Abstract
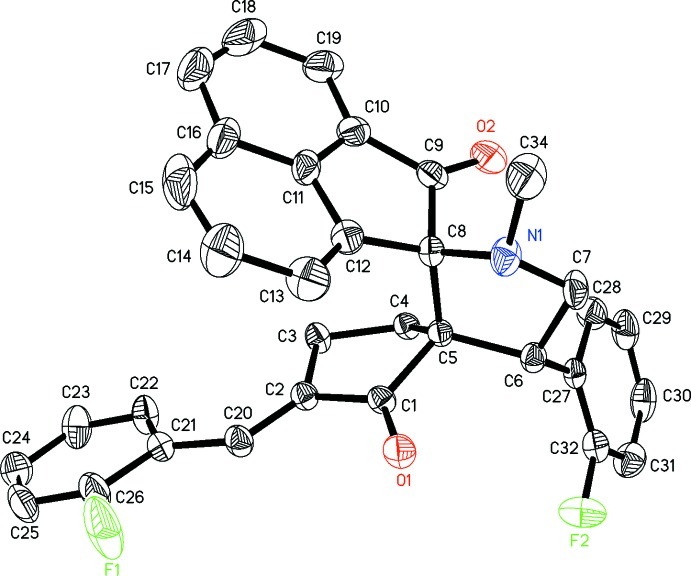
In the title compound, C33H25F2NO2, the acenaphthene ring system forms dihedral angles of 50.93 (14) and 36.89 (14)° with the benzene rings. The pyrrolidine and cyclopentanone rings adopt envelope (with the N atom as the flap) and twisted conformations, respectively. In the crystal, C—H⋯O and C—H⋯F interactions link the molecules.
Related literature
For related structures, see: Abdul Ajees et al. (2002 ▶); Usha et al. (2003 ▶). For background to the biological properties of spiro-pyrrolidine derivatives, see: Chande et al. (2005 ▶); Dandia et al. (2003 ▶); Cravotto et al. (2001 ▶); Winfred et al. (2000 ▶); Metwally et al. (1998 ▶); Suenaga et al. (2001 ▶). For the synthesis of the title compound, see: Kumar et al. (2008a
▶,b
▶); Liang et al. (2009 ▶).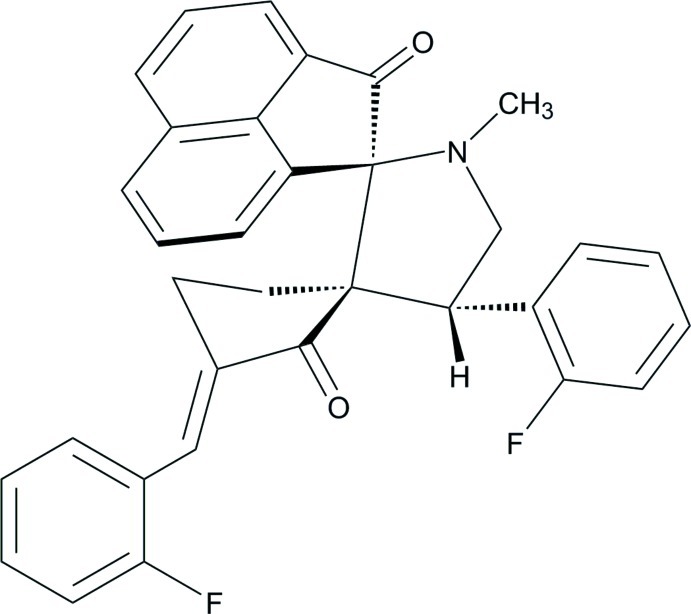
Experimental
Crystal data
C33H25F2NO2
M r = 505.54
Orthorhombic,
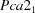
a = 17.728 (13) Å
b = 12.272 (9) Å
c = 12.094 (8) Å
V = 2631 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 293 K
0.45 × 0.38 × 0.27 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2002 ▶) T min = 0.641, T max = 1.000
12188 measured reflections
2500 independent reflections
2072 reflections with I > 2σ(I)
R int = 0.123
Refinement
R[F 2 > 2σ(F 2)] = 0.055
wR(F 2) = 0.126
S = 1.02
2500 reflections
344 parameters
13 restraints
H-atom parameters constrained
Δρmax = 0.24 e Å−3
Δρmin = −0.32 e Å−3
Data collection: SMART (Bruker, 2002 ▶); cell refinement: SAINT (Bruker, 2002 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812051550/pk2458sup1.cif
Structure factors: contains datablock(s) cd20184. DOI: 10.1107/S1600536812051550/pk2458Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3—H3B⋯O1i | 0.97 | 2.35 | 3.317 (5) | 172 |
| C14—H14A⋯F2ii | 0.93 | 2.43 | 3.212 (7) | 141 |
| C25—H25⋯O2iii | 0.93 | 2.58 | 3.458 (7) | 158 |
Symmetry codes: (i) 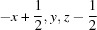 ; (ii)
; (ii) 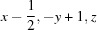 ; (iii)
; (iii)  .
