Abstract
In the title compound, C21H20BrNO5S, the thiophene group is located above the mean plane of the indole ring and displays rotational disorder (i.e. rotation through 180°). The site occupancy of the major component is 0.902 (2), while that of the minor component is 0.098 (2). In the crystal, pairs of weak C—H⋯O interactions link the molecules into centrosymmetric dimers.
Related literature
For background to the use of indoles as scaffolds in the synthesis of HIV-agents, see: Hassam et al. (2012 ▶) and for a recent review on stages of non-nucleoside reverse transcriptase inhibitors, see: Reynolds et al. (2012 ▶). For the crystal structures of closely related compounds, see: Beddoes et al. (1986 ▶), Hassam & Smith (2012 ▶).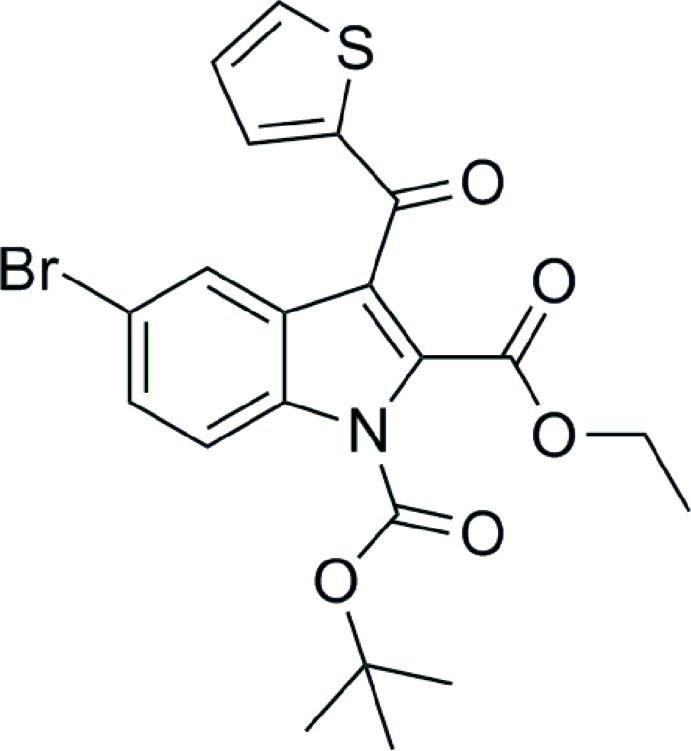
Experimental
Crystal data
C21H20BrNO5S
M r = 478.35
Monoclinic,
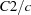
a = 16.220 (3) Å
b = 15.361 (3) Å
c = 18.224 (4) Å
β = 113.792 (2)°
V = 4154.7 (15) Å3
Z = 8
Mo Kα radiation
μ = 2.11 mm−1
T = 100 K
0.34 × 0.21 × 0.17 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan [symmetry-related measurements (SADABS; Bruker, 2009 ▶)] T min = 0.537, T max = 0.721
23562 measured reflections
4855 independent reflections
4101 reflections with I > 2σ(I)
R int = 0.039
Refinement
R[F 2 > 2σ(F 2)] = 0.029
wR(F 2) = 0.066
S = 1.05
4855 reflections
279 parameters
13 restraints
H-atom parameters constrained
Δρmax = 0.38 e Å−3
Δρmin = −0.38 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: X-SEED (Barbour, 2001 ▶; Atwood & Barbour, 2003 ▶); software used to prepare material for publication: X-SEED.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813000809/jj2160sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813000809/jj2160Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813000809/jj2160Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C12A—H12A⋯O3i | 0.95 | 2.48 | 3.418 (5) | 169 |
Symmetry code: (i) 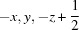 .
.
Acknowledgments
MH thanks Professor Willem A. L. van Otterlo and Dr S. C. Pelly for their valuable input and research oversight. Stellenbosch University’s Science Faculty is also acknowledged for providing laboratory space and financial research support (Subcommittee B). The South African National Research Foundation (NRF), Pretoria, is also acknowledged for providing research funds.
supplementary crystallographic information
Comment
Ethyl-5-bromo-1H-indole-2-carboxylate has been employed as a building block in the synthesis of various anti-HIV active molecules, particularly in the search for novel non-nucleoside reverse transcriptase inhibitors (Hassam et al. 2012). Protection on the indole NH of ethyl 5-bromo-3-(2- thiophenoyl)-1H-indole-2-carboxylate was carried out with di-tert -butyl-dicarbonate using 4-dimethylaminopyridine as a catalytic base.
