C1—C7A | 103.77 (19) | C7A—C8—H8C | 109.5 |
| C2'—C1—C7A | 120.50 (19) | H8A—C8—H8C | 109.5 |
| C2—C1—H1 | 106.8 | H8B—C8—H8C | 109.5 |
| C2'—C1—H1 | 106.8 | C2'—C1'—H1'1 | 109.5 |
| C7A—C1—H1 | 106.8 | C2'—C1'—H1'2 | 109.5 |
| C1—C2—C3 | 107.2 (2) | H1'1—C1'—H1'2 | 109.5 |
| C1—C2—H2A | 110.3 | C2'—C1'—H1'3 | 109.5 |
| C3—C2—H2A | 110.3 | H1'1—C1'—H1'3 | 109.5 |
| C1—C2—H2B | 110.3 | H1'2—C1'—H1'3 | 109.5 |
| C3—C2—H2B | 110.3 | C3'—C2'—C1' | 109.5 (2) |
| H2A—C2—H2B | 108.5 | C3'—C2'—C1 | 108.5 (2) |
| C3A—C3—C2 | 103.0 (2) | C1'—C2'—C1 | 113.7 (2) |
| C3A—C3—H3A | 111.2 | C3'—C2'—H2' | 108.3 |
| C2—C3—H3A | 111.2 | C1'—C2'—H2' | 108.3 |
| C3A—C3—H3B | 111.2 | C1—C2'—H2' | 108.3 |
| C2—C3—H3B | 111.2 | C4'—C3'—C2' | 128.7 (3) |
| H3A—C3—H3B | 109.1 | C4'—C3'—H3' | 115.6 |
| C4—C3A—C3 | 119.2 (2) | C2'—C3'—H3' | 115.6 |
| C4—C3A—C7A | 111.7 (2) | C3'—C4'—C5' | 126.3 (2) |
| C3—C3A—C7A | 105.5 (2) | C3'—C4'—H4' | 116.8 |
| C4—C3A—H3A1 | 106.6 | C5'—C4'—H4' | 116.8 |
| C3—C3A—H3A1 | 106.6 | C4'—C5'—C6' | 110.6 (2) |
| C7A—C3A—H3A1 | 106.6 | C4'—C5'—C7' | 113.2 (2) |
| O4—C4—C5 | 123.5 (2) | C6'—C5'—C7' | 112.2 (3) |
| O4—C4—C3A | 123.4 (3) | C4'—C5'—H5' | 106.8 |
| C5—C4—C3A | 113.1 (2) | C6'—C5'—H5' | 106.8 |
| C4—C5—C6 | 112.5 (2) | C7'—C5'—H5' | 106.8 |
| C4—C5—H5A | 109.1 | C5'—C6'—H6'1 | 109.5 |
| C6—C5—H5A | 109.1 | C5'—C6'—H6'2 | 109.5 |
| C4—C5—H5B | 109.1 | H6'1—C6'—H6'2 | 109.5 |
| C6—C5—H5B | 109.1 | C5'—C6'—H6'3 | 109.5 |
| H5A—C5—H5B | 107.8 | H6'1—C6'—H6'3 | 109.5 |
| C5—C6—C7 | 113.5 (3) | H6'2—C6'—H6'3 | 109.5 |
| C5—C6—H6A | 108.9 | O7'—C7'—C9' | 110.5 (3) |
| C7—C6—H6A | 108.9 | O7'—C7'—C8' | 108.6 (3) |
| C5—C6—H6B | 108.9 | C9'—C7'—C8' | 109.6 (4) |
| C7—C6—H6B | 108.9 | O7'—C7'—C5' | 105.1 (2) |
| H6A—C6—H6B | 107.7 | C9'—C7'—C5' | 113.1 (3) |
| C7A—C7—C6 | 112.0 (2) | C8'—C7'—C5' | 109.7 (3) |
| C7A—C7—H7A | 109.2 | C7'—O7'—H7' | 109.5 |
| C6—C7—H7A | 109.2 | C7'—C8'—H8'1 | 109.5 |
| C7A—C7—H7B | 109.2 | C7'—C8'—H8'2 | 109.5 |
| C6—C7—H7B | 109.2 | H8'1—C8'—H8'2 | 109.5 |
| H7A—C7—H7B | 107.9 | C7'—C8'—H8'3 | 109.5 |
| C8—C7A—C7 | 111.0 (2) | H8'1—C8'—H8'3 | 109.5 |
| C8—C7A—C3A | 111.4 (2) | H8'2—C8'—H8'3 | 109.5 |
| C7—C7A—C3A | 106.98 (19) | C7'—C9'—H9'1 | 109.5 |
| C8—C7A—C1 | 110.87 (18) | C7'—C9'—H9'2 | 109.5 |
| C7—C7A—C1 | 117.3 (2) | H9'1—C9'—H9'2 | 109.5 |
| C3A—C7A—C1 | 98.54 (18) | C7'—C9'—H9'3 | 109.5 |