.
Acknowledgments
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (grant Nos. LY12H16003 and Y4110197) and the Project of Wenzhou Science & Technology Bureau (Y20100273). The X-ray crystallographic facility at the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, is gratefully acknowledged for the data collection.
supplementary crystallographic information
Comment
Spiro-pyrrolidine compounds find applications in the synthesis of biologically active compounds. The synthesis of spiro compounds has drawn considerable attention of chemists, in view of their wide spectrum of pharmacological properties (Chande et al., 2005; Dandia et al., 2003; Cravotto et al., 2001; Winfred et al., 2000; Metwally et al., 1998; Suenaga et al., 2001). In the present study, the 1,3-dipolar cycloaddition of an azomethine ylide generated in situ from acenaphthenequinone and sarcosine to novel mono-carbonyl analogue of curcumin containg cyclopentanone afforded the title compound (Kumar et al., 2008; Liang et al., 2009). With this background, and in continuation of our structural analysis of spiro-pyrrolidine derivatives, the X-ray crystal structure determination of the title compound, (I), was undertaken.
The bond lengths and angles in the pyrrolidine ring are slightly larger than normal values because of bulky substituents on the pyrrolidine moiety. A similar effect has been observed in related reported structures (Abdul Ajees et al., 2002; Usha et al., 2003). The sum of the angles at atom N1 [339.1 (11)°] is in accordance with sp3-hybridization. The dihedral angles between the acenaphthene ring system and phenyl rings C21—C26 and C27—C32 are 50.93 (14)° and 36.89 (14)° respectively, while that between the two phenyl-ring substituents is 87.55 (17)°. The pyrrolidine and cyclopentanone ring both adopt an envelope conformation. In addition to van der Waals interactions, the crystal structure is stabilized by C—H···O and C—H···F intramolecular interactions. In the present study, the 1,3-dipolar cycloaddition of an azomethine ylide generated in situ from acenaphthenequinone and sarcosine to novel mono-carbonyl analogue of curcumin containg cyclopentanone afforded title compound.
Experimental
A mixture of (2E,5E)-2,5-bis(2-fluorobenzylidene)cyclopentanone (1 mmol), (Liang et al., 2009), acenaphthenequinone (0.182 g, 1 mmol), and sarcosine (0.089 g, 1 mmol) was dissolved in methanol (10 mL) and refluxed for 1 h. After completion of the reaction as evident from TLC, the mixture was cooled to room temperature and poured into cold water (50 mL). The precipitate was filtered and washed with water to obtain pure product as a yellow solid (79.6% yield, mp 93.2–95.0°C). Single crystals were grown in an ethyl acetate/CH2Cl2 mixture (2:1ν/ν).
Refinement
The H(C) atom positions were calculated. The H atoms bound to C were positioned geometrically and allowed to ride on their parent atoms at distances of 0.96 Å (RCH3), 0.97 Å (R2CH2), 0.98 Å (R3CH), 0.93 Å (R2CH), and with Uiso(H) values set to either 1.2Ueq or 1.5Ueq (RCH3) of the attached atom.
