Abstract
HIV-1-associated nephropathy (HIVAN) is a major complication of HIV-1 infection with distinct pathologic features. Introduction of the HIV-1 genome into mice results in a renal disease with all of the histologic and clinical hallmarks of HIVAN on the FVB/N genetic background (TgFVB). We assessed the influence of genetic background on the development or progression of HIVAN by making F1 hybrids of TgFVB with five other inbred strains (CBA, DBA/2, CAST/Ei, C3H/He, BALB/c) and determining phenotypes relevant to renal failure among transgenic offspring (histology, blood urea nitrogen, proteinuria, serum albumin, and serum cholesterol). We found striking variation in phenotypes among F1s, ranging from severe renal disease to no renal disease whatsoever (P < 0.001 for ANOVA across all groups). To map genes responsible for this variation, we produced a backcross of TgFVB/CAST F1 × TgFVB. By genome-wide analysis of linkage in 185 heterozygous transgenic backcross mice, we identified a locus on chromosome 3A1–3, HIVAN1, that showed highly significant linkage to renal disease [logarithm of odds (lod) score 4.9 at D3Mit203, accounting for 15% of the variance in renal disease]. Other loci on chromosomes 11, 14, and 16 were suggestive of linkage to renal disease, and a locus on chromosome 9 influenced serum cholesterol but not nephropathy. Interestingly, HIVAN1 is syntenic to human chromosome 3q25–27, an interval showing suggestive evidence of linkage to various nephropathies. These findings demonstrate a strong genetic influence on HIVAN and demonstrate a major renal disease susceptibility locus on mouse chromosome 3A1–3.
End-stage renal disease (ESRD) is a major public health problem, with a prevalence of nearly 1 per 700 in the United States (1). Only a small minority of affected subjects receive kidney transplantation (1), and the annual mortality on dialysis is extremely high, 23% per year (1). Although a number of risk factors for ESRD have been identified, including hypertension, diabetes mellitus, and infection with HIV-1, only a minority of individuals with these susceptibilities develop renal complications; the factors that determine whether renal failure will develop and/or progress are poorly understood (2–4). As for other areas of pathophysiology, genetic approaches have the capacity to provide insight into the fundamental causes of this important medical problem.
HIV-associated nephropathy (HIVAN) is a complication of HIV-1 infection with distinctive clinical and pathological features (5). Clinically, it is characterized by the development of proteinuria usually followed by rapid progression to renal failure; pathological features include focal and segmental glomerulosclerosis with characteristic glomerular collapse, podocyte hyperplasia, pronounced tubulo-interstitial inflammation and microcystic tubular dilation, and epithelial dedifferentiation, proliferation, and apoptosis (6–9). HIVAN results from direct epithelial infection (10–13); however, the mechanisms underlying disease susceptibility and progression remain unknown.
Epidemiology of HIVAN has revealed a striking finding: this complication of HIV-1 infection is found almost exclusively among subjects of African ancestry (14). Thus, whereas HIVAN is the third most common cause of ESRD among African-American males, it is very rare among Caucasians and Asians (1). Of the 4,218 patients reported by the U.S. Renal Data System for the period of 1997–2001, 90.2% were African American (1). This observation has been replicated in studies in Europe and Asia and is not explained by the mode of viral transmission (15–21). Moreover, relatives of index cases with HIVAN show an increased prevalence of ESRD from all causes,†† with only a small fraction of these additional cases attributable to HIV-1 (22). Estimates of the relative risk of ESRD to siblings of an index case with HIVAN are as high as 10.†† These findings suggest the existence of alleles that increase susceptibility to ESRD from a variety of insults such as HIV infection or diabetes; they also are consistent with genetic factors contributing to the specific outcomes of HIV-1 infection and its complications. Consequently, identification of these putative genetic factors could prove relevant not only to this form of ESRD, but to others as well.
Investigation of the genetic basis of HIVAN in humans is complicated by the rarity of multiplex families with HIVAN, making it very difficult to perform adequately powered genetic linkage studies. In such settings, animal models have the potential to contribute important insight. Mice transgenic for the HIV-1 genome provide an excellent animal model of HIVAN. In multiple independent transgenic constructs, mice and rats develop a renal disease with all of the hallmarks of HIVAN (24–28). Among these, a mouse strain transgenic for a replication defective HIV-1 proviral construct has been extensively characterized (24, 29–31). Heterozygous transgenic animals develop renal lesions indistinguishable from human HIVAN, with focal segmental glomerulosclerosis, mirocystic tubular dilatation, epithelial proliferation, and dedifferentiation of the mature podocyte phenotype (7, 24, 25). Affected mice develop proteinuria and ESRD similar to humans with HIVAN. The homozygous transgenic mice develop cachexia and die by 2 months, a phenotype reminiscent of the AIDS-associated wasting syndrome (7, 24, 29, 31). Development of HIVAN in this model depends on the presence of the HIV-1 genome in the kidney because wild-type kidneys transplanted into the HIV-1 trangenic background remain free of disease whereas transgenic kidneys transplanted into a wild-type background develop HIVAN (32).
We demonstrate herein that, in the presence of the HIV-1 transgenes, development of HIVAN is highly dependent on genetic background, with the phenotype ranging from fulminant disease culminating in early death to no detectable renal disease. This finding establishes unequivocally that development of HIVAN has a strong genetic influence. By production of a backcross between strains with high and low susceptibility to HIVAN, we have sought to map genes that contribute to this outcome by a genome-wide analysis of linkage. The results have identified a locus with a major effect on risk of development of HIVAN.