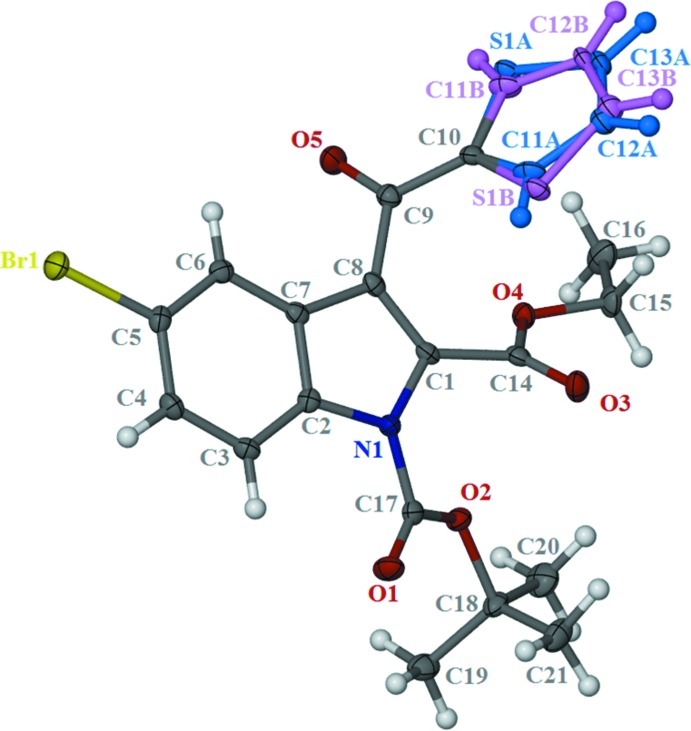
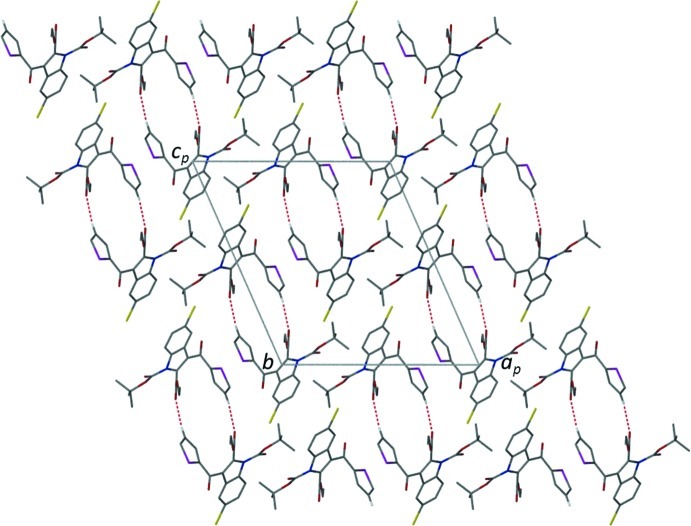
The title compound, C21H20BrNO5S, crystallizes with one molecule in the asymmetric unit (Fig. 1). The thiophene moiety is disordered over two positions with major (A) and minor (B) components in a 0.9021 (19):0.0979 (19)(2) ratio. The dihedral angles between the mean planes of the 5-bromo indole ring (Br1/N1/C1-C8) and the disordered thiophene rings (S1A/C10/C11A/C13A and S1B/C10/C11B/C13B) are 59.67 (9)° and 60.20 (76)°, respectively. The angles between the mean planes of the indole ring and the N-tert- butyloxy, ethyl ester and the ketone groups are 31.72 (7)°, 45.08 (6)° and 47.88 (7)°, respectively. The torsion angles of O5/C9/C10/S1A and O5/C9/C10/S1B are -20.67 (24)° and 159.92 (34)°, respectively, thereby describing the major component in a cis conformation and the minor component in a trans conformation. Molecular packing shows the molecules forming centrosymmetric dimers linked via weak C12A—H12A···O3 intermolecular interactions (Fig. 2, Table 1).
Experimental
4-dimethylaminopyridine (0.0100 g, 0.0818 mmol) was added to a solution of ethyl 5-bromo-3-(2-thiophenoyl)-1H-indole-2-carboxylate (1.10 g, 2.91 mmol) in THF (20 ml), followed by the addition of di-tert-butyl dicarbonate (1.16 g, 5.32 mmol). The reaction mixture was stirred at 298.15 K for 2 h. Colourless crystals were obtained from a hexane/dichloromethane solvent mixture (4:1) (1.15 g, 83%).