Figures
Fig. 1.
The molecular structure of the title compound, showing 30% displacement ellipsoids for the non-hydrogen atoms. Hydrogen atoms are drawn as spheres of arbitrary radius.
Crystal data
| C33H25F2NO2 | F(000) = 1052 |
| Mr = 505.54 | Dx = 1.274 Mg m−3 |
| Orthorhombic, Pca21 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2c -2ac | Cell parameters from 3224 reflections |
| a = 17.728 (13) Å | θ = 4.6–41.9° |
| b = 12.272 (9) Å | µ = 0.09 mm−1 |
| c = 12.094 (8) Å | T = 293 K |
| V = 2631 (3) Å3 | Prismatic, yellow |
| Z = 4 | 0.45 × 0.38 × 0.27 mm |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2500 independent reflections |
| Radiation source: fine-focus sealed tube | 2072 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.123 |
| φ and ω scans | θmax = 25.5°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2002) | h = −21→20 |
| Tmin = 0.641, Tmax = 1.000 | k = −14→10 |
| 12188 measured reflections | l = −14→13 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.055 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.126 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0837P)2] where P = (Fo2 + 2Fc2)/3 |
| 2500 reflections | (Δ/σ)max < 0.001 |
| 344 parameters | Δρmax = 0.24 e Å−3 |
| 13 restraints | Δρmin = −0.32 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F1 | 0.1562 (3) | 0.1946 (2) | 0.9893 (3) | 0.1212 (15) | |
| F2 | 0.37848 (15) | 0.7382 (2) | 1.1356 (2) | 0.0723 (8) | |
| O1 | 0.20514 (17) | 0.54755 (19) | 1.1294 (2) | 0.0503 (7) | |
| O2 | 0.08624 (18) | 0.9035 (2) | 0.9244 (3) | 0.0656 (9) | |
| N1 | 0.0970 (2) | 0.7803 (3) | 1.1430 (3) | 0.0494 (8) | |
| C1 | 0.1917 (2) | 0.5827 (3) | 1.0368 (3) | 0.0351 (8) | |
| C2 | 0.1848 (2) | 0.5181 (3) | 0.9340 (3) | 0.0367 (8) | |
| C3 | 0.1787 (2) | 0.5964 (3) | 0.8387 (3) | 0.0390 (8) | |
| H3A | 0.1272 | 0.6007 | 0.8120 | 0.047* | |
| H3B | 0.2113 | 0.5745 | 0.7783 | 0.047* | |
| C4 | 0.2041 (2) | 0.7055 (2) | 0.8874 (3) | 0.0351 (8) | |
| H4A | 0.1802 | 0.7655 | 0.8486 | 0.042* | |
| H4B | 0.2583 | 0.7135 | 0.8812 | 0.042* | |
| C5 | 0.18001 (19) | 0.7045 (2) | 1.0097 (3) | 0.0329 (7) | |
| C6 | 0.2244 (2) | 0.7800 (3) | 1.0916 (3) | 0.0376 (8) | |
| H6 | 0.2461 | 0.7324 | 1.1483 | 0.045* | |
| C7 | 0.1640 (2) | 0.8486 (3) | 1.1487 (3) | 0.0515 (10) | |
| H7A | 0.1777 | 0.8636 | 1.2249 | 0.062* | |
| H7B | 0.1562 | 0.9170 | 1.1103 | 0.062* | |
| C8 | 0.0945 (2) | 0.7332 (3) | 1.0318 (3) | 0.0376 (8) | |
| C9 | 0.0633 (2) | 0.8121 (3) | 0.9403 (3) | 0.0460 (9) | |
| C10 | 0.0007 (2) | 0.7571 (4) | 0.8828 (3) | 0.0520 (11) | |
| C11 | −0.0125 (2) | 0.6583 (3) | 0.9373 (3) | 0.0501 (10) | |
| C12 | 0.0375 (2) | 0.6408 (3) | 1.0259 (3) | 0.0467 (9) | |
| C13 | 0.0260 (3) | 0.5518 (4) | 1.0922 (4) | 0.0690 (13) | |
| H14 | 0.0563 | 0.5400 | 1.1538 | 0.083* | |
| C14 | −0.0322 (4) | 0.4789 (5) | 1.0659 (6) | 0.0884 (17) | |