Methods
Animal Breeding. The HI V-1 transgenic mouse line TgN(pNL43d14)26Lom 26 (hereafter referred to as TgFVB) was produced on the inbred FVB/N genetic background and has been extensively characterized (24, 25). F1 hybrids were produced by breeding TgFVB mice heterozygous for the transgene to other inbred strains and identifying transgene carriers by PCR. The strains used for F1 production were BALB/cJ, CBA/J, C3H/HeJ, DBA/2J, and CAST/EiJ (The Jackson Laboratory, abbreviated as BALB, CBA, C3H, DBA, and CAST in the text). The F1 progeny were characterized at 3–6 months of age for phenotypes as described below.
A backcross was produced by crossing the (TgFVB × CAST) F1 to the starting TgFVB line. Transgene heterozygous progeny were characterized at 12–16 weeks as described below. Animals that were moribund were killed before this end point. Before killing, mice were anesthetized with Avertin, a sample of blood was taken by cardiac puncture, and spontaneously voided urine was collected. The protocol was approved by the Institutional Animal Care and Use Committee at the Mt. Sinai Medical Center.
Phenotypic Characterization. Five primary phenotypes related to renal disease were characterized: renal histology, blood urea nitrogen (BUN), proteinuria, serum cholesterol, and serum albumin. Proteinuria was measured by spot urine dipsticks (Roche), which quantitated levels as 0, 0.5 (trace), +1, +2, or +3; venous blood was obtained by cardiac puncture for measurement of BUN, serum albumin, and serum cholesterol levels. Kidneys were harvested, fixed in 10% formalin, and embedded in paraffin. Three-micrometer sections were prepared and stained with hematoxylin-eosin and periodic-acid schiff. Renal histology was scored independently by two investigators (A.G.G. and V.D.D.) blinded to genetic background and other traits. Four traits related to tubulo-interstitial disease (epithelial regeneration/degeneration, tubular casts, tubular dilatation, and interstitial infiltrates) and three related to glomerular injury (glomerular sclerosis, podocyte hyperplasia and collapse) were scored by using a semiquantitative scale: 0 = no disease, 1 = 1–25% of tissue showing abnormalities, 2 = 26–50% and 3 = >50% of tissue affected. These seven histology scores were highly correlated with one another (r = 0.68–0.95) and were averaged to obtain the total histologic score. The correlations between the scores of the two readers were very high (r = 0.92–0.96 for the different traits) and the average of the scores of the two readers was used for analysis.
In addition, given the strong correlations found among primary renal phenotypes (Table 3, which is published as supporting information on the PNAS web site) and their physiologic correlation in nephrotic syndrome, principal component analysis was performed to objectively derive composite phenotypes. Primary phenotypes were standardized, and factors were extracted by using Eigenvalue thresholds of >1. This analysis identified two principal components, one that accounted for a large fraction of the variance in total histology score (r = 0.83), urine protein (r = 0.80), BUN (r = 0.75), and serum cholesterol (r = 0.87), but not serum albumin (r < 0.01). The second principal component was highly correlated with serum albumin (r = 0.99) only. These two principal components accounted for 65% of the total variance in the backcross. The equation for the first principal component (composite score 1, or CS1) is as follows: CS1 = (0.33 × histo score) + (0.34 × cholesterol) + (0.29 × BUN) + (0.29 × proteinuria) + (0.01 × albumin). Because the second principal component was indistinguishable from serum albumin alone, it did not add to the analysis. We used all transgenic animals together (TgFVB, F1 hybrids, and backcross) to derive composite scores; however, composite scores derived from the backcross cohort alone did not significantly alter the linkage results.
Genotyping and Analysis of Linkage. Concerning transgene status, HIV-1 transgene carriers were identified in F1s by PCR of tail DNA by using primers and probes specific for the HIV-1 transgene (24, 25). To distinguish transgene homozygotes and heterozygotes in the backcross, we identified the transgene integration site in the TgFVB mouse by using a modification of the bubble PCR method (33). This method identified the junction of mouse genomic DNA and the HIV transgene sequence and localized the mouse genomic sequence to chromosome 8 band C2, a locus shown to contain the integration site by fluorescence in situ hybridization analysis (P.E.K., unpublished results). PCR using primers specific for the HIV-1 genome and the flanking mouse sequence demonstrated the presence of this junction in uncloned genomic DNA. The genomic sequence of this interval identified a 62-bp repetitive sequence tightly linked to the transgene integration site that is polymorphic (sense primer, TGTGAAGATGAAAGACCTTGTC; antisense primer, AAGCCAGGTATGGTGATATACAC). Alleles of this polymorphism cosegregated with the transgene in backcross litters, permitting inference of transgene zygosity. In the backcross, we found 185 transgene heterozygotes and 69 transgene homozygotes.
For genome-wide analysis of linkage, genotyping was performed by using polymorphic marker loci. Fluorescent primers were used to direct PCR from genomic DNA, and products were analyzed on an ABI 3700 DNA sequencer by using genotyper 3.7 software. Marker positions were obtained from the Whitehead Institute and Mouse Genome Informatics web sites. Alleles were coded as deriving from the TgFVB (1) or CAST/Ei (2) parent.