Refinement
All non-hydrogen atoms were refined anisotropically. H atoms were placed geometrically [C—H = 0.95 - 0.99 Å; with Uiso(H) = 1.2 - 1.5Ueq(C)] and constrained to ride on their parent atoms. The site-occupancy factors of the disordered thiophene moieties were initially set to 0.5 and then refined, leading to an occupancy of 0.9021 (19) and 0.0979 (19)(2) for the major and minor components, respectively. Bond lengths for the thiophene and phenyl moieties were restrained using the SHELXL SADI command (s.u. = 0.002 Å). Atom displacement parameters for overlaping atoms of the disordered models were constrained using EADP.
Figures
Fig. 1.
Molecular structure of the title compound with atom displacement ellipsoidsdrawn at the 50% probability level. Disordered components (0.9021 (19) = blue) and (0.0979 (19) = purple).
Fig. 2.
Molecular Packing of the title compound viewed along the b axis. Centrosymmetric dimers are linked via weak C—H···O intermolecular interactions (dashed lines).
Crystal data
| C21H20BrNO5S | F(000) = 1952 |
| Mr = 478.35 | Dx = 1.530 Mg m−3 |
| Monoclinic, C2/c | Melting point: 370.13 K |
| Hall symbol: -C 2yc | Mo Kα radiation, λ = 0.71073 Å |
| a = 16.220 (3) Å | Cell parameters from 6586 reflections |
| b = 15.361 (3) Å | θ = 2.4–27.6° |
| c = 18.224 (4) Å | µ = 2.11 mm−1 |
| β = 113.792 (2)° | T = 100 K |
| V = 4154.7 (15) Å3 | Rectangular prisms, colourless |
| Z = 8 | 0.34 × 0.21 × 0.17 mm |
Data collection
| Bruker APEXII CCD diffractometer | 4855 independent reflections |
| Radiation source: fine-focus sealed tube, Bruker SMART Apex | 4101 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.039 |
| φ and ω scans | θmax = 28.2°, θmin = 1.9° |
| Absorption correction: multi-scan [symmetry-related measurements (SADABS; Bruker, 2009)] | h = −20→20 |
| Tmin = 0.537, Tmax = 0.721 | k = −19→19 |
| 23562 measured reflections | l = −24→24 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.029 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.066 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0296P)2 + 2.6317P] where P = (Fo2 + 2Fc2)/3 |
| 4855 reflections | (Δ/σ)max = 0.001 |
| 279 parameters | Δρmax = 0.38 e Å−3 |
| 13 restraints | Δρmin = −0.38 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Br1 | −0.187401 (12) | −0.022969 (12) | −0.284234 (11) | 0.02154 (6) | |
| O1 | 0.21832 (9) | 0.04217 (8) | 0.06003 (8) | 0.0220 (3) | |
| N1 | 0.09114 (9) | 0.12572 (9) | 0.02039 (8) | 0.0140 (3) | |
| C1 | 0.04334 (12) | 0.19832 (11) | 0.02894 (10) | 0.0143 (4) | |
| O2 | 0.22623 (8) | 0.18743 (8) | 0.08505 (7) | 0.0168 (3) | |
| C2 | 0.03540 (10) | 0.08118 (9) | −0.04949 (10) | 0.0143 (3) | |
