SUMMARY
The insulin/insulin-like peptides (ILPs) regulate key events in physiology, including neural plasticity. However, the cellular and circuit mechanisms whereby ILPs regulate learning remain largely unknown. Here, we characterize two ILPs that play antagonistic roles in aversive olfactory learning of C. elegans. We show that the ILP ins-6 acts from ASI sensory neurons to enable learning by repressing the transcription of another ILP, ins-7, specifically in URX neurons. A high level of INS-7 from URX disrupts learning by antagonizing the insulin receptor-like homolog DAF-2 in the postsynaptic neurons RIA, which play an essential role in the neural circuit underlying olfactory learning. We also show that increasing URX-generated INS-7 and loss of INS-6, both of which abolish learning, alter RIA neuronal property. Together, our results reveal an “ILP-to-ILP” pathway that links environment-sensing neurons, ASI and URX, to the key neuron, RIA, of a network that underlies olfactory plasticity and modulates its activity.
INTRODUCTION
In both invertebrates and vertebrates, insulin and insulin-like peptides (ILPs) play key roles in physiology and have been shown to act through insulin/insulin-like growth factor (IGF) receptors and a conserved intracellular kinase cascade that regulates the activity of the transcription factor FOXO. Many organisms encode multiple ILPs in their genomes (Brogiolo et al., 2001; Ikeya et al., 2002; Liu and Lovenberg, 2008; Pierce et al., 2001), which suggests complexity and diversity in their signaling mechanisms, as well as potential functional interactions among ILPs. Indeed, studies in C. elegans, Drosophila and mammals (Cornils et al., 2011; Gronke et al., 2010; Hwangbo et al., 2004; Kenyon et al., 1993; Kulkarni et al., 1999; Murphy et al., 2007) reveal that insulin and ILPs regulate not only developmental time and plasticity but also reproduction, metabolism, stress responses and/or life span, which could entail a multi-step “ILP-to-ILP” signaling pattern. This signaling pattern engages different tissues and cell types to generate physiological responses (Gronke et al., 2010; Murphy et al., 2007).
Interestingly, insulin and ILPs have also been implicated in experience-dependent neural plasticity (Chen et al., 2011; Kauffman et al., 2010; Kodama et al., 2006; Lin et al., 2001a; Man et al., 2000; Tomioka et al., 2006; Zhao et al., 1999). Neuropeptides play important roles in modulating the properties of neural networks that underlie context or experience-dependent changes (Marder, 2012). Similarly, ILP signaling may also tune the activity of neural circuits to enable plasticity. However, the underlying signaling mechanism remains to be elucidated. Because the large number of ILPs that exist in many animals have diverse physiological roles, which can be combinatorial in nature (Cornils et al., 2011; Gronke et al., 2010), it further raises the possibility that a combination of ILP activities regulates experience-dependent plasticity. Yet, the neural circuits regulated by ILP signals and the effect of ILP signaling on their properties remain largely unknown.
Caenorhabditis elegans provides an opportunity to address these questions. While there are ten members in the human insulin/ILP family (Liu and Lovenberg, 2008) and seven in Drosophila (Brogiolo et al., 2001; Ikeya et al., 2002), C. elegans has 40 putative ILPs (Li et al., 2003; Pierce et al., 2001). C. elegans also has an insulin receptor-like homolog DAF-2 that acts through a PI-3-kinase pathway to regulate the FOXO transcription factor DAF-16 (Kenyon et al., 1993; Kimura et al., 1997; Lin et al., 1997; Lin et al., 2001b; Morris et al., 1996; Ogg et al., 1997). Importantly, the wiring diagram of the C. elegans nervous system is defined (White et al., 1986), which has previously enabled us to map and characterize the properties of a neural network underlying a form of olfactory learning, whereby C. elegans learns to avoid the smell of pathogenic bacteria (Ha et al., 2010; Hendricks et al., 2012; Zhang et al., 2005). Thus, this system should allow us to mechanistically analyze the role of the ILP pathway in olfactory learning.
Here we report that two C. elegans ILPs, ins-6 and ins-7, play antagonistic roles in a conserved “ILP-to-ILP” signaling pattern to regulate the ability to learn to avoid the smell of pathogenic bacteria after ingestion. INS-6 produced by the environment-sensing neuron ASI enables learning, while INS-7 generated from another sensory neuron, URX, inhibits learning. These spatial patterns are essential for the signaling activities of these ILPs. Mechanistically, to promote learning, INS-6 represses ins-7 expression specifically in URX, likely through a paracrine manner. In turn, the learning inhibitory function of URX-produced INS-7 antagonizes DAF-2 receptor activity in the RIA interneurons and appropriate signaling of INS-6 and INS-7 are required for normal RIA neuronal activity. Because RIA plays an essential role in regulating aversive olfactory learning (Ha et al., 2010; Zhang et al., 2005), our results elucidate the molecular and circuit mechanisms for an inhibitory neuropeptide pathway in regulating learning. Together, our findings reveal that INS-6 and INS-7 employ a feedforward “ILP-to-ILP” signaling pathway that acts within a neural circuit that links the environment to a learning network, and thereby modulates the network’s activity (Figure 7E).
