Abstract
In the title salt, C26H27F2N2 +·C4H5O4 −, the piperazine N atom bearing the vinylic substituent is protonated. The piperazine ring adopts a chair conformation. In ther crystal, the succinate monoanions are connected via short O—H⋯O hydrogen bonds between the carboxylic acid and carboxylate groups into undulating chains extending along [001] and the flunarizinium monocations are attached to these chains via N+—H⋯O− hydrogen bonds. C—H⋯O interactions connect these chains into a three-dimensional network. The shortest centroid–centroid distance of 3.7256 (10) Å was found between one of the fluorinated benzene rings and the non-fluorinated phenyl ring in the neighbouring molecule related by a glide plane.
Related literature
For pharmaceutical properties of flunarizine, see: Holmes et al. (1984 ▶); Amery (1983 ▶) and of piperazine derivatives, see: Brockunier et al. (2004 ▶); Bogatcheva et al. (2006 ▶); Elliott (2011 ▶). For related structures, see: Betz et al. (2011a
▶,b
▶); Dayananda et al. (2012a
▶,b
▶); Fillers & Hawkinson (1982 ▶); Vanier & Brisse (1983 ▶). For puckering analysis of six-membered rings, see: Cremer & Pople (1975 ▶); Boeyens (1978 ▶). For graph-set analysis of hydrogen bonds, see: Etter et al. (1990 ▶); Bernstein et al. (1995 ▶).
Experimental
Crystal data
C26H27F2N2 +·C4H5O4 −
M r = 522.58
Monoclinic,
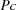
a = 10.7824 (2) Å
b = 10.6270 (2) Å
c = 11.2364 (2) Å
β = 91.678 (1)°
V = 1286.97 (4) Å3
Z = 2
Mo Kα radiation
μ = 0.10 mm−1
T = 200 K
0.56 × 0.29 × 0.16 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.947, T max = 0.985
16870 measured reflections
3619 independent reflections
3524 reflections with I > 2σ(I)
R int = 0.014
Refinement
R[F 2 > 2σ(F 2)] = 0.029
wR(F 2) = 0.081
S = 1.03
3619 reflections
348 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.25 e Å−3
Δρmin = −0.15 e Å−3
Data collection: APEX2 (Bruker, 2010 ▶); cell refinement: SAINT (Bruker, 2010 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 2012 ▶) and Mercury (Macrae et al., 2008 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813000706/gk2534sup1.cif
Supplementary material file. DOI: 10.1107/S1600536813000706/gk2534Isup2.cdx
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813000706/gk2534Isup3.hkl
Supplementary material file. DOI: 10.1107/S1600536813000706/gk2534Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H72⋯O1 | 0.96 (2) | 1.73 (2) | 2.6795 (16) | 174 (2) |
| O3—H3⋯O1i | 0.84 | 1.84 | 2.6564 (16) | 162 |
| C4—H4A⋯O4ii | 0.99 | 2.58 | 3.287 (3) | 128 |
| C4—H4B⋯O2iii | 0.99 | 2.39 | 3.3167 (19) | 155 |
| C25—H25⋯O2iv | 0.95 | 2.53 | 3.4168 (19) | 155 |
| C12—H12⋯Cg v | 0.95 | 2.81 | 3.7511 (17) | 170 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Acknowledgments
CNK thanks the University of Mysore for research facilities and the Principal of Maharani’s Science College for Women for permission to do research.
supplementary crystallographic information
Comment
Flunarizine is a drug classified as a calcium channel blocker (Amery, 1983). A review of its pharmacodynamic and pharmacokinetic properties and therapeutic use has been published (Holmes et al., 1984). Piperazines are among the most important building blocks in today's drug discovery and are found in biologically active compounds in a number of different therapeutic areas (Brockunier et al., 2004; Bogatcheva et al., 2006), and a review about the current pharmacological and toxicological information for piperazine derivatives is available (Elliott, 2011). The crystal structures of several related compounds are apparent in the literature (Fillers & Hawkinson, 1982; Vanier & Brisse, 1983; Betz et al., 2011a,b; Dayananda et al., 2012a,b). In continuation of our research about the salts of pharmacologically active compounds the title compound was synthesized and its crystal structure was determined.
Protonation of the flunarizine scaffold occurred on the nitrogen atom bearing the vinylic substituent. According to a puckering analysis (Cremer & Pople, 1975; Boeyens, 1978), the central 1,4-diazacyclohexane ring adopts a 4C1 conformation with both nitrogen atoms acting as the flap atoms (N2CN1). The C=C bond in the vinylic substituent is (E)-configured. The least-squares planes defined by the individual carbon atoms of the fluorinated phenyl moieties enclose an angle of 78.09 (8) °. The plane defined by the carbon atoms of the non-halogenated phenyl ring intersects at angles of 11.08 (8) ° and 87.51 (8) ° with the two aforementioned planes. The succinate monoanion is essentially flat (r.m.s. of all fitted non-hydrogen atoms = 0.0955 Å) with one of the carbon atoms of a methylene group deviating most from the common plane by 0.160 (1) Å (Fig. 1). The succinate monoanion adopts a zigzag conformation.