| O3 | 0.11295 (9) | 0.22588 (8) | 0.16936 (7) | 0.0223 (3) | |
| C3 | 0.05264 (11) | 0.00587 (10) | −0.08360 (9) | 0.0165 (4) | |
| H3 | 0.1083 | −0.0243 | −0.0598 | 0.020* | |
| O4 | 0.06607 (8) | 0.33759 (8) | 0.08185 (7) | 0.0167 (3) | |
| C4 | −0.01584 (11) | −0.02275 (11) | −0.15432 (10) | 0.0173 (4) | |
| H4 | −0.0072 | −0.0740 | −0.1795 | 0.021* | |
| O5 | −0.14693 (9) | 0.30149 (9) | −0.11372 (7) | 0.0229 (3) | |
| C5 | −0.09682 (11) | 0.02238 (10) | −0.18879 (10) | 0.0167 (4) | |
| C6 | −0.11345 (11) | 0.09814 (10) | −0.15555 (9) | 0.0159 (4) | |
| H6 | −0.1687 | 0.1289 | −0.1802 | 0.019* | |
| C7 | −0.04550 (10) | 0.12713 (11) | −0.08437 (9) | 0.0143 (3) | |
| C8 | −0.03952 (12) | 0.20088 (11) | −0.03284 (10) | 0.0147 (4) | |
| C9 | −0.11022 (12) | 0.26847 (12) | −0.04742 (11) | 0.0165 (4) | |
| C10 | −0.13346 (11) | 0.29232 (11) | 0.01992 (10) | 0.0147 (4) | |
| C14 | 0.07984 (11) | 0.25376 (11) | 0.10226 (11) | 0.0158 (4) | |
| C15 | 0.08485 (13) | 0.39806 (12) | 0.14842 (11) | 0.0217 (4) | |
| H15A | 0.1485 | 0.3932 | 0.1869 | 0.026* | |
| H15B | 0.0457 | 0.3853 | 0.1771 | 0.026* | |
| C16 | 0.06589 (15) | 0.48791 (13) | 0.11250 (13) | 0.0294 (5) | |
| H16C | 0.0026 | 0.4918 | 0.0748 | 0.044* | |
| H16A | 0.1047 | 0.4994 | 0.0840 | 0.044* | |
| H16B | 0.0780 | 0.5310 | 0.1553 | 0.044* | |
| C17 | 0.18541 (12) | 0.11267 (11) | 0.05822 (10) | 0.0159 (4) | |
| C18 | 0.32643 (12) | 0.19106 (12) | 0.13180 (11) | 0.0190 (4) | |
| C19 | 0.37308 (13) | 0.16063 (14) | 0.07930 (12) | 0.0249 (4) | |
| H19C | 0.3471 | 0.1907 | 0.0275 | 0.037* | |
| H19A | 0.3648 | 0.0977 | 0.0708 | 0.037* | |
| H19B | 0.4376 | 0.1738 | 0.1056 | 0.037* | |
| C20 | 0.34088 (14) | 0.28765 (13) | 0.14976 (13) | 0.0293 (5) | |
| H20A | 0.3061 | 0.3063 | 0.1802 | 0.044* | |
| H20B | 0.3209 | 0.3202 | 0.0993 | 0.044* | |
| H20C | 0.4050 | 0.2988 | 0.1814 | 0.044* | |
| C21 | 0.35137 (14) | 0.13828 (14) | 0.20765 (11) | 0.0278 (5) | |
| H21A | 0.3417 | 0.0763 | 0.1940 | 0.042* | |
| H21C | 0.3137 | 0.1561 | 0.2357 | 0.042* | |
| H21B | 0.4149 | 0.1481 | 0.2425 | 0.042* | |
| S1A | −0.18632 (4) | 0.39061 (3) | 0.01797 (3) | 0.01650 (16) | 0.9021 (19) |
| C11A | −0.12031 (18) | 0.24830 (18) | 0.08752 (18) | 0.0212 (5) | 0.902 (2) |
| H11A | −0.0924 | 0.1927 | 0.0987 | 0.025* | 0.9021 (19) |
| C12A | −0.1506 (2) | 0.2902 (3) | 0.1402 (3) | 0.0196 (4) | 0.9021 (19) |
| H12A | −0.1462 | 0.2667 | 0.1899 | 0.024* | 0.9021 (19) |
| C13A | −0.1874 (3) | 0.3695 (3) | 0.11084 (15) | 0.0161 (6) | 0.9021 (19) |
| H13A | −0.2108 | 0.4084 | 0.1382 | 0.019* | 0.9021 (19) |
| S1B | −0.1088 (5) | 0.2256 (4) | 0.1026 (3) | 0.01650 (16) | 0.0979 (19) |
| C11B | −0.1782 (13) | 0.3643 (9) | 0.0245 (15) | 0.0212 (5) | 0.0979 (19) |