We used the total histology score, urine protein, BUN, serum cholesterol, albumin, and composite score 1 as traits for analysis of linkage. Each trait was transformed to approximate a normal distribution (square root transformation for histologic score, urine protein, and serum albumin; log10 transformation for BUN and cholesterol; the composite score conformed to a normal distribution without transformation).
We first genotyped 138 informative markers (average spacing 13 cM) in 84 transgene heterozygous mice of the backcross selected from the extremes of the phenotypic distribution of histology score. Pairwise and multipoint analysis of linkage compared the inheritance of trait and marker loci. For all intervals that showed lod scores of 1 or greater for linkage to 1 or more phenotypes, we genotyped all transgene heterozygous animals in the backcross with markers spaced at an average of 5 cM (53 markers on 8 chromosomes in 185 mice). Pairwise and multipoint lod scores and the proportion of the variance of each trait explained by each trait locus were calculated by using the mapmanager qtx program (34) under an additive model. Traditional thresholds for significance in the backcross were used (lod score >3.3) (35). We also conducted permutation tests in which the observed sets of phenotypes for each animal were randomized on the genotypes; 10,000 replicates were performed to determine the empirical significance of the linkage findings. In secondary analyses, we also performed composite interval mapping and two-locus interaction analyses to attempt to detect additional trait loci.
Measurement of HIV-1 Gene Expression. We compared the expression of the HIV-1 envelope transcript and β-actin in kidneys from transgenic F1 hybrids by using real-time PCR as described in Fig. 4, which is published as supporting information on the PNAS web site. Quadruplicate samples of kidneys from four different mice in each F1 were analyzed and compared with a reference sample. Comparison of the level of expression of the HIV-1 transgene among F1 hybrids was performed by ANOVA.
Results
Variable Phenotype in Transgenic F1 Hybrids. To test the role of genetic background on the HIVAN phenotype, heterozygous TgFVB mice were crossed to the starting FVB strain and five other inbred strains (BALB, CBA, C3H, DBA, and CAST). Phenotypes observed in transgene-bearing F1s were compared with those in the transgenic FVB background.
There was a striking variation in the quantitative severity of renal disease among the transgenic F1s although, when present, the renal disease in all of the F1s was qualitatively indistinguishable from the disease seen in TgFVB (Table 1). The mean histology score was highest for the TgFVB; this cohort also had the highest composite score, the highest proteinuria, and the second highest BUN (Table 1). In contrast to the severe disease found in TgFVB, some F1 hybrids, such as the TgFVB/CAST and the TgFVB/BALB, showed no renal disease whatsoever, with mean phenotypic values that were not significantly different from nontransgenic wild-type controls (Table 1). The other F1s had intermediate values. The difference in the severity of disease between TgFVB and other strains was highly significant; the composite renal disease score for TgFVB was significantly higher than each of the other F1s, and the same applies for most other primary phenotypes (Table 1). In addition, there were significant differences in severity among many other F1s, and the global analysis of variance in severity among all strains tested was highly significant for each trait (P < 0.001). This striking variation in phenotypic expression of HIVAN strongly supports the presence of alleles in these strains that influence development or progression of renal disease.
Table 1. Variable severity of renal disease among HIV-1 transgenic mice.
| Groups | N | Composite score | Total histo score | BUN, mg/dl | Cholesterol, mg/dl | Albumin, mg/dl | Proteinuria |
|---|---|---|---|---|---|---|---|
| Nontransgenic | 36 | 0 ± 0.28‡ | 0.11 ± 0.2* | 23 ± 6† | 94 ± 20 | 2.7 ± 0.3* | 0.7 ± 0.3‡ |
| TgFVB/BALB F1 | 26 | 0 ± 0.33‡ | 0.09 ± 0.19* | 22 ± 4† | 104 ± 12 | 2.5 ± 0.4 | 0.8 ± 0.3‡ |
| TgFVB/CAST F1 | 43 | -0.42 ± 0.66‡ | 0.08 ± 0.14* | 19 ± 7† | 64 ± 19† | 2.5 ± 0.7 | 0.7 ± 0.4‡ |
| TgFVB/C3H F1 | 47 | 0.04 ± 0.96‡ | 0.3 ± 0.44* | 22 ± 9† | 76 ± 27† | 2.5 ± 0.5 | 1.2 ± 0.3‡ |
| TgFVB/CBA F1 | 35 | 0.12 ± 0.64‡ | 0.50 ± 0.44 | 20 ± 7† | 189 ± 167† | 2.3 ± 0.7 | 1.3 ± 0.4* |
| TgFVB/DBA F1 | 23 | 0.21 ± 1.0† | 0.52 ± 0.73 | 33 ± 26 | 167 ± 111† | 2.2 ± 0.4* | 1.4 ± 0.8 |
| TgFVB | 133 | 0.69 ± 0.71 | 0.73 ± 0.81 | 31 ± 12 | 98 ± 47 | 2.5 ± 0.5 | 1.85 ± 1.3 |
Values are mean ± SD; all transgenic mice are heterozygous for the transgene. Global ANOVA, P < 0.0001 for all variables. N, Number of animals; histo, histology. *, P < 0.05; †, P < 0.01; ‡, P < 0.001 vs. TgFVB.