In the crystal, C–H···O contacts whose range falls by up to more than 0.3 Å below the sum of van-der-Waals radii of the atoms participating are observed next to classical hydrogen bonds of the O–H···O and N–H···O type. The C–H···O contacts are supported by both hydrogen atoms of an intracyclic methylene group bonded to the protonated nitrogen atom as well as one hydrogen atom in ortho-position to a fluorine atom in one of the fluorophenyl moieties as donors. The protonated carboxyl group forms a hydrogen bond to the deprotonated carboxyl group, the latter one also serving as acceptor for the N–H···O type hydrogen bonds. In addition, a C–H···π interaction involving one of the hydrogen atoms in meta-position to the fluorine atoms on the fluorophenyl moiety that does not contribute to the C–H···O contacts as described above as the donor and the aromtic system of the non-halogenated phenyl ring as the acceptor is apparent. Metrical parameters as well as information about the symmetry of these contacts are summarized in Table 1. In total, the entities of the title compound are connected to a three-dimensional network. In terms of graph-set analysis (Etter et al., 1990; Bernstein et al., 1995), the descriptor for these contacts is DC11(7) for the classical hydrogen bonds on the unary level. The C–H···O contacts necessitate a DDD descriptor on the same level. The shortest intercentroid distance between twomoiety aromatic systems was found at 3.7256 (10) Å and is apparent between one of the fluorinated and the non-halogenated phenyl moiety in neighbouring cations.
The packing of the title compound in the crystal structure is shown in Figure 2.
Experimental
Flunarizine (4.05 g, 0.01 mol) and succinic acid (1.18 g, 0.01 mol) were dissolved in hot N,N-dimethylformamide and reacted for 30 minutes. The resulting solution was allowed to cool slowly at room temperature upon which crystals of the title compound appeared in the course of several days. The latter were of sufficient quality for the X-ray diffraction studies.
Refinement
Carbon-bound H atoms were placed in calculated positions (C–H 0.95 Å for aromatic and vinylic carbon atoms, C–H 0.99 Å for methylene groups and C–H 1.00 Å for the methine group) and were included in the refinement in the riding model approximation, with U(H) set to 1.2Ueq(C). The H atom of the hydroxyl group was allowed to rotate with a fixed angle around the C–O bond to best fit the experimental electron density [HFIX 147 in the SHELX program suite (Sheldrick, 2008)] with U(H) set to 1.5Ueq(O). The nitrogen-bound H atom was located on a difference Fourier map and refined freely.
Figures
Fig. 1.
The molecular structure of the title compound, with atom labels and anisotropic displacement ellipsoids drawn at 50% probability level.
Fig. 2.
Molecular packing of the title compound, viewed along [0 0 1] (anisotropic displacement ellipsoids drawn at the 50% probability level).
Crystal data
| C26H27F2N2+·C4H5O4− | F(000) = 552 |