| H11B | −0.1971 | 0.4082 | −0.0157 | 0.025* | 0.0979 (19) |
| C12B | −0.194 (3) | 0.368 (3) | 0.0944 (18) | 0.0161 (6) | 0.0979 (19) |
| H12B | −0.2251 | 0.4149 | 0.1065 | 0.019* | 0.0979 (19) |
| C13B | −0.161 (2) | 0.298 (2) | 0.143 (3) | 0.0196 (4) | 0.0979 (19) |
| H13B | −0.1655 | 0.2895 | 0.1930 | 0.024* | 0.0979 (19) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.01920 (10) | 0.02324 (11) | 0.01899 (10) | −0.00231 (8) | 0.00438 (7) | −0.00483 (7) |
| O1 | 0.0186 (7) | 0.0174 (7) | 0.0266 (7) | 0.0045 (5) | 0.0056 (6) | −0.0011 (5) |
| N1 | 0.0133 (7) | 0.0144 (7) | 0.0138 (7) | 0.0008 (6) | 0.0051 (6) | −0.0017 (6) |
| C1 | 0.0166 (9) | 0.0130 (8) | 0.0164 (9) | 0.0025 (7) | 0.0099 (7) | 0.0014 (7) |
| O2 | 0.0136 (6) | 0.0167 (6) | 0.0178 (6) | −0.0006 (5) | 0.0040 (5) | −0.0017 (5) |
| C2 | 0.0147 (8) | 0.0139 (8) | 0.0145 (8) | −0.0028 (7) | 0.0061 (7) | 0.0000 (7) |
| O3 | 0.0296 (7) | 0.0216 (7) | 0.0148 (7) | 0.0044 (6) | 0.0078 (6) | 0.0005 (5) |
| C3 | 0.0165 (9) | 0.0145 (9) | 0.0199 (9) | 0.0022 (7) | 0.0087 (7) | 0.0006 (7) |
| O4 | 0.0196 (6) | 0.0139 (6) | 0.0158 (6) | −0.0004 (5) | 0.0062 (5) | −0.0028 (5) |
| C4 | 0.0213 (9) | 0.0141 (8) | 0.0189 (9) | −0.0002 (7) | 0.0106 (8) | −0.0022 (7) |
| O5 | 0.0266 (7) | 0.0251 (7) | 0.0167 (7) | 0.0076 (6) | 0.0084 (6) | 0.0043 (5) |
| C5 | 0.0176 (9) | 0.0186 (9) | 0.0138 (8) | −0.0040 (7) | 0.0065 (7) | −0.0013 (7) |
| C6 | 0.0152 (9) | 0.0179 (9) | 0.0158 (9) | 0.0003 (7) | 0.0075 (7) | 0.0019 (7) |
| C7 | 0.0158 (9) | 0.0143 (8) | 0.0158 (9) | 0.0009 (7) | 0.0093 (7) | 0.0016 (7) |
| C8 | 0.0185 (9) | 0.0143 (8) | 0.0146 (9) | 0.0008 (7) | 0.0103 (7) | 0.0010 (7) |
| C9 | 0.0154 (9) | 0.0165 (9) | 0.0183 (9) | −0.0005 (7) | 0.0074 (7) | −0.0011 (7) |
| C10 | 0.0116 (8) | 0.0154 (8) | 0.0172 (9) | 0.0019 (7) | 0.0059 (7) | −0.0003 (7) |
| C14 | 0.0131 (8) | 0.0175 (9) | 0.0193 (9) | 0.0013 (7) | 0.0091 (7) | −0.0015 (7) |
| C15 | 0.0258 (10) | 0.0202 (10) | 0.0167 (9) | −0.0030 (8) | 0.0060 (8) | −0.0082 (7) |
| C16 | 0.0334 (12) | 0.0202 (10) | 0.0264 (11) | 0.0014 (9) | 0.0034 (9) | −0.0064 (8) |
| C17 | 0.0172 (9) | 0.0186 (9) | 0.0124 (8) | 0.0006 (7) | 0.0068 (7) | 0.0005 (7) |
| C18 | 0.0128 (8) | 0.0260 (10) | 0.0161 (9) | −0.0030 (7) | 0.0036 (7) | −0.0009 (7) |
| C19 | 0.0187 (10) | 0.0322 (11) | 0.0250 (10) | 0.0017 (8) | 0.0100 (8) | 0.0047 (9) |
| C20 | 0.0249 (11) | 0.0277 (11) | 0.0309 (11) | −0.0083 (9) | 0.0068 (9) | −0.0066 (9) |
| C21 | 0.0235 (10) | 0.0381 (12) | 0.0178 (10) | −0.0044 (9) | 0.0043 (8) | 0.0046 (9) |
| S1A | 0.0180 (3) | 0.0143 (3) | 0.0192 (3) | 0.0051 (2) | 0.0095 (2) | 0.0010 (2) |