To test whether some of the variation in renal disease is the consequence of varied expression of the HIV-1 genome, levels of expression of the HIV-1 envelope gene were measured by real-time PCR in kidneys of TgFVB and the different F1s. The results demonstrate that all of the F1s have levels of HIV-1 gene expression at least as high as TgFVB, indicating that reduced severity of HIVAN is not accounted for by intrinsically reduced expression of the HIV-1 genome (Fig. 4). There was no significant correlation of the level of expression of HIV-1 with the severity of HIVAN among different strains.
Characterization of (TgFVB/Cast) F1 × TgFVB Backcross. The striking contrast in the severity of renal disease between transgene-bearing TgFVB vs. either TgFVB/CAST or TgFVB/BALB provides strong evidence that genes affect susceptibility to the development and/or progression of HIVAN. Genes that influence this susceptibility can potentially be mapped in crosses segregating these alleles. Because renal disease is completely suppressed in the TgFVB/CAST and TgFVB/BALB F1s, the most informative cross might be a backcross of these F1s to TgFVB. Because the TgFVB and TgFVB/CAST F1 had very similar levels of renal HIV-1 expression, we produced a TgFVB/CAST F1 × TgFVB backcross and generated 375 backcross mice, 185 of which were transgene heterozygotes (see Methods).
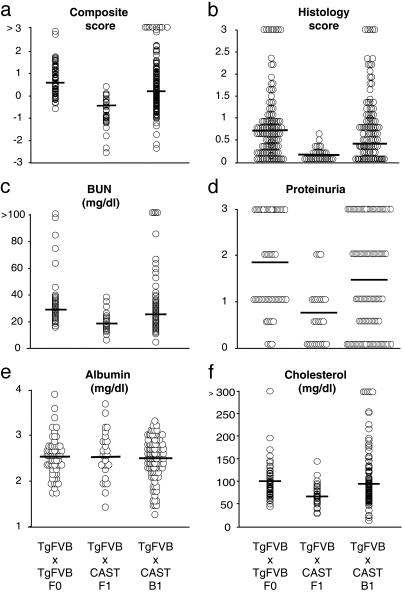
Phenotypic characterization of the heterozygous transgenic backcross mice demonstrated that renal disease, which was absent in the F1s, was recovered in some, but not all, backcross mice (Figs. 1 and 2). Moreover, there was a marked increase in phenotypic variance in the backcross compared with the TgFVB and F1 cohorts (Fig. 2), confirming that loci conferring susceptibility to renal disease loci segregated in this cross. Interestingly, there was evidence of transgressive segregation in the backcross (Fig. 2), suggesting either that alleles contributing to renal disease are being contributed by CAST or that protective alleles are contributed by TgFVB.
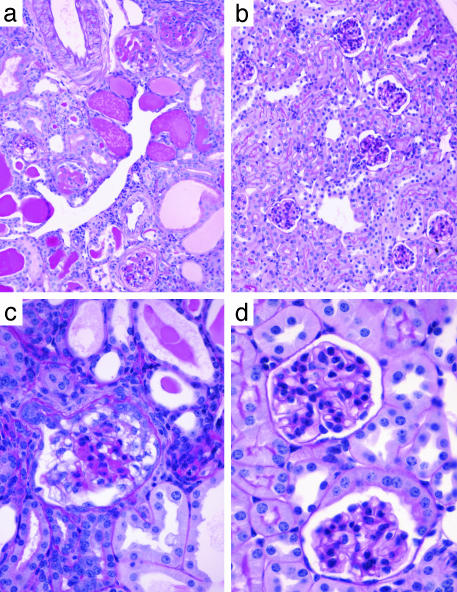
Fig. 1.
Renal pathology in transgenic HIV-1 backcross mice. Periodic-acid Schiff-stained kidney sections of HIV-1 transgenic mice at 3 months of age from the TgFVB × TgFVB/CAST backcross are shown. (a and c) A mouse with marked renal disease. (b and d) A mouse with no detectable disease. (a and b) Low-power views. The affected kidney shows microcystic tubular dilatation, tubular casts, interstitial infiltrate, and focal segmental glomerulosclerosis. (c and d) Higher-power magnification of glomeruli. The affected kidney shows a collapsing glomerulopathy reminiscent of human HIVAN.
Fig. 2.
Distribution of phenotypes in F0, F1, and backcross cohort of HIV-1 transgenic mice. A TgFVB/CAST F1 was backcrossed to TgFVB, and the transgene heterozygote progeny were phenotyped as described in Methods. The phenotypic values for each transgene heterozygote mouse in the parental and backcross cohorts are shown as follows: composite score (a), histology score (b), BUN (c), dipstick proteinuria (d), albumin (e), and cholesterol (f). The mean value for each group is indicated by the horizontal bars.