| Mr = 522.58 | Dx = 1.349 Mg m−3 |
| Monoclinic, Pc | Melting point = 383–378 K |
| Hall symbol: P -2yc | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.7824 (2) Å | Cell parameters from 9972 reflections |
| b = 10.6270 (2) Å | θ = 2.6–29.6° |
| c = 11.2364 (2) Å | µ = 0.10 mm−1 |
| β = 91.678 (1)° | T = 200 K |
| V = 1286.97 (4) Å3 | Rectangular, yellow |
| Z = 2 | 0.56 × 0.29 × 0.16 mm |
Data collection
| Bruker APEXII CCD diffractometer | 3619 independent reflections |
| Radiation source: fine-focus sealed tube | 3524 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.014 |
| φ and ω scans | θmax = 29.6°, θmin = 2.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −14→14 |
| Tmin = 0.947, Tmax = 0.985 | k = −14→14 |
| 16870 measured reflections | l = −15→15 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.029 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.081 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0532P)2 + 0.1548P] where P = (Fo2 + 2Fc2)/3 |
| 3619 reflections | (Δ/σ)max < 0.001 |
| 348 parameters | Δρmax = 0.25 e Å−3 |
| 2 restraints | Δρmin = −0.15 e Å−3 |
Special details
| Refinement. Due to the absence of a strong anomalous scatterer, the Flack parameter is meaningless. Thus, Friedel opposites (3016 pairs) have been merged and the item was removed from the CIF. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F1 | 0.60945 (15) | −0.00547 (14) | 0.96672 (13) | 0.0575 (3) | |
| F2 | 0.76769 (12) | 0.30206 (12) | 0.22338 (10) | 0.0453 (3) | |
| N1 | 0.39495 (10) | 0.42901 (11) | 0.63218 (10) | 0.0199 (2) | |
| N2 | 0.16828 (10) | 0.57409 (11) | 0.64797 (10) | 0.0205 (2) | |
| H72 | 0.192 (2) | 0.640 (2) | 0.7023 (19) | 0.028 (5)* | |
| C1 | 0.52807 (12) | 0.40104 (13) | 0.65297 (11) | 0.0204 (2) | |
| H1 | 0.5681 | 0.4768 | 0.6903 | 0.024* | |
| C2 | 0.32435 (13) | 0.43027 (13) | 0.74174 (12) | 0.0223 (2) | |
| H2A | 0.3558 | 0.4980 | 0.7950 | 0.027* | |
| H2B | 0.3351 | 0.3490 | 0.7839 | 0.027* | |
| C3 | 0.18784 (13) | 0.45191 (13) | 0.71164 (12) | 0.0222 (2) | |
| H3A | 0.1560 | 0.3822 | 0.6609 | 0.027* | |
| H3B | 0.1406 | 0.4521 | 0.7859 | 0.027* | |
| C4 | 0.24729 (12) | 0.57850 (13) | 0.54108 (11) | 0.0214 (2) | |
| H4A | 0.2400 | 0.6623 | 0.5031 | 0.026* | |
| H4B | 0.2181 | 0.5146 | 0.4825 | 0.026* | |
| C5 | 0.38119 (13) | 0.55330 (13) | 0.57621 (12) | 0.0220 (2) | |
| H5A | 0.4325 | 0.5572 | 0.5047 | 0.026* | |
| H5B | 0.4111 | 0.6190 | 0.6325 | 0.026* | |
| C6 | 0.03255 (13) | 0.58877 (15) | 0.61516 (13) | 0.0268 (3) | |
| H6A | 0.0047 | 0.5155 | 0.5669 | 0.032* | |
| H6B | −0.0159 | 0.5900 | 0.6886 | 0.032* | |
| C7 | 0.00768 (14) | 0.70704 (15) | 0.54620 (14) | 0.0282 (3) | |
| H7 | 0.0228 | 0.7860 | 0.5836 | 0.034* | |
| C8 | −0.03506 (14) | 0.70463 (14) | 0.43376 (14) | 0.0268 (3) | |
| H8 | −0.0467 | 0.6237 | 0.3993 | 0.032* | |
| C11 | 0.54868 (12) | 0.29049 (13) | 0.73680 (12) | 0.0216 (2) | |
| C12 | 0.60202 (14) | 0.31075 (15) | 0.84953 (13) | 0.0274 (3) | |
| H12 | 0.6246 | 0.3936 | 0.8733 | 0.033* | |