| C11A | 0.0184 (13) | 0.0143 (13) | 0.0291 (14) | 0.0058 (10) | 0.0080 (11) | 0.0014 (10) |
| C12A | 0.0206 (13) | 0.0209 (13) | 0.0189 (10) | 0.0016 (10) | 0.0096 (10) | 0.0000 (8) |
| C13A | 0.0188 (12) | 0.0188 (9) | 0.0142 (15) | 0.0031 (9) | 0.0102 (14) | −0.0018 (14) |
| S1B | 0.0180 (3) | 0.0143 (3) | 0.0192 (3) | 0.0051 (2) | 0.0095 (2) | 0.0010 (2) |
| C11B | 0.0184 (13) | 0.0143 (13) | 0.0291 (14) | 0.0058 (10) | 0.0080 (11) | 0.0014 (10) |
| C12B | 0.0188 (12) | 0.0188 (9) | 0.0142 (15) | 0.0031 (9) | 0.0102 (14) | −0.0018 (14) |
| C13B | 0.0206 (13) | 0.0209 (13) | 0.0189 (10) | 0.0016 (10) | 0.0096 (10) | 0.0000 (8) |
Geometric parameters (Å, º)
| Br1—C5 | 1.8993 (17) | C15—H15A | 0.9900 |
| O1—C17 | 1.202 (2) | C15—H15B | 0.9900 |
| N1—C1 | 1.402 (2) | C16—H16C | 0.9800 |
| N1—C2 | 1.406 (2) | C16—H16A | 0.9800 |
| N1—C17 | 1.415 (2) | C16—H16B | 0.9800 |
| C1—C8 | 1.362 (2) | C18—C21 | 1.510 (3) |
| C1—C14 | 1.491 (2) | C18—C19 | 1.514 (3) |
| O2—C17 | 1.317 (2) | C18—C20 | 1.517 (3) |
| O2—C18 | 1.501 (2) | C19—H19C | 0.9800 |
| C2—C3 | 1.3940 (15) | C19—H19A | 0.9800 |
| C2—C7 | 1.3968 (15) | C19—H19B | 0.9800 |
| O3—C14 | 1.199 (2) | C20—H20A | 0.9800 |
| C3—C4 | 1.3903 (16) | C20—H20B | 0.9800 |
| C3—H3 | 0.9500 | C20—H20C | 0.9800 |
| O4—C14 | 1.334 (2) | C21—H21A | 0.9800 |
| O4—C15 | 1.459 (2) | C21—H21C | 0.9800 |
| C4—C5 | 1.3909 (16) | C21—H21B | 0.9800 |
| C4—H4 | 0.9500 | S1A—C13A | 1.730 (3) |
| O5—C9 | 1.221 (2) | C11A—C12A | 1.400 (3) |
| C5—C6 | 1.3878 (15) | C11A—H11A | 0.9500 |
| C6—C7 | 1.3935 (16) | C12A—C13A | 1.367 (3) |
| C6—H6 | 0.9500 | C12A—H12A | 0.9500 |
| C7—C8 | 1.450 (2) | C13A—H13A | 0.9500 |
| C8—C9 | 1.489 (2) | S1B—C13B | 1.730 (3) |
| C9—C10 | 1.468 (2) | C11B—C12B | 1.400 (4) |
| C10—C11B | 1.344 (4) | C11B—H11B | 0.9500 |
| C10—C11A | 1.345 (3) | C12B—C13B | 1.367 (3) |
| C10—S1A | 1.7297 (17) | C12B—H12B | 0.9500 |
| C10—S1B | 1.730 (2) | C13B—H13B | 0.9500 |
| C15—C16 | 1.505 (3) | ||
| C1—N1—C2 | 107.77 (13) | H16C—C16—H16A | 109.5 |
| C1—N1—C17 | 126.56 (15) | C15—C16—H16B | 109.5 |
| C2—N1—C17 | 123.07 (13) | H16C—C16—H16B | 109.5 |
| C8—C1—N1 | 109.67 (15) | H16A—C16—H16B | 109.5 |
| C8—C1—C14 | 128.83 (16) | O1—C17—O2 | 128.62 (17) |
| N1—C1—C14 | 121.09 (15) | O1—C17—N1 | 121.44 (16) |
| C17—O2—C18 | 120.67 (14) | O2—C17—N1 | 109.86 (15) |
| C3—C2—C7 | 122.48 (14) | O2—C18—C21 | 109.18 (15) |
| C3—C2—N1 | 129.51 (13) | O2—C18—C19 | 109.43 (14) |
| C7—C2—N1 | 108.00 (12) | C21—C18—C19 | 113.28 (17) |
| C4—C3—C2 | 116.55 (15) | O2—C18—C20 | 101.33 (14) |
| C4—C3—H3 | 121.7 | C21—C18—C20 | 111.51 (16) |
| C2—C3—H3 | 121.7 | C19—C18—C20 | 111.42 (16) |
| C14—O4—C15 | 115.31 (14) | C18—C19—H19C | 109.5 |