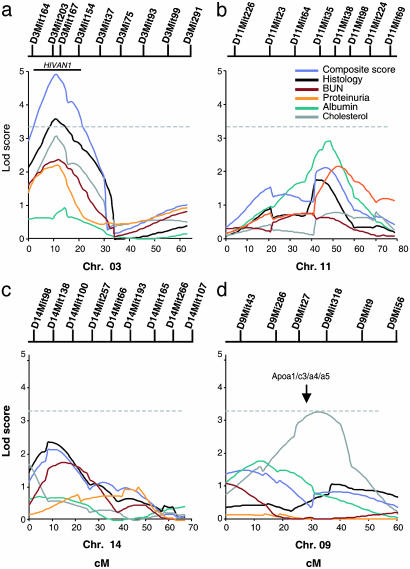
Analysis of Linkage in Backcross. Analysis of linkage was performed by genotyping 138 informative markers distributed across the mouse genome and comparing the inheritance of each segment of each chromosome to the inheritance of each quantitative trait in transgene heterozygote members of the backcross as described in Methods. The strongest evidence for linkage was to chromosome 3 at the location of D3Mit203 (located 9.8 cM from the top of the chromosome 3 linkage group in the MIT F2 Intercross). The composite score showed a multipoint lod score of 4.9 for linkage to this location (Fig. 3a). Similarly, most of the individual primary phenotypes also showed evidence of linkage to this segment: renal histology showed a lod score of 3.5, cholesterol lod score 3.1, BUN 2.4, and urinary protein 2.1 (Fig. 3a). By using traditional criteria for evaluation of linkage results in a backcross, the lod score for the composite phenotype is highly significant, while the total histology is also significant, and the lod scores for cholesterol, BUN, and urinary protein are suggestive of linkage.
Fig. 3.
Linkage in the backcross cohort. Multipoint lod score plots for phenotypes on chromosomes 3 (a), 11 (b), 14 (c), and 9 (d) for the six primary phenotypes are shown. Map distance is presented in cM. The positions of informative markers genotyped are shown above each plot. The dashed horizontal bar indicates the threshold (3.3) for genome-wide significance for a backcross. The solid bar above the chromosome 3 lod plot indicates the 95% confidence interval for HIVAN1 computed by bootstrap resampling (composite score). The location of the Apoa1/c3/a4/a5 gene cluster is shown above the lod plot for chromosome 9.
The empiric significance of the observed results was assessed by randomly permuting the sets of phenotypes on the genotypes and determining how frequently lod scores as high as or higher than observed lod scores were obtained in 10,000 replicates. A lod score of 4.9 or greater was found in 8 of 10,000 permutations of the composite scores (P = 0.0008). Similarly, a lod score of ≥3.5 was obtained in 190 of 10,000 permutations of renal histology score (P = 0.019). Evaluation of the 95% confidence interval for this trait locus was determined by bootstrap resampling, which localized it to a 30-cM interval delimited by D3Mit164 to D3Mit154. We have named this locus HIVAN1. HIVAN1 explains 15% of the variance in the composite renal score and 10% of the variance in histology and cholesterol level in the backcross. Interestingly, it is the CAST allele that confers increased severity (Table 2).
Table 2. Phenotypic scores by genotypes at HIVAN1 (D3Mit203).
| D3Mit 203 genotype | N | Composite score† | Histology score† | BUN, mg/dl* | Cholesterol, mg/dl* | Albumin, mg/dl | Proteinuria† |
|---|---|---|---|---|---|---|---|
| TgFVB/TgFVB | 96 | –0.01 ± 0.94 | 0.38 ± 0.33 | 26 ± 9 | 87 ± 34 | 2.5 ± 0.4 | 1.2 ± 0.4 |
| TgFVB/CAST | 89 | 0.66 ± 1.5 | 0.66 ± 0.73 | 30 ± 12 | 121 ± 96 | 2.6 ± 0.3 | 1.8 ± 0.3 |
TgFVB, TgFVB alleles; CAST, CAST/Ei alleles; N, number of animals. *, P<0.01; †, P<0.001 for comparison between genotypic classes at D3Mit203.
In addition to HIVAN1, three other intervals on chromosomes 11, 14, and 16 showed suggestive evidence of linkage to the composite renal phenotype and at least one additional phenotype, with the FVB allele in each case contributing to increased disease severity (Table 4, which is published as supporting information on the PNAS web site, and Fig. 3 b–d). For example, an interval on chromosome 11 bounded by loci D11Mit64 and D11Mit224 showed a maximum lod score of 2.2 for composite renal score, 2.9 for serum albumin, 2.1 for urine protein, and 1.7 for histology. Determining whether these putative linkages are true positives or are chance occurrences will require replication studies or derivation of congenic strains.
In addition to these intervals influencing renal disease, an interval on chromosome 9 provided significant evidence of linkage to cholesterol (lod score 3.3). This locus, delimited by loci D9Mit286 and D9Mit9, explained 10% of the variance in cholesterol levels. Evidence for linkage of this locus to the other primary phenotypes was weak or absent (lod scores 0.1–0.9). These findings strongly suggest that this locus contributes directly to cholesterol level and not renal disease. Support for this contention comes from recognition that the linked interval contains the Apoa1-Apoc3-Apoa4-Apoa5 gene cluster, whose protein products are all involved in cholesterol homeostasis. Moreover, this interval has been previously implicated in linkage studies of serum high-density lipoprotein levels in a cross involving CAST/Ei (36). This locus shows an additive effect with HIVAN1 on cholesterol levels.
In secondary analyses, we searched for two locus interactions and performed composite interval mapping by rescanning the genome after controlling for the effects of HIVAN1 but did not detect additional loci contributing to renal disease. Similarly, analysis of linkage by using phenotype residuals after accounting for the effects of correlated phenotypes did not reveal evidence for other loci.