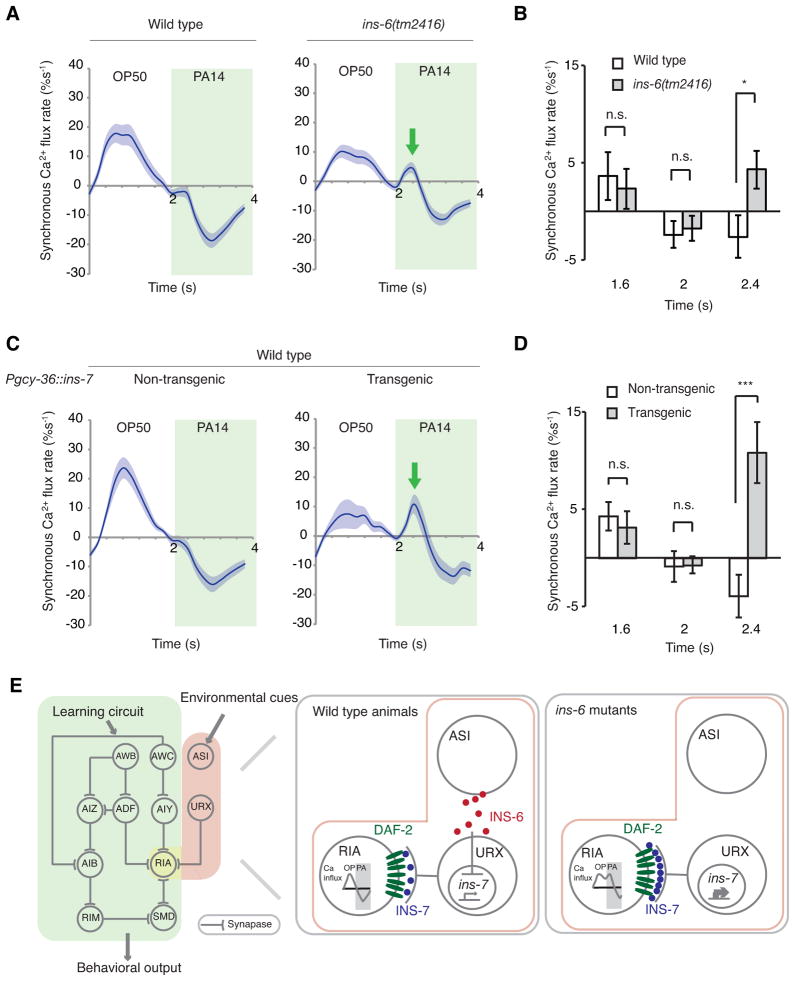
Figure 7. The pathway of INS-6 and INS-7 regulates RIA neuronal activity.
(A, C) Histogram of synchronized GCaMP3 signals in RIA in response to alternating OP50- and PA14-conditioned media in wild type and ins-6(tm2416) mutants (A) or in wild-type animals that overexpress INS-7 in URX and their non-transgenic siblings (C). Solid lines denote mean values and shaded lines denote SEM. Arrows point to ectopic peaks.
(B, D) Bar charts of the RIA synchronized GCaMP3 signals in A and C, respectively, at three different time points.
For B and D, Student’s t-test, n ≥ 13 animals, ***P < 0.001, *P < 0.05, n.s.: not significant; mean +/− SEM
(E) A working model for the pathway of INS-6 and INS-7 in aversive olfactory learning. In wild type, INS-6 in ASI inhibits ins-7 expression in URX; but in ins-6(tm2416) mutants, ins-7 expression is upregulated, which in turn leads to inhibition of DAF-2 activity in the RIA neuron, alteration of RIA neuronal properties and disruption in learning.
RESULTS
C. elegans ILPs play distinct roles in aversive olfactory learning
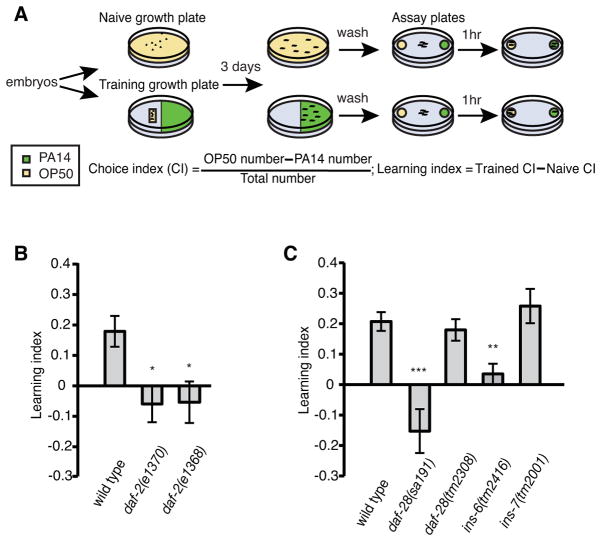
Previously, we have shown that naive animals that are never exposed to pathogenic bacteria, such as Pseudomonas aeruginosa PA14, slightly prefer or are indifferent to the smell of the pathogen. In contrast, trained animals that have ingested the pathogen learn to avoid its smell (Ha et al., 2010; Zhang et al., 2005). We use chemotaxis assays to measure the olfactory preference between PA14 and a standard bacterial food source, Escherichia coli OP50. We compare the olfactory preference of trained animals, which have been exposed to PA14, with that of naive animals, which have been grown only on OP50. The difference between the naive and trained olfactory preferences indicates the learned olfactory aversion [(Ha et al., 2010; Zhang et al., 2005); Figure 1A].