| C3—C4—C5 | 121.11 (15) | C18—C19—H19A | 109.5 |
| C3—C4—H4 | 119.4 | H19C—C19—H19A | 109.5 |
| C5—C4—H4 | 119.4 | C18—C19—H19B | 109.5 |
| C6—C5—C4 | 122.36 (15) | H19C—C19—H19B | 109.5 |
| C6—C5—Br1 | 119.51 (11) | H19A—C19—H19B | 109.5 |
| C4—C5—Br1 | 118.13 (11) | C18—C20—H20A | 109.5 |
| C5—C6—C7 | 117.02 (15) | C18—C20—H20B | 109.5 |
| C5—C6—H6 | 121.5 | H20A—C20—H20B | 109.5 |
| C7—C6—H6 | 121.5 | C18—C20—H20C | 109.5 |
| C6—C7—C2 | 120.47 (14) | H20A—C20—H20C | 109.5 |
| C6—C7—C8 | 132.37 (15) | H20B—C20—H20C | 109.5 |
| C2—C7—C8 | 107.16 (14) | C18—C21—H21A | 109.5 |
| C1—C8—C7 | 107.38 (15) | C18—C21—H21C | 109.5 |
| C1—C8—C9 | 126.85 (16) | H21A—C21—H21C | 109.5 |
| C7—C8—C9 | 125.72 (15) | C18—C21—H21B | 109.5 |
| O5—C9—C10 | 122.43 (16) | H21A—C21—H21B | 109.5 |
| O5—C9—C8 | 119.94 (16) | H21C—C21—H21B | 109.5 |
| C10—C9—C8 | 117.62 (15) | C10—S1A—C13A | 90.96 (18) |
| C11B—C10—C11A | 104.3 (12) | C10—C11A—C12A | 115.3 (3) |
| C11B—C10—C9 | 125.8 (12) | C10—C11A—H11A | 122.3 |
| C11A—C10—C9 | 129.86 (16) | C12A—C11A—H11A | 122.3 |
| C11A—C10—S1A | 110.60 (15) | C13A—C12A—C11A | 111.1 (4) |
| C9—C10—S1A | 119.53 (12) | C13A—C12A—H12A | 124.5 |
| C11B—C10—S1B | 112.0 (12) | C11A—C12A—H12A | 124.5 |
| C9—C10—S1B | 122.2 (3) | C12A—C13A—S1A | 112.1 (4) |
| S1A—C10—S1B | 118.3 (3) | C12A—C13A—H13A | 124.0 |
| O3—C14—O4 | 125.80 (16) | S1A—C13A—H13A | 124.0 |
| O3—C14—C1 | 124.23 (16) | C13B—S1B—C10 | 90.7 (19) |
| O4—C14—C1 | 109.90 (15) | C10—C11B—C12B | 113 (3) |
| O4—C15—C16 | 106.54 (15) | C10—C11B—H11B | 123.4 |
| O4—C15—H15A | 110.4 | C12B—C11B—H11B | 123.4 |
| C16—C15—H15A | 110.4 | C13B—C12B—C11B | 113 (4) |
| O4—C15—H15B | 110.4 | C13B—C12B—H12B | 123.6 |
| C16—C15—H15B | 110.4 | C11B—C12B—H12B | 123.6 |
| H15A—C15—H15B | 108.6 | C12B—C13B—S1B | 111 (4) |
| C15—C16—H16C | 109.5 | C12B—C13B—H13B | 124.4 |
| C15—C16—H16A | 109.5 | S1B—C13B—H13B | 124.4 |
| C2—N1—C1—C8 | −0.25 (19) | C8—C9—C10—S1B | −19.6 (4) |
| C17—N1—C1—C8 | 161.74 (16) | C15—O4—C14—O3 | −6.9 (2) |
| C2—N1—C1—C14 | 173.05 (15) | C15—O4—C14—C1 | 170.13 (14) |
| C17—N1—C1—C14 | −25.0 (3) | C8—C1—C14—O3 | 127.0 (2) |
| C1—N1—C2—C3 | 179.52 (17) | N1—C1—C14—O3 | −44.9 (3) |
| C17—N1—C2—C3 | 16.8 (3) | C8—C1—C14—O4 | −50.1 (2) |
| C1—N1—C2—C7 | 1.06 (19) | N1—C1—C14—O4 | 138.06 (16) |
| C17—N1—C2—C7 | −161.70 (15) | C14—O4—C15—C16 | 179.51 (15) |
| C7—C2—C3—C4 | −1.0 (3) | C18—O2—C17—O1 | −6.5 (3) |
| N1—C2—C3—C4 | −179.25 (17) | C18—O2—C17—N1 | 176.60 (14) |
| C2—C3—C4—C5 | 0.3 (3) | C1—N1—C17—O1 | 165.06 (17) |
| C3—C4—C5—C6 | 0.7 (3) | C2—N1—C17—O1 | −35.5 (3) |
| C3—C4—C5—Br1 | −178.72 (14) | C1—N1—C17—O2 | −17.8 (2) |
| C4—C5—C6—C7 | −1.1 (3) | C2—N1—C17—O2 | 141.64 (15) |