| H14A | −0.0390 | 0.4182 | 1.1109 | 0.106* | |
| C15 | −0.0788 (3) | 0.4923 (4) | 0.9792 (6) | 0.093 (2) | |
| H15 | −0.1158 | 0.4407 | 0.9646 | 0.112* | |
| C16 | −0.0713 (3) | 0.5851 (4) | 0.9099 (4) | 0.0718 (14) | |
| C17 | −0.1165 (3) | 0.6177 (6) | 0.8195 (6) | 0.094 (2) | |
| H17 | −0.1559 | 0.5731 | 0.7967 | 0.113* | |
| C18 | −0.1034 (3) | 0.7133 (7) | 0.7648 (5) | 0.098 (2) | |
| H18 | −0.1339 | 0.7309 | 0.7048 | 0.118* | |
| C19 | −0.0455 (3) | 0.7867 (4) | 0.7954 (4) | 0.0719 (15) | |
| H19 | −0.0387 | 0.8523 | 0.7582 | 0.086* | |
| C20 | 0.1819 (2) | 0.4093 (3) | 0.9397 (3) | 0.0430 (8) | |
| H21 | 0.1849 | 0.3811 | 1.0109 | 0.052* | |
| C21 | 0.1747 (2) | 0.3280 (3) | 0.8532 (3) | 0.0452 (9) | |
| C22 | 0.1803 (3) | 0.3482 (3) | 0.7384 (3) | 0.0562 (11) | |
| H22 | 0.1859 | 0.4196 | 0.7140 | 0.067* | |
| C23 | 0.1776 (3) | 0.2659 (4) | 0.6621 (4) | 0.0689 (14) | |
| H23 | 0.1820 | 0.2824 | 0.5873 | 0.083* | |
| C24 | 0.1685 (3) | 0.1603 (4) | 0.6939 (5) | 0.0726 (14) | |
| H24 | 0.1678 | 0.1048 | 0.6415 | 0.087* | |
| C25 | 0.1604 (4) | 0.1365 (4) | 0.8058 (5) | 0.0829 (18) | |
| H25 | 0.1528 | 0.0652 | 0.8294 | 0.099* | |
| C26 | 0.1639 (3) | 0.2197 (3) | 0.8797 (4) | 0.0676 (14) | |
| C27 | 0.2887 (2) | 0.8456 (3) | 1.0442 (3) | 0.0374 (8) | |
| C28 | 0.2768 (3) | 0.9352 (3) | 0.9739 (3) | 0.0533 (11) | |
| H28 | 0.2278 | 0.9556 | 0.9559 | 0.064* | |
| C29 | 0.3368 (3) | 0.9934 (3) | 0.9313 (4) | 0.0632 (12) | |
| H29 | 0.3274 | 1.0522 | 0.8849 | 0.076* | |
| C30 | 0.4090 (3) | 0.9666 (4) | 0.9558 (4) | 0.0647 (13) | |
| H30 | 0.4485 | 1.0070 | 0.9263 | 0.078* | |
| C31 | 0.4239 (3) | 0.8797 (4) | 1.0239 (4) | 0.0614 (11) | |
| H31 | 0.4731 | 0.8598 | 1.0412 | 0.074* | |
| C32 | 0.3623 (2) | 0.8225 (3) | 1.0661 (3) | 0.0457 (9) | |
| C33 | 0.0275 (3) | 0.8304 (4) | 1.1823 (4) | 0.0793 (15) | |
| H33A | 0.0325 | 0.8479 | 1.2593 | 0.119* | |
| H33B | −0.0136 | 0.7805 | 1.1724 | 0.119* | |
| H33C | 0.0179 | 0.8958 | 1.1410 | 0.119* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.251 (5) | 0.0477 (15) | 0.0652 (18) | −0.031 (2) | −0.035 (2) | 0.0173 (15) |
| F2 | 0.0645 (19) | 0.0727 (17) | 0.0798 (18) | 0.0172 (13) | 0.0031 (14) | 0.0249 (15) |
| O1 | 0.078 (2) | 0.0374 (14) | 0.0354 (13) | −0.0033 (12) | −0.0117 (13) | 0.0106 (11) |
| O2 | 0.073 (2) | 0.0410 (16) | 0.082 (2) | 0.0067 (14) | −0.0102 (17) | 0.0123 (16) |
| N1 | 0.050 (2) | 0.060 (2) | 0.0389 (16) | −0.0045 (15) | 0.0072 (14) | −0.0143 (15) |
| C1 | 0.041 (2) | 0.0303 (17) | 0.0343 (18) | −0.0005 (14) | 0.0016 (15) | 0.0056 (15) |
| C2 | 0.043 (2) | 0.0327 (18) | 0.0347 (17) | −0.0015 (14) | 0.0011 (15) | 0.0035 (15) |
| C3 | 0.057 (3) | 0.0307 (18) | 0.0296 (16) | −0.0015 (15) | 0.0021 (16) | 0.0000 (14) |
| C4 | 0.045 (2) | 0.0305 (17) | 0.0303 (16) | 0.0003 (14) | 0.0002 (14) | 0.0042 (14) |
| C5 | 0.039 (2) | 0.0298 (17) | 0.0298 (16) | 0.0002 (14) | −0.0024 (14) | −0.0003 (14) |
| C6 | 0.046 (2) | 0.0338 (17) | 0.0326 (16) | −0.0041 (14) | −0.0044 (16) | −0.0004 (14) |