| C13 | 0.62266 (16) | 0.21099 (19) | 0.92779 (14) | 0.0351 (3) | |
| H13 | 0.6591 | 0.2248 | 1.0047 | 0.042* | |
| C14 | 0.58933 (17) | 0.09282 (18) | 0.89143 (16) | 0.0365 (4) | |
| C15 | 0.53618 (18) | 0.06793 (16) | 0.78097 (16) | 0.0364 (3) | |
| H15 | 0.5139 | −0.0154 | 0.7584 | 0.044* | |
| C16 | 0.51610 (15) | 0.16833 (14) | 0.70341 (14) | 0.0291 (3) | |
| H16 | 0.4797 | 0.1534 | 0.6267 | 0.035* | |
| C21 | 0.58957 (12) | 0.37651 (12) | 0.53494 (12) | 0.0212 (2) | |
| C22 | 0.52566 (13) | 0.32091 (13) | 0.43931 (13) | 0.0243 (2) | |
| H22 | 0.4402 | 0.3010 | 0.4460 | 0.029* | |
| C23 | 0.58533 (15) | 0.29394 (14) | 0.33373 (13) | 0.0279 (3) | |
| H23 | 0.5418 | 0.2559 | 0.2684 | 0.033* | |
| C24 | 0.70925 (15) | 0.32415 (15) | 0.32721 (14) | 0.0301 (3) | |
| C25 | 0.77684 (14) | 0.37818 (16) | 0.41996 (15) | 0.0317 (3) | |
| H25 | 0.8626 | 0.3964 | 0.4132 | 0.038* | |
| C26 | 0.71468 (13) | 0.40504 (15) | 0.52386 (13) | 0.0269 (3) | |
| H26 | 0.7586 | 0.4437 | 0.5886 | 0.032* | |
| C31 | −0.06628 (13) | 0.81225 (14) | 0.35691 (13) | 0.0257 (3) | |
| C32 | −0.05674 (15) | 0.93704 (15) | 0.39524 (15) | 0.0310 (3) | |
| H32 | −0.0249 | 0.9550 | 0.4731 | 0.037* | |
| C33 | −0.09348 (17) | 1.03489 (17) | 0.32034 (17) | 0.0369 (3) | |
| H33 | −0.0861 | 1.1194 | 0.3471 | 0.044* | |
| C34 | −0.14092 (17) | 1.00983 (18) | 0.20662 (17) | 0.0379 (4) | |
| H34 | −0.1675 | 1.0769 | 0.1562 | 0.046* | |
| C35 | −0.14940 (18) | 0.88675 (19) | 0.16704 (15) | 0.0378 (4) | |
| H35 | −0.1810 | 0.8693 | 0.0890 | 0.045* | |
| C36 | −0.11186 (16) | 0.78902 (16) | 0.24134 (14) | 0.0326 (3) | |
| H36 | −0.1172 | 0.7048 | 0.2132 | 0.039* | |
| O1 | 0.21803 (11) | 0.76463 (10) | 0.79808 (9) | 0.0295 (2) | |
| O2 | 0.09155 (11) | 0.65696 (11) | 0.91333 (10) | 0.0310 (2) | |
| O3 | 0.20616 (15) | 1.03647 (13) | 1.15502 (12) | 0.0423 (3) | |
| H3 | 0.2229 | 1.0913 | 1.2069 | 0.063* | |
| O4 | 0.35531 (19) | 1.13282 (17) | 1.06159 (18) | 0.0694 (6) | |
| C41 | 0.16219 (13) | 0.74667 (13) | 0.89574 (12) | 0.0226 (2) | |
| C42 | 0.18904 (14) | 0.83825 (13) | 0.99780 (12) | 0.0254 (3) | |
| H42A | 0.1093 | 0.8623 | 1.0328 | 0.030* | |
| H42B | 0.2387 | 0.7939 | 1.0602 | 0.030* | |
| C43 | 0.25742 (15) | 0.95766 (14) | 0.96455 (13) | 0.0279 (3) | |
| H43A | 0.3386 | 0.9337 | 0.9325 | 0.034* | |
| H43B | 0.2094 | 1.0003 | 0.8999 | 0.034* | |
| C44 | 0.27975 (16) | 1.05070 (14) | 1.06472 (14) | 0.0296 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.0687 (8) | 0.0515 (7) | 0.0524 (7) | 0.0115 (6) | 0.0015 (6) | 0.0322 (6) |
| F2 | 0.0511 (6) | 0.0496 (6) | 0.0364 (5) | 0.0085 (5) | 0.0228 (5) | 0.0009 (5) |
| N1 | 0.0200 (5) | 0.0214 (5) | 0.0185 (5) | 0.0035 (4) | 0.0028 (4) | 0.0027 (4) |
| N2 | 0.0216 (5) | 0.0220 (5) | 0.0178 (5) | 0.0036 (4) | 0.0005 (4) | −0.0015 (4) |
| C1 | 0.0204 (5) | 0.0204 (5) | 0.0203 (5) | 0.0017 (4) | 0.0001 (4) | 0.0002 (4) |
| C2 | 0.0239 (6) | 0.0246 (6) | 0.0184 (5) | 0.0038 (5) | 0.0023 (4) | 0.0026 (4) |