Figure 1. C. elegans ILPs play distinct roles in aversive olfactory learning.
(A) Schematics for the aversive olfactory learning assay. “.”: embryos, “~”: adults.
(B, C) The aversive olfactory learning abilities of daf-2 mutants and several ILP mutants (ANOVA with multiple comparisons corrected with the Dunnett’s test, ***P < 0.001, **P < 0.01, *P < 0.05; n ≥ 5 assays; mean +/− SEM).
To examine the potential involvement of C. elegans ILPs in aversive olfactory learning, we first tested the learning ability of animals that carry mutations in daf-2, which encodes the only known worm homolog of the mammalian insulin/IGF-1 receptor (Kimura et al., 1997). We found that two reduction-of-function alleles, e1370 and e1368, were both significantly defective in learning to avoid the smell of PA14 (Figure 1B). Using standard chemotaxis assays, we found that daf-2(e1370) mutants responded well to a panel of olfactory attractants and repellents over a series of concentrations (Figure S1), indicating that the learning defect of daf-2 mutants does not result from defective olfactory sensation. These results suggest that the C. elegans ILP pathway is required for aversive olfactory learning.
Next, we sought to identify which ILPs regulate this learning ability. First, we determined whether the semi-dominant mutation for the ILP daf-28, sa191, abolished learning. sa191 is thought to disrupt the cleavage and folding of the DAF-28 peptide and other ILPs that are expressed in the same neurons as daf-28 and have a similar structure (Li et al., 2003). Like daf-2 mutants, daf-28(sa191) did not learn to avoid the smell of PA14 (Figure 1C), further suggesting that ILPs are involved in this learning ability. However, a null mutation for daf-28, tm2308, had little or no effect (Figure 1C), implying that ILPs other than DAF-28 mediate learning. We then measured the learning abilities of deletion mutants, ins-6(tm2416) and ins-7(tm2001), for two ILPs that have a predicted structure similar to that of DAF-28 (Li et al., 2003). Interestingly, we found that ins-6(tm2416) mutants were severely defective in learning to avoid the smell of PA14, whereas ins-7(tm2001) mutants were normal (Figure 1C). Together these results indicate that different ILPs play distinct roles in aversive olfactory learning.
INS-6 generated by the sensory neurons ASI enables olfactory learning
INS-6 belongs to the C. elegans ILP superfamily type-β class, which is predicted to form a distinct set of intra-molecular disulfide bonds (Pierce et al., 2001). Previous biochemical analysis showed that INS-6 can bind to the human insulin receptor and act as an agonist (Hua et al., 2003). Despite the strong learning defect, the ins-6(tm2416) mutants are normal in sensing various odorant attractants and repellents over a series of concentrations (Figure S1) and in their resistance to the pathogenic bacteria PA14 (Figure S2). In addition, we fully rescued the learning defect of ins-6(tm2416) animals with the wild-type ins-6 genomic DNA (Pins-6::ins-6), which contains both upstream and downstream cis-regulatory regions (Figure 2A), confirming that wild-type ins-6 has a positive role in learning.
Figure 2. INS-6 in the ASI neurons enables aversive olfactory learning.
(A) Wild-type ins-6 genomic DNA (Pins-6::ins-6) restored the learning ability to ins-6(tm2416) mutants.
(B, C) Expression of ins-6 in ASI sensory neurons (B, Pstr-3::ins-6), but not in ASJ neurons (C, Ptrx-1::ins-6), rescued the learning defect of ins-6(tm2416) mutants.
In A–C, the learning ability of transgenic animals was compared with that of non-transgenic siblings using the paired Student’s t-test (**P < 0.01, *P < 0.05; n.s., not significant; n ≥ 5 assays; mean +/− SEM).
(D–F) The expression of Pins-6::mcherry transgene in wild-type animals under naive (D) and training (E) conditions and their quantification (F, Student’s t-test,***P < 0.001; n.s., not significant; n ≥ 50 animals; mean +/− SEM; AU, artificial unit). Arrowheads: ASI neurons.