| C7 | 0.058 (3) | 0.054 (2) | 0.043 (2) | −0.0043 (18) | 0.0014 (18) | −0.0198 (19) |
| C8 | 0.041 (2) | 0.0350 (18) | 0.0366 (17) | −0.0028 (14) | −0.0019 (16) | −0.0044 (15) |
| C9 | 0.052 (3) | 0.039 (2) | 0.046 (2) | 0.0113 (17) | 0.0017 (19) | −0.0034 (17) |
| C10 | 0.049 (3) | 0.063 (3) | 0.045 (2) | 0.0167 (19) | −0.0042 (19) | −0.0184 (19) |
| C11 | 0.044 (2) | 0.054 (2) | 0.053 (2) | −0.0005 (16) | 0.0025 (19) | −0.024 (2) |
| C12 | 0.043 (2) | 0.045 (2) | 0.052 (2) | −0.0027 (16) | 0.0068 (19) | −0.0049 (18) |
| C13 | 0.057 (3) | 0.068 (3) | 0.082 (3) | −0.012 (2) | 0.017 (2) | 0.017 (3) |
| C14 | 0.075 (4) | 0.076 (4) | 0.114 (5) | −0.032 (3) | 0.013 (4) | 0.007 (3) |
| C15 | 0.078 (4) | 0.069 (3) | 0.133 (5) | −0.038 (3) | 0.027 (4) | −0.028 (4) |
| C16 | 0.047 (3) | 0.086 (4) | 0.082 (3) | −0.005 (2) | 0.001 (2) | −0.043 (3) |
| C17 | 0.067 (4) | 0.115 (5) | 0.101 (5) | −0.001 (3) | −0.019 (3) | −0.060 (4) |
| C18 | 0.077 (4) | 0.149 (6) | 0.069 (4) | 0.031 (4) | −0.037 (3) | −0.055 (4) |
| C19 | 0.071 (4) | 0.094 (4) | 0.050 (2) | 0.031 (3) | −0.014 (2) | −0.018 (2) |
| C20 | 0.055 (2) | 0.0339 (19) | 0.0407 (18) | −0.0037 (15) | −0.0023 (17) | 0.0052 (16) |
| C21 | 0.048 (2) | 0.035 (2) | 0.053 (2) | −0.0003 (16) | −0.0060 (18) | −0.0006 (17) |
| C22 | 0.073 (3) | 0.043 (2) | 0.052 (2) | −0.016 (2) | 0.004 (2) | −0.0028 (19) |
| C23 | 0.080 (4) | 0.068 (3) | 0.058 (3) | −0.020 (2) | 0.009 (2) | −0.018 (2) |
| C24 | 0.081 (4) | 0.057 (3) | 0.080 (4) | 0.011 (2) | −0.018 (3) | −0.032 (3) |
| C25 | 0.131 (5) | 0.032 (2) | 0.085 (4) | 0.004 (2) | −0.040 (3) | −0.004 (3) |
| C26 | 0.109 (4) | 0.035 (2) | 0.059 (3) | −0.004 (2) | −0.024 (3) | 0.008 (2) |
| C27 | 0.048 (2) | 0.0332 (18) | 0.0313 (16) | −0.0048 (14) | −0.0008 (15) | −0.0074 (15) |
| C28 | 0.067 (3) | 0.040 (2) | 0.052 (2) | −0.0045 (18) | −0.006 (2) | 0.0060 (18) |
| C29 | 0.090 (4) | 0.038 (2) | 0.061 (3) | −0.022 (2) | −0.003 (3) | 0.002 (2) |
| C30 | 0.083 (4) | 0.061 (3) | 0.050 (3) | −0.037 (2) | 0.009 (2) | −0.011 (2) |
| C31 | 0.050 (3) | 0.073 (3) | 0.061 (3) | −0.005 (2) | 0.004 (2) | −0.009 (2) |
| C32 | 0.053 (3) | 0.040 (2) | 0.044 (2) | −0.0025 (17) | 0.0018 (18) | −0.0059 (17) |
| C33 | 0.058 (3) | 0.106 (4) | 0.074 (3) | 0.000 (3) | 0.014 (3) | −0.039 (3) |
Geometric parameters (Å, º)
| F1—C26 | 1.368 (6) | C14—H14A | 0.9300 |
| F2—C32 | 1.362 (5) | C15—C16 | 1.421 (8) |
| O1—C1 | 1.224 (4) | C15—H15 | 0.9300 |
| O2—C9 | 1.209 (5) | C16—C17 | 1.413 (9) |
| N1—C7 | 1.455 (5) | C17—C18 | 1.367 (9) |
| N1—C33 | 1.456 (6) | C17—H17 | 0.9300 |
| N1—C8 | 1.465 (5) | C18—C19 | 1.415 (8) |
| C1—C2 | 1.478 (5) | C18—H18 | 0.9300 |
| C1—C5 | 1.545 (5) | C19—H19 | 0.9300 |
| C2—C20 | 1.338 (5) | C20—C21 | 1.451 (5) |
| C2—C3 | 1.505 (5) | C20—H21 | 0.9300 |
| C3—C4 | 1.530 (5) | C21—C26 | 1.380 (6) |
| C3—H3A | 0.9700 | C21—C22 | 1.414 (6) |
| C3—H3B | 0.9700 | C22—C23 | 1.369 (6) |
| C4—C5 | 1.539 (5) | C22—H22 | 0.9300 |