| C3 | 0.0246 (6) | 0.0226 (6) | 0.0197 (5) | 0.0025 (5) | 0.0035 (4) | 0.0017 (4) |
| C4 | 0.0234 (6) | 0.0248 (6) | 0.0160 (5) | 0.0035 (5) | 0.0021 (4) | 0.0011 (4) |
| C5 | 0.0228 (6) | 0.0212 (6) | 0.0221 (6) | 0.0020 (5) | 0.0017 (4) | 0.0033 (4) |
| C6 | 0.0204 (6) | 0.0346 (7) | 0.0253 (6) | 0.0044 (5) | 0.0001 (5) | 0.0004 (5) |
| C7 | 0.0267 (6) | 0.0290 (7) | 0.0288 (7) | 0.0081 (5) | −0.0017 (5) | −0.0033 (5) |
| C8 | 0.0254 (6) | 0.0271 (6) | 0.0277 (6) | 0.0009 (5) | −0.0005 (5) | −0.0010 (5) |
| C11 | 0.0207 (5) | 0.0235 (6) | 0.0205 (6) | 0.0036 (5) | 0.0010 (4) | 0.0019 (5) |
| C12 | 0.0264 (6) | 0.0332 (7) | 0.0226 (6) | 0.0017 (5) | −0.0002 (5) | 0.0007 (5) |
| C13 | 0.0315 (7) | 0.0496 (9) | 0.0242 (7) | 0.0048 (7) | −0.0009 (5) | 0.0089 (6) |
| C14 | 0.0359 (8) | 0.0392 (8) | 0.0348 (8) | 0.0085 (7) | 0.0061 (6) | 0.0178 (7) |
| C15 | 0.0461 (9) | 0.0245 (7) | 0.0388 (8) | 0.0033 (6) | 0.0047 (7) | 0.0073 (6) |
| C16 | 0.0361 (7) | 0.0233 (6) | 0.0278 (7) | 0.0015 (5) | −0.0005 (5) | 0.0017 (5) |
| C21 | 0.0223 (6) | 0.0198 (6) | 0.0215 (5) | 0.0031 (4) | 0.0027 (4) | 0.0029 (4) |
| C22 | 0.0238 (6) | 0.0236 (6) | 0.0257 (6) | 0.0004 (5) | 0.0018 (5) | 0.0004 (5) |
| C23 | 0.0353 (7) | 0.0244 (6) | 0.0240 (6) | 0.0043 (5) | 0.0027 (5) | −0.0003 (5) |
| C24 | 0.0353 (8) | 0.0272 (7) | 0.0285 (7) | 0.0082 (6) | 0.0118 (6) | 0.0054 (5) |
| C25 | 0.0251 (6) | 0.0320 (8) | 0.0383 (8) | 0.0043 (5) | 0.0082 (6) | 0.0072 (6) |
| C26 | 0.0221 (6) | 0.0281 (7) | 0.0304 (7) | 0.0012 (5) | 0.0006 (5) | 0.0041 (5) |
| C31 | 0.0217 (6) | 0.0297 (7) | 0.0258 (6) | −0.0004 (5) | 0.0000 (5) | 0.0006 (5) |
| C32 | 0.0303 (7) | 0.0314 (7) | 0.0311 (7) | −0.0006 (6) | −0.0012 (6) | −0.0020 (6) |
| C33 | 0.0388 (8) | 0.0285 (7) | 0.0434 (9) | 0.0013 (6) | −0.0005 (7) | 0.0014 (6) |
| C34 | 0.0354 (8) | 0.0396 (9) | 0.0387 (8) | 0.0021 (7) | −0.0011 (6) | 0.0096 (7) |
| C35 | 0.0412 (8) | 0.0432 (9) | 0.0286 (7) | −0.0014 (7) | −0.0055 (6) | 0.0041 (6) |
| C36 | 0.0358 (8) | 0.0339 (8) | 0.0280 (7) | −0.0027 (6) | −0.0029 (6) | −0.0013 (6) |
| O1 | 0.0443 (6) | 0.0237 (5) | 0.0207 (4) | −0.0016 (4) | 0.0052 (4) | −0.0010 (4) |
| O2 | 0.0343 (5) | 0.0262 (5) | 0.0328 (5) | −0.0049 (4) | 0.0033 (4) | −0.0012 (4) |
| O3 | 0.0591 (8) | 0.0372 (6) | 0.0314 (6) | −0.0147 (6) | 0.0143 (5) | −0.0140 (5) |
| O4 | 0.0814 (12) | 0.0525 (9) | 0.0770 (11) | −0.0382 (9) | 0.0448 (10) | −0.0354 (9) |
| C41 | 0.0273 (6) | 0.0185 (5) | 0.0219 (5) | 0.0044 (5) | −0.0009 (4) | 0.0002 (4) |
| C42 | 0.0345 (7) | 0.0219 (6) | 0.0200 (5) | −0.0021 (5) | 0.0049 (5) | −0.0017 (4) |
| C43 | 0.0377 (7) | 0.0217 (6) | 0.0248 (6) | −0.0033 (5) | 0.0075 (5) | −0.0025 (5) |
| C44 | 0.0356 (7) | 0.0221 (6) | 0.0312 (7) | −0.0006 (5) | 0.0061 (6) | −0.0041 (5) |
Geometric parameters (Å, º)
| F1—C14 | 1.3576 (18) | C15—C16 | 1.391 (2) |
| F2—C24 | 1.3625 (17) | C15—H15 | 0.9500 |
| N1—C2 | 1.4663 (16) | C16—H16 | 0.9500 |
| N1—C5 | 1.4686 (17) | C21—C22 | 1.3911 (19) |
| N1—C1 | 1.4776 (16) | C21—C26 | 1.3918 (19) |