Discussion
Host factors influence the outcome of many infectious diseases; these factors include genes that confer protection from malaria as well as genes that increase susceptibility to tuberculosis, leprosy, and schistosomiasis (37–40). For HIV-1, genetic variations at HLA and chemokine receptor loci influence the outcome of infection, and mutation in the coreceptor CCR5 protects against primary infection (41–43).
In an effort to map loci influencing the development and progression of an important complication of HIV-1 infection, HIVAN, we have used a well-established mouse model of this disease. For multifactorial traits, inbred mouse models have a number of distinct advantages. These advantages include the ability to produce large numbers of animals segregating no more than two alleles at any locus, the ability to virtually eliminate environmental variation, and the ability to examine relevant tissue from all members of a large cohort (44, 45).
A key assumption in such animal studies is that the disease in question is a valid model of its human counterpart. In the present study, the contributing factor in both human and mouse HIVAN is the genome of the identical virus, HIV-1. Moreover, the disease produced in mouse is histologically indistinguishable from its human counterpart and follows the same clinical course (24, 25, 29).
In this mouse model, we show that genetic factors impart a large effect on the development and/or progression of HIVAN, with F1 hybrids varying from severe renal disease to no renal disease. Moreover, the marked increase in phenotypic variance seen in the backcross animals vs. the genetically homogeneous F0 or F1 cohorts further supports the role of genetic variation on development of this trait. This evidence for genetic background effects on HIVAN in mouse supports the possibility that the striking ethnic variation in human HIVAN may also be attributable to genetic factors.
Mapping studies in a large informative backcross cohort demonstrated highly significant linkage of HIVAN to a segment of mouse chromosome 3. These results do not distinguish whether this linkage is accounted for by increased susceptibility to development of HIVAN or its progression. This linkage was strongest with the composite phenotype objectively derived from principal component analysis and was also significant for independent measures contributing to this trait. It is of interest that the interval containing this locus, HIVAN1, is syntenic to human chromosome 3q25–27. This human region has shown suggestive evidence of linkage to diabetic nephropathy in independent studies of Pima-Indian with type 2 diabetes (46), Caucasians with type I diabetes (47), and to serum creatinine clearance in hypertensive African Americans (48). Given the observation that renal failure of diverse causes can cluster within the same kindred, it is possible that HIVAN1 and this putative human locus are orthologous, affording the speculation that HIVAN1 may prove relevant to the development or progression of many forms of ESRD.
The 95% confidence interval for HIVAN1 corresponds to ≈60 Mb and contains many potential positional candidates, such as the angiotensin type 1b receptor, genes involved in neoplasia and cell-cycle regulation (SKI proto-oncogene, protein kinase Cλ), and genes associated with HIV-1 complications such as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL; tumor necrosis factor ligand superfamily, member 10), a gene implicated in potentiation of neuronal apoptosis via HIV-1 infected macrophages (23). Progress from this initial linkage screen can be pursued in several fashions. First, production of congenic mice in which HIVAN1 is introgressed from CAST/EiJ into the TgFVB background can be performed, and mapping studies can then proceed to narrow the location of the underlying gene. Alternatively, positional candidate genes can be screened, and the functional significance of identified variants can be implicated by further in vitro and in vivo studies.
The complexity of genetic analysis of complex traits is underscored by the finding that, although the CAST genome completely suppresses HIVAN in the F1 hybrid, the CAST allele at HIVAN1 actually increases HIVAN severity (Table 2). Consistent with this finding, 16 of the 19 animals that comprise the top decile of composite renal score severity inherited the CAST allele at HIVAN1. Although at first blush this result is surprising, the possibility of such alleles was foreshadowed by the evidence for transgressive segregation in the backcross. Given that the inbred mouse strains studied were not selectively bred for susceptibility to HIVAN, it is not surprising that starting strains harbor both protective and permissive alleles for development of renal disease.
In addition to HIVAN1, we found suggestive evidence for linkage of HIVAN to segments of chromosomes 11, 14, and 16. Although HIVAN1 explained 15% of the variance in the composite phenotype, these other loci contributed small fractions of the variance. Together, these loci account for less than half of the phenotypic variation in the backcross and suggest that many loci with small effects account for most of the remaining phenotypic variation in the backcross.
Finally, these studies validate the ability to use mouse models to map genetic modifiers of HIVAN. Given the wide variation in disease severity in the F1s studied, it is likely that mapping by using different counterstrains can identify additional disease loci that may help define the pathways that confer susceptibility to and protection from renal disease.
Supplementary Material
Acknowledgments
We thank Glen Markowitz and Kirk Foster for assistance with preparation of histology figures. This work was supported by National Institutes of Health Grant DK56492-01. A.G.G. was supported by National Institutes of Health Grant K08 DK02610-01 and an award from the Emerald Foundation. R.P.L. is an investigator of the Howard Hughes Medical Institute.
Abbreviations: HIVAN, HIV-1-associated nephropathy; ESRD, end-stage renal disease; TgFVB, HIV-1 transgenic mice; BUN, blood urea nitrogen; lod, logarithm of odds.
Footnotes
Simon, D. B., Farhi, A., Mahnensmith, R. & Lifton, R. P. (1996) J. Am. Soc. Nephrol. 7, 1343 (abstr.).
References
- 1.U.S. Renal Data System (2003) Annual Data Report: Atlas of End-Stage Renal Disease in the United States (National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD).