| C4—H4A | 0.9700 | C23—C24 | 1.361 (7) |
| C4—H4B | 0.9700 | C23—H23 | 0.9300 |
| C5—C6 | 1.568 (5) | C24—C25 | 1.392 (8) |
| C5—C8 | 1.580 (5) | C24—H24 | 0.9300 |
| C6—C27 | 1.508 (5) | C25—C26 | 1.359 (6) |
| C6—C7 | 1.527 (5) | C25—H25 | 0.9300 |
| C6—H6 | 0.9800 | C27—C32 | 1.362 (5) |
| C7—H7A | 0.9700 | C27—C28 | 1.405 (5) |
| C7—H7B | 0.9700 | C28—C29 | 1.381 (6) |
| C8—C12 | 1.520 (5) | C28—H28 | 0.9300 |
| C8—C9 | 1.571 (5) | C29—C30 | 1.353 (7) |
| C9—C10 | 1.473 (6) | C29—H29 | 0.9300 |
| C10—C19 | 1.386 (6) | C30—C31 | 1.373 (7) |
| C10—C11 | 1.400 (6) | C30—H30 | 0.9300 |
| C11—C12 | 1.408 (6) | C31—C32 | 1.395 (6) |
| C11—C16 | 1.415 (6) | C31—H31 | 0.9300 |
| C12—C13 | 1.370 (6) | C33—H33A | 0.9600 |
| C13—C14 | 1.402 (7) | C33—H33B | 0.9600 |
| C13—H14 | 0.9300 | C33—H33C | 0.9600 |
| C14—C15 | 1.344 (10) | ||
| C7—N1—C33 | 115.6 (3) | C13—C14—H14A | 118.3 |
| C7—N1—C8 | 107.2 (3) | C14—C15—C16 | 120.1 (5) |
| C33—N1—C8 | 116.1 (4) | C14—C15—H15 | 120.0 |
| O1—C1—C2 | 126.6 (3) | C16—C15—H15 | 120.0 |
| O1—C1—C5 | 124.1 (3) | C17—C16—C11 | 114.8 (6) |
| C2—C1—C5 | 109.3 (3) | C17—C16—C15 | 129.1 (5) |
| C20—C2—C1 | 119.7 (3) | C11—C16—C15 | 116.0 (5) |
| C20—C2—C3 | 132.3 (3) | C18—C17—C16 | 121.4 (5) |
| C1—C2—C3 | 107.9 (3) | C18—C17—H17 | 119.3 |
| C2—C3—C4 | 104.1 (3) | C16—C17—H17 | 119.3 |
| C2—C3—H3A | 110.9 | C17—C18—C19 | 122.9 (5) |
| C4—C3—H3A | 110.9 | C17—C18—H18 | 118.6 |
| C2—C3—H3B | 110.9 | C19—C18—H18 | 118.6 |
| C4—C3—H3B | 110.9 | C10—C19—C18 | 117.5 (6) |
| H3A—C3—H3B | 109.0 | C10—C19—H19 | 121.3 |
| C3—C4—C5 | 106.3 (3) | C18—C19—H19 | 121.3 |
| C3—C4—H4A | 110.5 | C2—C20—C21 | 130.8 (3) |
| C5—C4—H4A | 110.5 | C2—C20—H21 | 114.6 |
| C3—C4—H4B | 110.5 | C21—C20—H21 | 114.6 |
| C5—C4—H4B | 110.5 | C26—C21—C22 | 113.9 (4) |
| H4A—C4—H4B | 108.7 | C26—C21—C20 | 120.5 (4) |
| C4—C5—C1 | 100.0 (3) | C22—C21—C20 | 125.5 (4) |
| C4—C5—C6 | 117.6 (3) | C23—C22—C21 | 122.1 (4) |
| C1—C5—C6 | 111.8 (3) | C23—C22—H22 | 119.0 |
| C4—C5—C8 | 115.3 (3) | C21—C22—H22 | 119.0 |
| C1—C5—C8 | 108.0 (3) | C24—C23—C22 | 121.0 (5) |
| C6—C5—C8 | 104.1 (3) | C24—C23—H23 | 119.5 |
| C27—C6—C7 | 114.1 (3) | C22—C23—H23 | 119.5 |
| C27—C6—C5 | 117.0 (3) | C23—C24—C25 | 119.2 (4) |
| C7—C6—C5 | 105.1 (3) | C23—C24—H24 | 120.4 |
| C27—C6—H6 | 106.7 | C25—C24—H24 | 120.4 |
| C7—C6—H6 | 106.7 | C26—C25—C24 | 118.4 (5) |
| C5—C6—H6 | 106.7 | C26—C25—H25 | 120.8 |
| N1—C7—C6 | 103.5 (3) | C24—C25—H25 | 120.8 |
| N1—C7—H7A | 111.1 | C25—C26—F1 | 117.6 (4) |
| C6—C7—H7A | 111.1 | C25—C26—C21 | 125.3 (4) |
| N1—C7—H7B | 111.1 | F1—C26—C21 | 117.1 (4) |
| C6—C7—H7B | 111.1 | C32—C27—C28 | 115.1 (3) |
| H7A—C7—H7B | 109.0 | C32—C27—C6 | 122.6 (3) |
| N1—C8—C12 | 110.9 (3) | C28—C27—C6 | 122.2 (3) |
| N1—C8—C9 | 114.5 (3) | C29—C28—C27 | 120.9 (4) |
| C12—C8—C9 | 101.2 (3) | C29—C28—H28 | 119.5 |
| N1—C8—C5 | 102.4 (3) | C27—C28—H28 | 119.5 |
| C12—C8—C5 | 117.6 (3) | C30—C29—C28 | 121.4 (4) |
| C9—C8—C5 | 110.9 (3) | C30—C29—H29 | 119.3 |