In well-fed animals, ins-6 is specifically expressed in the ASI sensory neurons. In animals that arrest at or exit from a diapause stage, called dauer, which animals enter under harsh environments, ins-6 expression is downregulated in ASI and upregulated in another pair of sensory neurons, ASJ (Cornils et al., 2011). To identify the release site of INS-6 for its role in olfactory learning, we expressed wild-type ins-6 selectively in either ASI or ASJ in ins-6(tm2416) mutants. We found that wild-type ins-6 in ASI (Pstr-3::ins-6) fully rescued the learning defect of ins-6(tm2416) mutants (Figure 2B), whereas expression of ins-6 in ASJ (Ptrx-1::ins-6) did not rescue (Figure 2C). In contrast, ins-6 expression from either ASI or ASJ, using the same transgenes, could rescue ins-6(tm2416) defects in both dauer entry and exit (Cornils et al., 2011), suggesting that INS-6 regulates learning and the dauer program using different mechanisms. Since the ASJ-specific promoter, Ptrx-1 (Cornils et al., 2011; Miranda-Vizuete et al., 2006), is not weaker than the ASI-specific promoter, Pstr-3 (Peckol et al., 2001), the lack of ins-6 rescue from ASJ should not be due to lower ins-6 expression from ASJ versus ASI. Rather, it likely results from potential differences in the cellular properties of ASI and ASJ, which will be discussed below. Together our results reveal a specific role for the INS-6 signal generated by ASI neurons in aversive olfactory learning.
We next examined whether pathogen exposure would alter ins-6 expression in ASI to regulate learning, similar to what we previously observed for the expression of the serotonin biosynthetic enzyme tryptophan hydroxylase TPH-1 (Sze et al., 2000) in the ADF serotonergic neurons (Zhang et al., 2005). We quantified the expression of a transcriptional reporter Pins-6::mcherry in both naive and trained animals and used the Ptph-1::gfp transcriptional reporter in parallel as a control for the training procedure. We observed no significant change either in the intensity or the site of Pins-6::mcherry expression (Figures 2D to 2F), which is again different from the switch in ins-6 expression from ASI to ASJ in dauer and post-dauer animals (Cornils et al., 2011). Together our results again indicate that INS-6 regulates developmental and neural plasticity through different cellular mechanisms.
INS-7 antagonizes INS-6 in regulating olfactory learning
Because INS-6 positively regulates aversive olfactory learning, we examined whether increasing ins-6 expression in wild type would enhance learning ability. We found that wild-type animals that were transformed with ins-6 genomic DNA (Pins-6::ins-6) displayed comparable learning ability as their non-transgenic siblings (Figure 3A), suggesting that increasing INS-6 activity alone is insufficient in enhancing learning and that INS-6 may regulate learning by interacting with other factors.
Figure 3. The loss of INS-7 function suppresses the learning defect of ins-6 mutants.
(A) Increased expression of ins-6 is insufficient to enhance learning ability.
(B) The loss of function in ins-7, but not in daf-28, suppresses the learning defect of ins-6(tm2416) mutants. The learning abilities of double mutants and ins-6(tm2416) single mutant were compared using ANOVA with multiple comparison corrected with the Dunnett’s test (***P < 0.001, *P < 0.05; n.s., not significant; n ≥ 5 assays; mean +/− SEM).
(C) Wild-type ins-7 genomic DNA (Pins-7::ins-7) restored the learning defect to the ins-6(tm2416); ins-7(tm2001) double mutants.
In A and C, the learning ability of transgenic animals was compared with that of non-transgenic siblings using the paired Student’s t-test (**P < 0.01; n.s., not significant; n ≥ 5 assays; mean +/− SEM).
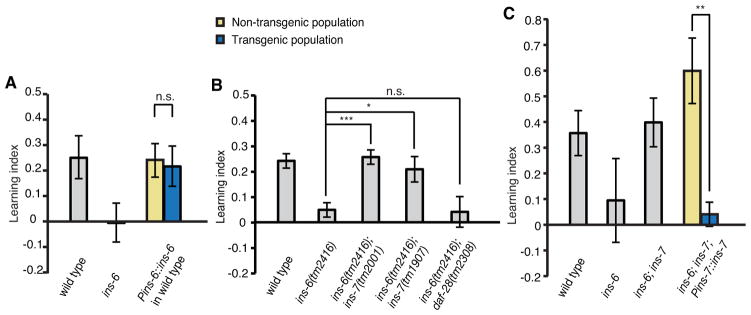
Given the previously observed functional interactions among ILPs in regulating the dauer program (Cornils et al., 2011; Li et al., 2003), we examined possible genetic interactions among ins-6, ins-7 and daf-28 in regulating learning. Intriguingly, we found that two different deletion alleles of ins-7, tm2001 and tm1907, completely suppressed the learning defect of ins-6(tm2416) mutants, as demonstrated by the normal learning ability of both ins-6(tm2416); ins-7(tm2001) and ins-6(tm2416); ins-7(tm1907) double mutants (Figure 3B). In contrast, the deletion in daf-28(tm2308) did not alter the learning phenotype of ins-6(tm2416) (Figure 3B). Because daf-28 encodes an ILP of the same class as those encoded by ins-6 and ins-7 (Li et al., 2003), these results indicate that the suppression of the ins-6(tm2416) learning defect by ins-7(tm2001) or ins-7(tm1907) is not likely due to a general decrease in the strength of ILP signaling, but to a genetic interaction between ins-6 and ins-7.