| N2—C4 | 1.4934 (16) | C22—C23 | 1.3957 (19) |
| N2—C3 | 1.4944 (17) | C22—H22 | 0.9500 |
| N2—C6 | 1.5069 (17) | C23—C24 | 1.378 (2) |
| N2—H72 | 0.96 (2) | C23—H23 | 0.9500 |
| C1—C11 | 1.5181 (18) | C24—C25 | 1.379 (2) |
| C1—C21 | 1.5225 (18) | C25—C26 | 1.393 (2) |
| C1—H1 | 1.0000 | C25—H25 | 0.9500 |
| C2—C3 | 1.5182 (19) | C26—H26 | 0.9500 |
| C2—H2A | 0.9900 | C31—C36 | 1.397 (2) |
| C2—H2B | 0.9900 | C31—C32 | 1.397 (2) |
| C3—H3A | 0.9900 | C32—C33 | 1.388 (2) |
| C3—H3B | 0.9900 | C32—H32 | 0.9500 |
| C4—C5 | 1.5094 (19) | C33—C34 | 1.388 (3) |
| C4—H4A | 0.9900 | C33—H33 | 0.9500 |
| C4—H4B | 0.9900 | C34—C35 | 1.384 (3) |
| C5—H5A | 0.9900 | C34—H34 | 0.9500 |
| C5—H5B | 0.9900 | C35—C36 | 1.385 (2) |
| C6—C7 | 1.497 (2) | C35—H35 | 0.9500 |
| C6—H6A | 0.9900 | C36—H36 | 0.9500 |
| C6—H6B | 0.9900 | O1—C41 | 1.2813 (17) |
| C7—C8 | 1.332 (2) | O2—C41 | 1.2398 (19) |
| C7—H7 | 0.9500 | O3—C44 | 1.3150 (19) |
| C8—C31 | 1.466 (2) | O3—H3 | 0.8400 |
| C8—H8 | 0.9500 | O4—C44 | 1.195 (2) |
| C11—C12 | 1.3924 (19) | C41—C42 | 1.5254 (19) |
| C11—C16 | 1.394 (2) | C42—C43 | 1.520 (2) |
| C12—C13 | 1.391 (2) | C42—H42A | 0.9900 |
| C12—H12 | 0.9500 | C42—H42B | 0.9900 |
| C13—C14 | 1.366 (3) | C43—C44 | 1.512 (2) |
| C13—H13 | 0.9500 | C43—H43A | 0.9900 |
| C14—C15 | 1.377 (3) | C43—H43B | 0.9900 |
| C2—N1—C5 | 107.63 (10) | C13—C14—C15 | 123.04 (14) |
| C2—N1—C1 | 113.26 (10) | C14—C15—C16 | 118.06 (16) |
| C5—N1—C1 | 109.52 (10) | C14—C15—H15 | 121.0 |
| C4—N2—C3 | 109.66 (10) | C16—C15—H15 | 121.0 |
| C4—N2—C6 | 111.89 (10) | C15—C16—C11 | 120.84 (15) |
| C3—N2—C6 | 109.24 (11) | C15—C16—H16 | 119.6 |
| C4—N2—H72 | 110.2 (13) | C11—C16—H16 | 119.6 |
| C3—N2—H72 | 107.3 (13) | C22—C21—C26 | 118.88 (12) |
| C6—N2—H72 | 108.5 (13) | C22—C21—C1 | 121.80 (12) |
| N1—C1—C11 | 112.18 (11) | C26—C21—C1 | 119.25 (12) |
| N1—C1—C21 | 110.04 (10) | C21—C22—C23 | 120.90 (13) |
| C11—C1—C21 | 110.39 (11) | C21—C22—H22 | 119.6 |
| N1—C1—H1 | 108.0 | C23—C22—H22 | 119.6 |
| C11—C1—H1 | 108.0 | C24—C23—C22 | 117.97 (14) |
| C21—C1—H1 | 108.0 | C24—C23—H23 | 121.0 |
| N1—C2—C3 | 109.77 (10) | C22—C23—H23 | 121.0 |
| N1—C2—H2A | 109.7 | F2—C24—C23 | 118.61 (15) |
| C3—C2—H2A | 109.7 | F2—C24—C25 | 118.12 (14) |
| N1—C2—H2B | 109.7 | C23—C24—C25 | 123.26 (13) |
| C3—C2—H2B | 109.7 | C24—C25—C26 | 117.53 (14) |
| H2A—C2—H2B | 108.2 | C24—C25—H25 | 121.2 |
| N2—C3—C2 | 111.13 (11) | C26—C25—H25 | 121.2 |
| N2—C3—H3A | 109.4 | C21—C26—C25 | 121.46 (14) |
| C2—C3—H3A | 109.4 | C21—C26—H26 | 119.3 |
| N2—C3—H3B | 109.4 | C25—C26—H26 | 119.3 |
| C2—C3—H3B | 109.4 | C36—C31—C32 | 118.35 (14) |
| H3A—C3—H3B | 108.0 | C36—C31—C8 | 118.57 (14) |
| N2—C4—C5 | 110.36 (10) | C32—C31—C8 | 123.04 (13) |
| N2—C4—H4A | 109.6 | C33—C32—C31 | 120.47 (15) |
| C5—C4—H4A | 109.6 | C33—C32—H32 | 119.8 |
| N2—C4—H4B | 109.6 | C31—C32—H32 | 119.8 |
| C5—C4—H4B | 109.6 | C32—C33—C34 | 120.35 (17) |
| H4A—C4—H4B | 108.1 | C32—C33—H33 | 119.8 |