| O2—C9—C10 | 127.2 (4) | C28—C29—H29 | 119.3 |
| O2—C9—C8 | 124.4 (4) | C29—C30—C31 | 120.1 (4) |
| C10—C9—C8 | 108.4 (3) | C29—C30—H30 | 119.9 |
| C19—C10—C11 | 119.2 (5) | C31—C30—H30 | 119.9 |
| C19—C10—C9 | 133.2 (5) | C30—C31—C32 | 117.4 (4) |
| C11—C10—C9 | 107.5 (3) | C30—C31—H31 | 121.3 |
| C10—C11—C12 | 112.7 (3) | C32—C31—H31 | 121.3 |
| C10—C11—C16 | 124.2 (4) | C27—C32—F2 | 118.7 (3) |
| C12—C11—C16 | 123.1 (4) | C27—C32—C31 | 125.0 (4) |
| C13—C12—C11 | 118.3 (4) | F2—C32—C31 | 116.3 (4) |
| C13—C12—C8 | 131.8 (4) | N1—C33—H33A | 109.5 |
| C11—C12—C8 | 109.9 (3) | N1—C33—H33B | 109.5 |
| C12—C13—C14 | 119.1 (5) | H33A—C33—H33B | 109.5 |
| C12—C13—H14 | 120.5 | N1—C33—H33C | 109.5 |
| C14—C13—H14 | 120.5 | H33A—C33—H33C | 109.5 |
| C15—C14—C13 | 123.4 (5) | H33B—C33—H33C | 109.5 |
| C15—C14—H14A | 118.3 | ||
| O1—C1—C2—C20 | 10.5 (6) | C16—C11—C12—C13 | −3.8 (6) |
| C5—C1—C2—C20 | −170.2 (3) | C10—C11—C12—C8 | −3.1 (5) |
| O1—C1—C2—C3 | −172.3 (4) | C16—C11—C12—C8 | 179.1 (4) |
| C5—C1—C2—C3 | 7.0 (4) | N1—C8—C12—C13 | −49.0 (5) |
| C20—C2—C3—C4 | −168.1 (4) | C9—C8—C12—C13 | −170.8 (4) |
| C1—C2—C3—C4 | 15.2 (4) | C5—C8—C12—C13 | 68.3 (6) |
| C2—C3—C4—C5 | −32.0 (4) | N1—C8—C12—C11 | 127.7 (3) |
| C3—C4—C5—C1 | 34.8 (4) | C9—C8—C12—C11 | 5.8 (4) |
| C3—C4—C5—C6 | 156.0 (3) | C5—C8—C12—C11 | −115.1 (3) |
| C3—C4—C5—C8 | −80.6 (3) | C11—C12—C13—C14 | 3.4 (6) |
| O1—C1—C5—C4 | 153.6 (4) | C8—C12—C13—C14 | 179.9 (5) |
| C2—C1—C5—C4 | −25.7 (3) | C12—C13—C14—C15 | −1.0 (9) |
| O1—C1—C5—C6 | 28.4 (5) | C13—C14—C15—C16 | −1.3 (10) |
| C2—C1—C5—C6 | −150.9 (3) | C10—C11—C16—C17 | 1.4 (6) |
| O1—C1—C5—C8 | −85.5 (4) | C12—C11—C16—C17 | 178.9 (4) |
| C2—C1—C5—C8 | 95.2 (3) | C10—C11—C16—C15 | −176.1 (4) |
| C4—C5—C6—C27 | −1.6 (4) | C12—C11—C16—C15 | 1.5 (6) |
| C1—C5—C6—C27 | 113.3 (3) | C14—C15—C16—C17 | −175.9 (6) |
| C8—C5—C6—C27 | −130.4 (3) | C14—C15—C16—C11 | 1.0 (8) |
| C4—C5—C6—C7 | 126.1 (3) | C11—C16—C17—C18 | −0.4 (8) |
| C1—C5—C6—C7 | −119.0 (3) | C15—C16—C17—C18 | 176.6 (6) |
| C8—C5—C6—C7 | −2.7 (3) | C16—C17—C18—C19 | −1.2 (9) |
| C33—N1—C7—C6 | −174.1 (4) | C11—C10—C19—C18 | −1.0 (6) |
| C8—N1—C7—C6 | −42.8 (4) | C9—C10—C19—C18 | −177.1 (4) |
| C27—C6—C7—N1 | 155.9 (3) | C17—C18—C19—C10 | 1.9 (8) |
| C5—C6—C7—N1 | 26.4 (4) | C1—C2—C20—C21 | 179.5 (4) |
| C7—N1—C8—C12 | 166.6 (3) | C3—C2—C20—C21 | 3.0 (7) |
| C33—N1—C8—C12 | −62.4 (5) | C2—C20—C21—C26 | −172.3 (4) |
| C7—N1—C8—C9 | −79.7 (4) | C2—C20—C21—C22 | 9.8 (7) |
| C33—N1—C8—C9 | 51.3 (5) | C26—C21—C22—C23 | −2.1 (7) |
| C7—N1—C8—C5 | 40.4 (4) | C20—C21—C22—C23 | 175.8 (4) |
| C33—N1—C8—C5 | 171.4 (4) | C21—C22—C23—C24 | 0.7 (7) |
| C4—C5—C8—N1 | −151.9 (3) | C22—C23—C24—C25 | 1.3 (8) |
| C1—C5—C8—N1 | 97.2 (3) | C23—C24—C25—C26 | −1.7 (9) |
| C6—C5—C8—N1 | −21.7 (3) | C24—C25—C26—F1 | −179.6 (6) |
| C4—C5—C8—C12 | 86.3 (4) | C24—C25—C26—C21 | 0.2 (9) |
| C1—C5—C8—C12 | −24.6 (4) | C22—C21—C26—C25 | 1.7 (7) |
| C6—C5—C8—C12 | −143.5 (3) | C20—C21—C26—C25 | −176.4 (5) |