The deletion in ins-7(tm1907) removes the entire coding region of ins-7; and the deletion in ins-7(tm2001) eliminates most of the predicted signal peptide required for the proper processing and folding of INS-7 and severely decreases ins-7 transcription (A. C. Reyes, D. F. de Abreu, J. Alcedo and Q. Ch’ng, personal communication). The ins-6(tm2416); ins-7(tm2001) double mutants exhibit pathogen resistance and odor responses that are comparable to wild type and ins-6(tm2416) single mutants (Figures S1 and S2), indicating that altered pathogen resistance or olfactory sensation does not account for the suppression of the ins-6(tm2416) learning phenotype by ins-7(tm2001). Moreover, expression of the wild-type ins-7 genomic DNA (Pins-7::ins-7) reverted the normal learning ability of ins-6(tm2416); ins-7(tm2001) double mutants back to the defective learning of ins-6(tm2416) single mutants (Figure 3C). Together these results indicate antagonistic roles for INS-6 and INS-7 in regulating aversive olfactory learning and suggest that wild-type INS-7 plays a negative role in this trait.
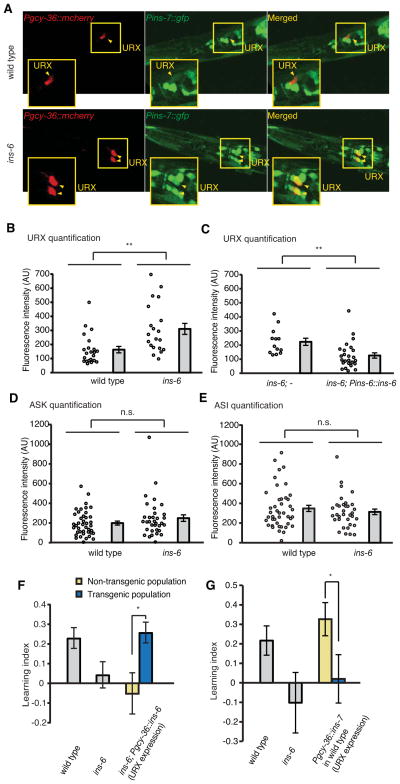
INS-7 from the URX neurons negatively regulates learning
Next, to characterize the function of ins-7 in learning, we examined the expression pattern of ins-7. We generated an integrated transgenic line that expressed a GFP transcriptional reporter flanked by both the 5′ and 3′ cis-regulatory sequences of ins-7 [yxIs13(Pins-7::gfp)]. We found that yxIs13(Pins-7::gfp) was expressed in multiple tissues, including several head neurons and the intestine (Figures 4A and 4B), consistent with previous findings (Murphy et al., 2007). Using a dye-filling procedure that labels a defined set of ciliated sensory neurons (Hedgecock et al., 1985) or reporters known to be expressed in specific cells, we found that the ins-7-expressing head neurons include ADF, ASI, ASK and URX (Figures 4C–4E) among others.
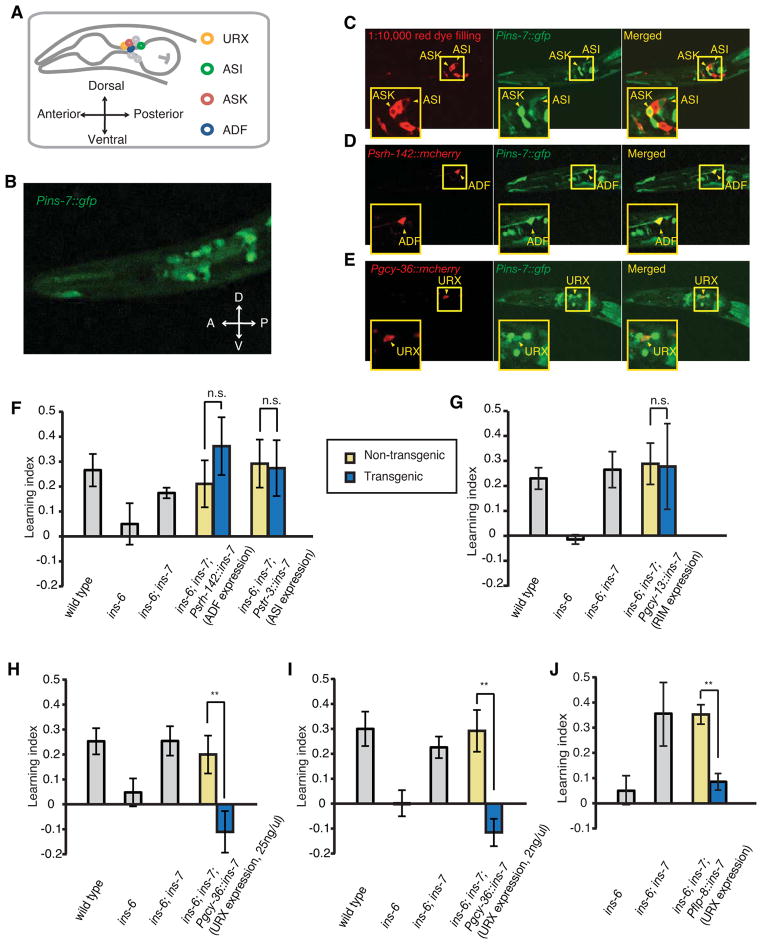
Figure 4. INS-7 from URX neurons antagonizes INS-6 in regulating aversive olfactory learning.