| C7A—C8—H8A | 109.5 | H9'1—C9'—H9'3 | 109.5 |
| C7A—C8—H8B | 109.5 | H9'2—C9'—H9'3 | 109.5 |
| C2'—C1—C2—C3 | 153.7 (2) | C2—C1—C7A—C8 | 75.8 (3) |
| C7A—C1—C2—C3 | 22.6 (3) | C2'—C1—C7A—C8 | −49.9 (3) |
| C1—C2—C3—C3A | 6.2 (3) | C2—C1—C7A—C7 | −155.3 (2) |
| C2—C3—C3A—C4 | −159.6 (3) | C2'—C1—C7A—C7 | 79.0 (3) |
| C2—C3—C3A—C7A | −33.2 (3) | C2—C1—C7A—C3A | −41.2 (2) |
| C3—C3A—C4—O4 | −0.6 (5) | C2'—C1—C7A—C3A | −166.8 (2) |
| C7A—C3A—C4—O4 | −124.0 (3) | C2—C1—C2'—C3' | 59.0 (3) |
| C3—C3A—C4—C5 | −179.5 (3) | C7A—C1—C2'—C3' | −179.0 (2) |
| C7A—C3A—C4—C5 | 57.1 (3) | C2—C1—C2'—C1' | −178.8 (3) |
| O4—C4—C5—C6 | 132.6 (3) | C7A—C1—C2'—C1' | −56.9 (3) |
| C3A—C4—C5—C6 | −48.5 (4) | C1'—C2'—C3'—C4' | 117.4 (3) |
| C4—C5—C6—C7 | 46.1 (4) | C1—C2'—C3'—C4' | −118.0 (3) |
| C5—C6—C7—C7A | −52.5 (4) | C2'—C3'—C4'—C5' | 178.7 (3) |
| C6—C7—C7A—C8 | −63.9 (3) | C3'—C4'—C5'—C6' | −122.4 (4) |
| C6—C7—C7A—C3A | 57.8 (3) | C3'—C4'—C5'—C7' | 110.6 (3) |
| C6—C7—C7A—C1 | 167.2 (2) | C4'—C5'—C7'—O7' | −177.4 (2) |
| C4—C3A—C7A—C8 | 61.0 (3) | C6'—C5'—C7'—O7' | 56.5 (3) |
| C3—C3A—C7A—C8 | −69.8 (3) | C4'—C5'—C7'—C9' | 62.0 (4) |
| C4—C3A—C7A—C7 | −60.5 (3) | C6'—C5'—C7'—C9' | −64.2 (4) |
| C3—C3A—C7A—C7 | 168.7 (2) | C4'—C5'—C7'—C8' | −60.8 (3) |
| C4—C3A—C7A—C1 | 177.5 (2) | C6'—C5'—C7'—C8' | 173.1 (3) |
| C3—C3A—C7A—C1 | 46.7 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7′—H7′···O4i | 0.82 | 2.08 | 2.876 (3) | 164 |
| C3A—H3A1···O7′ii | 0.98 | 2.56 | 3.523 (3) | 166 |
Symmetry codes: (i) x+1/2, y+1/2, z+1; (ii) −x+1, y, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PV2614).
References
- Bruker (1998). SMART and SAINT Bruker AXS Inc., Madinson, Wisconsin, USA.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Heaney, R. P. (2008). Clin. J. Am. Soc. Nephrol 3, 1535–1541. [DOI] [PMC free article] [PubMed]
- Henry, H. L. (2011). Best Pract. Res. Clin. Endocrinol. Metab. 25, 531–541. [DOI] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Maehr, H. & Uskokovic, M. R. (2004). Eur. J. Org. Chem. pp. 1703–1713.
- Sheldrick, G. M. (1996). SADABS. University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812051343/pv2614sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812051343/pv2614Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812051343/pv2614Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report