| C4—C5—C8—C9 | −29.4 (4) | C22—C21—C26—F1 | −178.5 (5) |
| C1—C5—C8—C9 | −140.2 (3) | C20—C21—C26—F1 | 3.4 (7) |
| C6—C5—C8—C9 | 100.8 (3) | C7—C6—C27—C32 | 128.5 (4) |
| N1—C8—C9—O2 | 51.8 (5) | C5—C6—C27—C32 | −108.3 (4) |
| C12—C8—C9—O2 | 171.1 (4) | C7—C6—C27—C28 | −51.5 (4) |
| C5—C8—C9—O2 | −63.4 (4) | C5—C6—C27—C28 | 71.7 (4) |
| N1—C8—C9—C10 | −125.9 (3) | C32—C27—C28—C29 | 0.3 (5) |
| C12—C8—C9—C10 | −6.6 (4) | C6—C27—C28—C29 | −179.7 (4) |
| C5—C8—C9—C10 | 118.9 (3) | C27—C28—C29—C30 | −0.2 (7) |
| O2—C9—C10—C19 | 4.0 (7) | C28—C29—C30—C31 | 0.2 (7) |
| C8—C9—C10—C19 | −178.4 (4) | C29—C30—C31—C32 | −0.4 (6) |
| O2—C9—C10—C11 | −172.4 (4) | C28—C27—C32—F2 | 178.6 (3) |
| C8—C9—C10—C11 | 5.2 (4) | C6—C27—C32—F2 | −1.4 (5) |
| C19—C10—C11—C12 | −178.4 (4) | C28—C27—C32—C31 | −0.6 (5) |
| C9—C10—C11—C12 | −1.4 (5) | C6—C27—C32—C31 | 179.5 (3) |
| C19—C10—C11—C16 | −0.7 (6) | C30—C31—C32—C27 | 0.6 (6) |
| C9—C10—C11—C16 | 176.4 (4) | C30—C31—C32—F2 | −178.5 (3) |
| C10—C11—C12—C13 | 174.0 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3—H3B···O1i | 0.97 | 2.35 | 3.317 (5) | 172 |
| C14—H14A···F2ii | 0.93 | 2.43 | 3.212 (7) | 141 |
| C25—H25···O2iii | 0.93 | 2.58 | 3.458 (7) | 158 |
Symmetry codes: (i) −x+1/2, y, z−1/2; (ii) x−1/2, −y+1, z; (iii) x, y−1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: PK2458).
References
- Abdul Ajees, A., Manikandan, S. & Raghunathan, R. (2002). Acta Cryst. E58, o802–o804.
- Bruker (2002). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Chande, M. S., Verma, R. S., Barve, P. A. & Khanwelkar, R. R. (2005). Eur. J. Med. Chem. 40, 1143–1148. [DOI] [PubMed]
- Cravotto, G., Giovenzana, G. B., Pilati, T., Sisti, M. & Palmisano, G. (2001). J. Org. Chem. 66, 8447–8453. [DOI] [PubMed]
- Dandia, A., Sati, M., Arya, K., Sharma, R. & Loupy, A. (2003). Chem. Pharm. Bull. 51, 1137–1141. [DOI] [PubMed]
- Kumar, R. R., Perumal, S., Senthilkumar, P., Yogeeswari, P. & Sriram, D. (2008a). Tetrahedron, 64, 2962–2971.
- Kumar, R. R., Perumal, S., Senthilkumar, P., Yogeeswari, P. & Sriram, D. (2008b). J. Med. Chem. 51, 5731–5735. [DOI] [PubMed]
- Liang, G., Shao, L. L., Wang, Y., Zhao, C. G., Chu, Y. H., Xiao, J., Zhao, Y., Li, X. K. & Yang, S. L. (2009). Bioorg. Med. Chem. 17, 2623–2631. [DOI] [PubMed]
- Metwally, K. A., Dukat, M. & Egan, C. T. (1998). J. Med. Chem. 41, 5084–5093. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Suenaga, K., Araki, K. & Sengoku, T. (2001). Org. Lett. 3, 527–529. [DOI] [PubMed]
- Usha, G., Selvanayagam, S., Yogavel, M., Velmurugan, D., Amalraj, A., Raghunathan, R., Shanmuga Sundara Raj, S. & Fun, H.-K. (2003). Acta Cryst. E59, o1572–o1574.
- Winfred, G. B., Rutger, M. & Fieseler, F. (2000). J. Org. Chem. 65, 8317–8320.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812051550/pk2458sup1.cif
Structure factors: contains datablock(s) cd20184. DOI: 10.1107/S1600536812051550/pk2458Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report