- 2.McClellan, W. M. & Flanders, W. D. (2003) J. Am. Soc. Nephrol. 14, S65-S70. [DOI] [PubMed] [Google Scholar]
- 3.Tarver-Carr, M. E., Powe, N. R., Eberhardt, M. S., LaVeist, T. A., Kington, R. S., Coresh, J. & Brancati, F. L. (2002) J. Am. Soc. Nephrol. 13, 2363-2370. [DOI] [PubMed] [Google Scholar]
- 4.Kramer, H. J., Nguyen, Q. D., Curhan, G. & Hsu, C. Y. (2003) J. Am. Med. Assoc. 289, 3273-3277. [DOI] [PubMed] [Google Scholar]
- 5.Klotman, P. E. (1999) Kidney Int. 56, 1161-1176. [DOI] [PubMed] [Google Scholar]
- 6.D'Agati, V. & Appel, G. B. (1997) J. Am. Soc. Nephrol. 8, 138-152. [DOI] [PubMed] [Google Scholar]
- 7.Barisoni, L., Bruggeman, L. A., Mundel, P., D'Agati, V. D. & Klotman, P. E. (2000) Kidney Int. 58, 173-181. [DOI] [PubMed] [Google Scholar]
- 8.Shankland, S. J., Eitner, F., Hudkins, K. L., Goodpaster, T., D'Agati, V. & Alpers, C. E. (2000) Kidney Int. 58, 674-683. [DOI] [PubMed] [Google Scholar]
- 9.D'Agati, V. & Appel, G. B. (1998) Semin. Nephrol. 18, 406-421. [PubMed] [Google Scholar]
- 10.Bruggeman, L. A., Ross, M. D., Tanji, N., Cara, A., Dikman, S., Gordon, R. E., Burns, G. C., D'Agati, V. D., Winston, J. A., Klotman, M. E. & Klotman, P. E. (2000) J. Am. Soc. Nephrol. 11, 2079-2087. [DOI] [PubMed] [Google Scholar]
- 11.Winston, J. A., Bruggeman, L. A., Ross, M. D., Jacobson, J., Ross, L., D'Agati, V. D., Klotman, P. E. & Klotman, M. E. (2001) N. Engl. J. Med. 344, 1979-1984. [DOI] [PubMed] [Google Scholar]
- 12.Ray, P. E., Liu, X. H., Henry, D., Dye, L., 3rd, Xu, L., Orenstein, J. M. & Schuztbank, T. E. (1998) Kidney Int. 53, 1217-1229. [DOI] [PubMed] [Google Scholar]
- 13.Kimmel, P. L., Ferreira-Centeno, A., Farkas-Szallasi, T., Abraham, A. A. & Garrett, C. T. (1993) Kidney Int. 43, 1347-1352. [DOI] [PubMed] [Google Scholar]
- 14.Monahan, M., Tanji, N. & Klotman, P. E. (2001) Semin. Nephrol. 21, 394-402. [DOI] [PubMed] [Google Scholar]
- 15.Laradi, A., Mallet, A., Beaufils, H., Allouache, M. & Martinez, F. (1998) J. Am. Soc. Nephrol. 9, 2327-2335. [DOI] [PubMed] [Google Scholar]
- 16.Nochy, D., Glotz, D., Dosquet, P., Pruna, A., Guettier, C., Weiss, L., Hinglais, N., Idatte, J. M., Mery, J. P., Kazatchkine, M., et al. (1993) Nephrol. Dial. Transplant. 8, 11-19. [DOI] [PubMed] [Google Scholar]
- 17.Connolly, J. O., Weston, C. E. & Hendry, B. M. (1995) Q. J. Med. 88, 627-634. [PubMed] [Google Scholar]
- 18.Williams, D. I., Williams, D. J., Williams, I. G., Unwin, R. J., Griffiths, M. H. & Miller, R. F. (1998) Sex. Transm. Infect. 74, 179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frassetto, L., Schoenfeld, P. Y. & Humphreys, M. H. (1991) Am. J. Kidney Dis. 18, 655-659. [DOI] [PubMed] [Google Scholar]
- 20.Hailemariam, S., Walder, M., Burger, H. R., Cathomas, G., Mihatsch, M., Binswanger, U. & Ambuhl, P. M. (2001) Swiss Med. Wkly. 131, 412-417. [DOI] [PubMed] [Google Scholar]
- 21.Praditpornsilpa, K., Napathorn, S., Yenrudi, S., Wankrairot, P., Tungsaga, K. & Sitprija, V. (1999) Am. J. Kidney Dis. 33, 282-286. [DOI] [PubMed] [Google Scholar]
- 22.Freedman, B. I., Soucie, J. M., Stone, S. M. & Pegram, S. (1999) Am. J. Kidney Dis. 34, 254-258. [DOI] [PubMed] [Google Scholar]
- 23.Miura, Y., Misawa, N., Kawano, Y., Okada, H., Inagaki, Y., Yamamoto, N., Ito, M., Yagita, H., Okumura, K., Mizusawa, H. & Koyanagi, Y. (2003) Proc. Natl. Acad. Sci. USA 100, 2777-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp, J. B., Klotman, M. E., Adler, S. H., Bruggeman, L. A., Dickie, P., Marinos, N. J., Eckhaus, M., Bryant, J. L., Notkins, A. L. & Klotman, P. E. (1992) Proc. Natl. Acad. Sci. USA 89, 1577-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickie, P., Felser, J., Eckhaus, M., Bryant, J., Silver, J., Marinos, N. & Notkins, A. L. (1991) Virology 185, 109-119. [DOI] [PubMed] [Google Scholar]