(A) Schematic for C. elegans head neurons.
(B) The expression reporter yxIs13[Pins-7::gfp] is expressed in head neurons.
(C–E) yxIs13[Pins-7::gfp] is expressed in ASI, ASK (C), ADF (D) and URX (E) neurons. Red fluorescence was generated from DiI-staining of several sensory neurons (C) or expression of neuron-specific reporters of mcherry (D, E).
(F, G) ins-7 expression in ADF or ASI (Psrh-142::ins-7 or Pstr-3::ins-7, respectively, in F) or in RIM (Pgcy-13::ins-7 in G) did not alter the learning ability of ins-6(tm2416); ins-7(tm2001) double mutants.
(H–J) ins-7 expression in URX using the gcy-36 promoter (at 25 ng/ul in H or 2 ng/ul in I) or using the flp-8 promoter (at 25 ng/ul in J) reverted the normal learning of ins-6(tm2416); ins-7(tm2001) double mutants back to the defective learning of ins-6(tm2416) single mutants.
In F–J, the learning ability of transgenic animals was compared with that of non-transgenic siblings using the paired Student’s t-test (**P < 0.01; n.s., not significant; n ≥ 5 assays; mean +/− SEM).
To identify the release sites of INS-7 for its antagonistic effect on ins-6 function in learning, we used neuronal-specific promoters to selectively express a wild-type ins-7 cDNA in specific sets of neurons in the ins-6(tm2416); ins-7(tm2001) double mutants. We found that expression of wild-type ins-7 in either the ADF (Psrh-142::ins-7) or ASI (Pstr-3::ins-7) sensory neurons failed to restore the learning defect to the double mutants (Figure 4F). Since we occasionally observed ins-7 expression in the RIM motor neurons using transcriptional reporters, we also tested the effect of expressing wild-type ins-7 in RIM and again found no rescuing effect (Figure 4G). In contrast, expression of wild-type ins-7 in URX, AQR and PQR neurons, using the gcy-36 promoter (Pgcy-36::ins-7), either at a standard concentration (25 ng/ul) or a low concentration (2 ng/ul), completely abolished the suppressive effect of ins-7(tm2001), leading to animals that were as defective in learning as the ins-6(tm2416) single mutants (Figures 4H and 4I). We also placed the ins-7 cDNA under a different promoter, flp-8, which drives expression only in the URX and AUA neurons (Macosko et al., 2009), and found a similar rescuing effect in the ins-6(tm2416); ins-7(tm2001) double mutants (Figure 4J). Because the expression patterns of the gcy-36 and flp-8 promoters overlap with ins-7 expression only in the URX neurons, our results show that the negative role of INS-7 in learning depends on its expression in URX.
INS-6 enables learning by repressing the expression of ins-7 in URX neurons
Next, to understand how INS-7 antagonizes the positive role of INS-6 in olfactory learning, we examined whether INS-6 from ASI neurons and INS-7 from URX neurons function in parallel or in a linear pathway. Hence, we compared the expression intensity of yxIs13(Pins-7::gfp) in wild type and ins-6(tm2416) mutants. We found that Pins-7::gfp expression was significantly enhanced in the URX neurons of ins-6(tm2416) mutants in comparison with that of wild type (Figures 5A and 5B). This enhancement was fully rescued by the wild-type ins-6 genomic DNA (Pins-6::ins-6) (Figure 5C). In contrast, the expression of Pins-7::gfp in several other ins-7-expressing neurons, such as ASK and ASI, was not different between wild-type and the ins-6 mutant animals (Figures 5D and 5E). Together these results demonstrate an INS-6-mediated cell-specific reduction of ins-7 expression in URX. Since the level of ins-6 transcription is not significantly altered by the ins-7(tm2001) or ins-7(tm1907) mutation (A. C. Reyes, D. F. de Abreu, J. Alcedo and Q. Ch’ng, personal communication), our findings support a linear pathway from INS-6 to INS-7 in regulating aversive olfactory learning.
Figure 5. ins-6 negatively regulates ins-7 transcription in URX neurons.
(A) Representative images for the expression of yxIs13[Pins-7::gfp] reporter in URX neurons in wild-type animals and ins-6(tm2416) mutants. Red fluorescence in URX: expression of the Pgcy-36::mcherry transgene.
(B, C) The expression of yxIs13[Pins-7::gfp] in URX neurons is increased in ins-6(tm2416) mutants (B) and this defect is rescued by the wild-type ins-6 genomic DNA (Pins-6::ins-6) (C).
(D, E) The expression of yxIs13[Pins-7::gfp] in ASK (D) and ASI (E) neurons was comparable between wild type and ins-6(tm2416) mutants.
In B – E, each circle indicates a measurement of yxIs13[Pins-7::gfp] fluorescence intensity of the specified neuron from one animal (Student’s t-test, **P < 0.01, n.s., not significant; mean +/- SEM; AU, artificial unit).