| N1—C5—C4 | 110.82 (11) | C34—C33—H33 | 119.8 |
| N1—C5—H5A | 109.5 | C35—C34—C33 | 119.75 (16) |
| C4—C5—H5A | 109.5 | C35—C34—H34 | 120.1 |
| N1—C5—H5B | 109.5 | C33—C34—H34 | 120.1 |
| C4—C5—H5B | 109.5 | C34—C35—C36 | 119.97 (16) |
| H5A—C5—H5B | 108.1 | C34—C35—H35 | 120.0 |
| C7—C6—N2 | 111.81 (12) | C36—C35—H35 | 120.0 |
| C7—C6—H6A | 109.3 | C35—C36—C31 | 121.09 (16) |
| N2—C6—H6A | 109.3 | C35—C36—H36 | 119.5 |
| C7—C6—H6B | 109.3 | C31—C36—H36 | 119.5 |
| N2—C6—H6B | 109.3 | C44—O3—H3 | 109.5 |
| H6A—C6—H6B | 107.9 | O2—C41—O1 | 123.88 (13) |
| C8—C7—C6 | 121.77 (14) | O2—C41—C42 | 118.34 (12) |
| C8—C7—H7 | 119.1 | O1—C41—C42 | 117.73 (12) |
| C6—C7—H7 | 119.1 | C43—C42—C41 | 115.51 (11) |
| C7—C8—C31 | 127.63 (14) | C43—C42—H42A | 108.4 |
| C7—C8—H8 | 116.2 | C41—C42—H42A | 108.4 |
| C31—C8—H8 | 116.2 | C43—C42—H42B | 108.4 |
| C12—C11—C16 | 118.89 (13) | C41—C42—H42B | 108.4 |
| C12—C11—C1 | 119.60 (13) | H42A—C42—H42B | 107.5 |
| C16—C11—C1 | 121.51 (12) | C44—C43—C42 | 115.52 (12) |
| C13—C12—C11 | 120.76 (15) | C44—C43—H43A | 108.4 |
| C13—C12—H12 | 119.6 | C42—C43—H43A | 108.4 |
| C11—C12—H12 | 119.6 | C44—C43—H43B | 108.4 |
| C14—C13—C12 | 118.41 (15) | C42—C43—H43B | 108.4 |
| C14—C13—H13 | 120.8 | H43A—C43—H43B | 107.5 |
| C12—C13—H13 | 120.8 | O4—C44—O3 | 122.28 (15) |
| F1—C14—C13 | 119.03 (17) | O4—C44—C43 | 123.32 (15) |
| F1—C14—C15 | 117.93 (18) | O3—C44—C43 | 114.37 (13) |
| C2—N1—C1—C11 | 45.82 (15) | C1—C11—C16—C15 | 179.64 (15) |
| C5—N1—C1—C11 | 165.97 (11) | N1—C1—C21—C22 | −32.40 (17) |
| C2—N1—C1—C21 | 169.14 (11) | C11—C1—C21—C22 | 91.95 (15) |
| C5—N1—C1—C21 | −70.72 (13) | N1—C1—C21—C26 | 150.61 (12) |
| C5—N1—C2—C3 | 62.13 (13) | C11—C1—C21—C26 | −85.03 (15) |
| C1—N1—C2—C3 | −176.65 (11) | C26—C21—C22—C23 | 0.0 (2) |
| C4—N2—C3—C2 | 54.54 (14) | C1—C21—C22—C23 | −177.01 (12) |
| C6—N2—C3—C2 | 177.49 (11) | C21—C22—C23—C24 | −0.1 (2) |
| N1—C2—C3—N2 | −59.43 (14) | C22—C23—C24—F2 | −177.93 (13) |
| C3—N2—C4—C5 | −54.17 (14) | C22—C23—C24—C25 | 0.7 (2) |
| C6—N2—C4—C5 | −175.54 (11) | F2—C24—C25—C26 | 177.40 (13) |
| C2—N1—C5—C4 | −62.90 (13) | C23—C24—C25—C26 | −1.3 (2) |
| C1—N1—C5—C4 | 173.58 (10) | C22—C21—C26—C25 | −0.5 (2) |
| N2—C4—C5—N1 | 59.60 (14) | C1—C21—C26—C25 | 176.52 (13) |
| C4—N2—C6—C7 | −55.86 (15) | C24—C25—C26—C21 | 1.2 (2) |
| C3—N2—C6—C7 | −177.48 (11) | C7—C8—C31—C36 | −179.03 (16) |
| N2—C6—C7—C8 | 115.58 (16) | C7—C8—C31—C32 | −1.4 (2) |
| C6—C7—C8—C31 | 178.37 (14) | C36—C31—C32—C33 | 0.9 (2) |
| N1—C1—C11—C12 | −111.85 (14) | C8—C31—C32—C33 | −176.78 (15) |
| C21—C1—C11—C12 | 125.03 (13) | C31—C32—C33—C34 | 0.4 (3) |
| N1—C1—C11—C16 | 68.57 (17) | C32—C33—C34—C35 | −1.2 (3) |
| C21—C1—C11—C16 | −54.55 (17) | C33—C34—C35—C36 | 0.7 (3) |
| C16—C11—C12—C13 | 0.0 (2) | C34—C35—C36—C31 | 0.6 (3) |
| C1—C11—C12—C13 | −179.58 (13) | C32—C31—C36—C35 | −1.4 (2) |
| C11—C12—C13—C14 | 0.0 (2) | C8—C31—C36—C35 | 176.36 (15) |
| C12—C13—C14—F1 | 179.71 (16) | O2—C41—C42—C43 | −169.74 (14) |