- 26.Hanna, Z., Kay, D. G., Rebai, N., Guimond, A., Jothy, S. & Jolicoeur, P. (1998) Cell 95, 163-175. [DOI] [PubMed] [Google Scholar]
- 27.Kajiyama, W., Kopp, J. B., Marinos, N. J., Klotman, P. E. & Dickie, P. (2000) Kidney Int. 58, 1148-1159. [DOI] [PubMed] [Google Scholar]
- 28.Reid, W., Sadowska, M., Denaro, F., Rao, S., Foulke, J., Jr., Hayes, N., Jones, O., Doodnauth, D., Davis, H., Sill, A., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 9271-9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruggeman, L. A., Thomson, M. M., Nelson, P. J., Kopp, J. B., Rappaport, J., Klotman, P. E. & Klotman, M. E. (1994) Virology 202, 940-948. [DOI] [PubMed] [Google Scholar]
- 30.Ray, P. E., Bruggeman, L. A., Weeks, B. S., Kopp, J. B., Bryant, J. L., Owens, J. W., Notkins, A. L. & Klotman, P. E. (1994) Kidney Int. 46, 759-772. [DOI] [PubMed] [Google Scholar]
- 31.Santoro, T. J., Bryant, J. L., Pellicoro, J., Klotman, M. E., Kopp, J. B., Bruggeman, L. A., Franks, R. R., Notkins, A. L. & Klotman, P. E. (1994) Virology 201, 147-151. [DOI] [PubMed] [Google Scholar]
- 32.Bruggeman, L. A., Dikman, S., Meng, C., Quaggin, S. E., Coffman, T. M. & Klotman, P. E. (1997) J. Clin. Invest. 100, 84-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munroe, D. J., Haas, M., Bric, E., Whitton, T., Aburatani, H., Hunter, K., Ward, D. & Housman, D. E. (1994) Genomics 19, 506-514. [DOI] [PubMed] [Google Scholar]
- 34.Manly, K. F., Cudmore, R. H., Jr., & Meer, J. M. (2001) Mamm. Genome 12, 930-932. [DOI] [PubMed] [Google Scholar]
- 35.Lander, E. & Kruglyak, L. (1995) Nat. Genet. 11, 241-247. [DOI] [PubMed] [Google Scholar]
- 36.Mehrabian, M., Wen, P. Z., Fisler, J., Davis, R. C. & Lusis, A. J. (1998) J. Clin. Invest. 101, 2485-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooke, G. S. & Hill, A. V. (2001) Nat. Rev. Genet. 2, 967-977. [DOI] [PubMed] [Google Scholar]
- 38.Siddiqui, M. R., Meisner, S., Tosh, K., Balakrishnan, K., Ghei, S., Fisher, S. E., Golding, M., Shanker Narayan, N. P., Sitaraman, T., Sengupta, U., et al. (2001) Nat. Genet. 27, 439-441. [DOI] [PubMed] [Google Scholar]
- 39.Bellamy, R., Beyers, N., McAdam, K. P., Ruwende, C., Gie, R., Samaai, P., Bester, D., Meyer, M., Corrah, T., Collin, M., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 8005-8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marquet, S., Abel, L., Hillaire, D., Dessein, H., Kalil, J., Feingold, J., Weissenbach, J. & Dessein, A. J. (1996) Nat. Genet. 14, 181-184. [DOI] [PubMed] [Google Scholar]
- 41.Smith, M. W., Dean, M., Carrington, M., Winkler, C., Huttley, G. A., Lomb, D. A., Goedert, J. J., O'Brien, T. R., Jacobson, L. P., Kaslow, R., et al. (1997) Science 277, 959-965. [DOI] [PubMed] [Google Scholar]
- 42.Dean, M., Carrington, M., Winkler, C., Huttley, G. A., Smith, M. W., Allikmets, R., Goedert, J. J., Buchbinder, S. P., Vittinghoff, E., Gomperts, E., et al. (1996) Science 273, 1856-1862. [DOI] [PubMed] [Google Scholar]
- 43.Moore, C. B., John, M., James, I. R., Christiansen, F. T., Witt, C. S. & Mallal, S. A. (2002) Science 296, 1439-1443. [DOI] [PubMed] [Google Scholar]
- 44.Darvasi, A. (1998) Nat. Genet. 18, 19-24. [DOI] [PubMed] [Google Scholar]
- 45.Lander, E. S. & Botstein, D. (1989) Genetics 121, 185-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imperatore, G., Hanson, R. L., Pettitt, D. J., Kobes, S., Bennett, P. H. & Knowler, W. C. (1998) Diabetes 47, 821-830. [DOI] [PubMed] [Google Scholar]
- 47.Moczulski, D. K., Rogus, J. J., Antonellis, A., Warram, J. H. & Krolewski, A. S. (1998) Diabetes 47, 1164-1169. [DOI] [PubMed] [Google Scholar]
- 48.DeWan, A. T., Arnett, D. K., Atwood, L. D., Province, M. A., Lewis, C. E., Hunt, S. C. & Eckfeldt, J. (2001) Am. J. Hum. Genet. 68, 136-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.