(F) Expression of ins-6 in URX using the gcy-36 promoter rescued the learning defect in ins-6(tm2416) mutants.
(G) Increasing URX expression of ins-7 (Pgcy-36::ins-7) disrupts learning in wild-type animals.
In F and G, the learning ability of transgenic animals was compared with that of non-transgenic siblings using the paired Student’s t-test (*P < 0.05; n ≥ 5 assays; mean +/− SEM).
In both naive and trained animals, ins-6 is expressed only in the ASI sensory neurons (Figures 2D and 2E), which have no synapses or gap junctions with URX (White et al., 1986). Thus, INS-6, like many neuropeptides, could function independently of synapses to directly regulate target neurons in a paracrine manner (Edwards, 1998; Richmond and Broadie, 2002) or regulate URX indirectly through secondary cells or signals. To test these possibilities, we selectively expressed ins-6 in the URX neurons of ins-6(tm2416) mutants using the gcy-36 promoter and assessed its rescuing effect on learning. We reasoned that if INS-6 directly acted on URX in a paracrine manner, expression of ins-6 in URX itself should release the INS-6 peptide into the immediate local environment of URX and rescue ins-6(tm2416) mutants. Indeed, similar to its expression in ASI, ins-6 expression in URX fully rescued the learning defect of ins-6(tm2416) animals (Figure 5F), consistent with the hypothesis that INS-6 diffuses to URX to regulate learning. These findings also suggest that INS-6 could act like a number of secreted molecules, such as TGF-β, Wnt and Hedgehog, which provide distinct positional information to target tissues that differ in distance from the localized sources of the signals (Drossopoulou et al., 2000; Nellen et al., 1996; Zecca et al., 1996). Thus, INS-6 generated from ASI, whose cell body is situated on the dorsal side of the head and close to URX, could provide different spatial information from INS-6 generated from ASJ, which is on the ventral side of the head (White et al., 1986). This would also be consistent with the difference between Pstr-3::ins-6 and Ptrx-1::ins-6 in rescuing the learning ability of ins-6 mutants (Figures 2B and 2C).
Because our results suggest that INS-6 from ASI neurons enables learning by selectively repressing ins-7 expression in URX, we next examined if increased ins-7 expression in URX, as seen in ins-6(tm2416) loss-of-function mutants, was sufficient to suppress learning. Strikingly, increasing ins-7 expression in URX through Pgcy-36::ins-7 disrupted the normal learning ability of wild type and produced a defect similar to that in ins-6(tm2416) mutants (Figure 5G), indicating that an enhanced signal of INS-7 from URX plays a strong inhibitory role in learning. Together these results reveal an “ILP-to-ILP” pathway that negatively regulates learning (“INS-6 in ASI ⊣INS-7 in URX ⊣learning”).
The INS-6 and INS-7 pathway regulates learning by antagonizing the DAF-2 receptor in the RIA interneurons
Next, we sought the mechanisms underlying the role of “INS-6 ⊣INS-7” pathway in learning. We have previously shown that serotonin signaling from the ADF sensory neurons, partly via increased transcription of the tryptophan hydroxylase tph-1, is required for animals to learn to avoid the smell of pathogenic bacteria (Zhang et al., 2005). However, the levels of tph-1 transcription in ADF of naive and trained animals were not obviously altered by mutations in ins-6(tm2416) or ins-7(tm2001) single mutants or in ins-6(tm2416); ins-7(tm2001) double mutants (Figure S3), suggesting that the role of INS-6 and INS-7 in learning is independent of transcriptional regulation of tph-1 in ADF. In addition, similar to ins-6 (Figures 2D–2F), ins-7 expression in URX is not altered by training (Figure S4), suggesting that the role of the “INS-6 ⊣INS-7” pathway in learning does not involve training-dependent transcriptional regulation of the ILPs.