| C12—C13—C14—C15 | −0.1 (3) | O1—C41—C42—C43 | 12.88 (19) |
| F1—C14—C15—C16 | −179.64 (16) | C41—C42—C43—C44 | 177.78 (13) |
| C13—C14—C15—C16 | 0.1 (3) | C42—C43—C44—O4 | 161.8 (2) |
| C14—C15—C16—C11 | −0.1 (3) | C42—C43—C44—O3 | −20.2 (2) |
| C12—C11—C16—C15 | 0.1 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H72···O1 | 0.96 (2) | 1.73 (2) | 2.6795 (16) | 174 (2) |
| O3—H3···O1i | 0.84 | 1.84 | 2.6564 (16) | 162 |
| C4—H4A···O4ii | 0.99 | 2.58 | 3.287 (3) | 128 |
| C4—H4B···O2iii | 0.99 | 2.39 | 3.3167 (19) | 155 |
| C25—H25···O2iv | 0.95 | 2.53 | 3.4168 (19) | 155 |
| C12—H12···Cgv | 0.95 | 2.81 | 3.7511 (17) | 170 |
Symmetry codes: (i) x, −y+2, z+1/2; (ii) x, −y+2, z−1/2; (iii) x, −y+1, z−1/2; (iv) x+1, −y+1, z−1/2; (v) x, −y+1, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: GK2534).
References
- Amery, W. K. (1983). Headache, 23, 70–74. [DOI] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Betz, R., Gerber, T., Hosten, E., Dayananda, A. S. & Yathirajan, H. S. (2011a). Acta Cryst. E67, o2783–o2784. [DOI] [PMC free article] [PubMed]
- Betz, R., Gerber, T., Hosten, E., Dayananda, A. S., Yathirajan, H. S. & Narayana, B. (2011b). Acta Cryst. E67, o2587–o2588. [DOI] [PMC free article] [PubMed]
- Boeyens, J. C. A. (1978). J. Cryst. Mol. Struct. 8, 317–320.
- Bogatcheva, E., Hanrahan, C., Nikonenko, B., Samala, R., Chen, P., Gearhart, J., Barbosa, F., Einck, L., Nacy, C. A. & Protopopova, M. (2006). J. Med. Chem. 49, 3045–3048. [DOI] [PMC free article] [PubMed]
- Brockunier, L. L., He, J., Colwell, L. F. Jr, Habulihaz, B., He, H., Leiting, B., Lyons, K. A., Marsilio, F., Patel, R. A., Teffera, Y., Wu, J. K., Thornberry, N. A., Weber, A. E. & Parmee, E. R. (2004). Bioorg. Med. Chem. Lett. 14, 4763–4766. [DOI] [PubMed]
- Bruker (2008). SADABS . Bruker Inc., Madison, Wisconsin, USA.
- Bruker (2010). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Dayananda, A. S., Dutkiewicz, G., Yathirajan, H. S., Ramesha, A. R. & Kubicki, M. (2012a). Acta Cryst. E68, o2817. [DOI] [PMC free article] [PubMed]
- Dayananda, A. S., Yathirajan, H. S., Keeley, A. C. & Jasinski, J. P. (2012b). Acta Cryst. E68, o2237. [DOI] [PMC free article] [PubMed]
- Elliott, S. (2011). Drug. Test. Anal. 3, 430–438. [DOI] [PubMed]
- Etter, M. C., MacDonald, J. C. & Bernstein, J. (1990). Acta Cryst. B46, 256–262. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Fillers, J. P. & Hawkinson, S. W. (1982). Acta Cryst. B38, 3041–3045.
- Holmes, B., Brogden, R. N., Heel, R. C., Speight, T. M. & Avery, G. S. (1984). Drugs, 27, 6–44. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Vanier, M. & Brisse, F. (1983). Acta Cryst. C39, 912–914.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813000706/gk2534sup1.cif
Supplementary material file. DOI: 10.1107/S1600536813000706/gk2534Isup2.cdx
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813000706/gk2534Isup3.hkl
Supplementary material file. DOI: 10.1107/S1600536813000706/gk2534Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