| Br1—C5—C6—C7 | 178.34 (12) | C17—O2—C18—C21 | −61.6 (2) |
| C5—C6—C7—C2 | 0.4 (2) | C17—O2—C18—C19 | 62.9 (2) |
| C5—C6—C7—C8 | −178.75 (17) | C17—O2—C18—C20 | −179.30 (15) |
| C3—C2—C7—C6 | 0.6 (3) | C11B—C10—S1A—C13A | 24 (8) |
| N1—C2—C7—C6 | 179.21 (15) | C11A—C10—S1A—C13A | 0.89 (19) |
| C3—C2—C7—C8 | 179.98 (16) | C9—C10—S1A—C13A | −179.27 (18) |
| N1—C2—C7—C8 | −1.43 (19) | S1B—C10—S1A—C13A | 0.2 (3) |
| N1—C1—C8—C7 | −0.63 (19) | C11B—C10—C11A—C12A | −3.2 (11) |
| C14—C1—C8—C7 | −173.26 (16) | C9—C10—C11A—C12A | 179.8 (2) |
| N1—C1—C8—C9 | −178.36 (16) | S1A—C10—C11A—C12A | −0.4 (3) |
| C14—C1—C8—C9 | 9.0 (3) | S1B—C10—C11A—C12A | 175 (3) |
| C6—C7—C8—C1 | −179.46 (18) | C10—C11A—C12A—C13A | −0.5 (3) |
| C2—C7—C8—C1 | 1.28 (19) | C11A—C12A—C13A—S1A | 1.2 (2) |
| C6—C7—C8—C9 | −1.7 (3) | C10—S1A—C13A—C12A | −1.20 (17) |
| C2—C7—C8—C9 | 179.04 (16) | C11B—C10—S1B—C13B | 0.0 (4) |
| C1—C8—C9—O5 | 131.0 (2) | C11A—C10—S1B—C13B | −2 (3) |
| C7—C8—C9—O5 | −46.3 (3) | C9—C10—S1B—C13B | −177.5 (11) |
| C1—C8—C9—C10 | −49.4 (3) | S1A—C10—S1B—C13B | 3.1 (10) |
| C7—C8—C9—C10 | 133.27 (18) | C11A—C10—C11B—C12B | 0.3 (6) |
| O5—C9—C10—C11B | −17.3 (12) | C9—C10—C11B—C12B | 177.4 (11) |
| C8—C9—C10—C11B | 163.2 (11) | S1A—C10—C11B—C12B | −157 (8) |
| O5—C9—C10—C11A | 159.1 (2) | S1B—C10—C11B—C12B | 0.0 (5) |
| C8—C9—C10—C11A | −20.4 (3) | C10—C11B—C12B—C13B | 0.0 (3) |
| O5—C9—C10—S1A | −20.7 (2) | C11B—C12B—C13B—S1B | 0.0 (2) |
| C8—C9—C10—S1A | 159.78 (13) | C10—S1B—C13B—C12B | 0.0 (3) |
| O5—C9—C10—S1B | 159.9 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C12A—H12A···O3i | 0.95 | 2.48 | 3.418 (5) | 169 |
Symmetry code: (i) −x, y, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: JJ2160).
References
- Atwood, J. L. & Barbour, L. J. (2003). Cryst. Growth Des. 3, 3–8.
- Barbour, L. J. (2001). J. Supramol. Chem. 1, 189–191.
- Beddoes, R. L., Dalton, L., Joule, J. A., Mills, O. S., Street, J. D. & Watt, C. I. F. (1986). J. Chem. Soc. Perkin Trans. 2, pp. 787–797.
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Hassam, M., Basson, A. E., Liotta, D. C., Morris, L., van Otterlo, W. A. L. & Pelly, S. C. (2012). ACS Med. Chem. Lett. 3, 470–475. [DOI] [PMC free article] [PubMed]
- Hassam, M. & Smith, V. J. (2012). Acta Cryst. E68, o3357. [DOI] [PMC free article] [PubMed]
- Reynolds, C., Koning, C. B., Pelly, S. C., van Otterlo, W. A. L. & Bode, M. L. (2012). Chem. Soc. Rev. 41, 4657–4670. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813000809/jj2160sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813000809/jj2160Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813000809/jj2160Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report