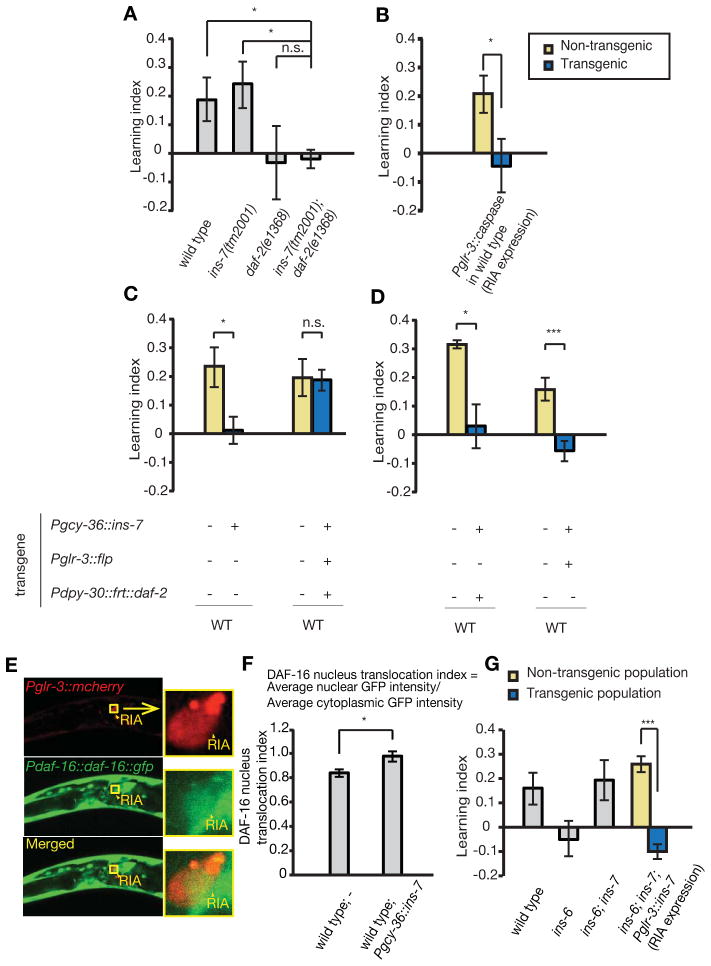
To address the function of the “INS-6 ⊣INS-7” pathway in learning, we sought the target neuron(s) of INS-7. First, we characterized the functional interaction between DAF-2, the only known C. elegans insulin/IGF receptor, and INS-7 in regulating olfactory learning. Since ins-6; ins-7 double mutants and ins-7 single mutants showed normal learning ability (Figures 1C and 3B), but both daf-2 reduction-of-function mutants, e1370 and e1368, were severely defective in learning (Figure 1B), we measured the learning ability of ins-7(tm2001); daf-2(e1368) double mutants. Similar to daf-2(e1368) single mutants, ins-7(tm2001); daf-2(e1368) double mutants are defective in learning (Figure 6A), indicating that the positive role of wild-type DAF-2 in learning is epistatic and antagonistic to INS-7 (Figure 7E). In addition, the complete suppression of the normal learning ability of ins-7(tm2001) animals by daf-2(e1368) (Figure 6A) is consistent with the possibility of a linear pathway, i.e., INS-6 ⊣INS-7 ⊣DAF-2 → learning.
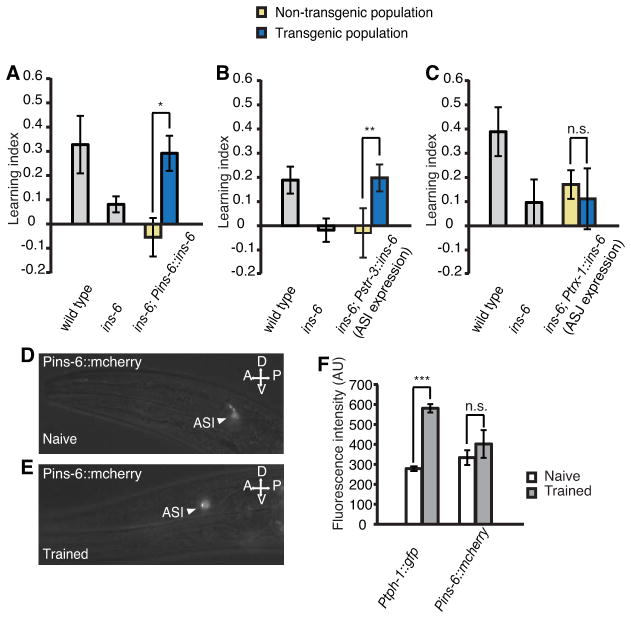
Figure 6. INS-7 from URX neurons inhibits learning by antagonizing DAF-2 signaling in the RIA interneuron.
(A) The learning abilities of ins-7(tm2001) and daf-2(e1368) single mutants, and the learning ability of ins-7; daf-2 double mutants (ANOVA with multiple comparisons corrected with Bonfferoni correction, n ≥ 5 assays).
(B) Genetic ablation of RIA interneurons (Pglr-3::caspase) disrupts learning in wild-type animals (Paired Student’s t-test, n ≥ 5 assays).
(C, D) While overexpression of ins-7 in the URX neurons (Pgcy-36::ins-7) of wild-type animals disrupts learning, increasing expression of daf-2 cDNA in RIA, through co-expression of the Pdpy-30-FRT-mcherry-terminator-FRT-gfp-SL2-daf-2 (abbreviated as Pdpy-30::frt::daf-2 in C and D) and Pglr-3::flp transgenes, suppresses this negative effect (C); but expression of either transgene alone does not suppress the effect (D) (Paired Student’s t-test, n ≥ 5 assays).
(E) Expression of a daf-16::gfp translational fusion in the RIA neuron. The red fluorescence is produced by a RIA-specific transcriptional reporter Pglr-3::mcherry. Arrowheads and boxes: RIA.
(F) RIA nuclear localization of daf-16::gfp translational fusion increases in wild-type animals that overexpress ins-7 in URX neurons (Student’s t-test, n ≥ 15 animals).
(G) Expression of ins-7 in RIA is sufficient to revert the learning ability of ins-6(tm2416); ins-7(tm2001) double mutants back to the defective learning of ins-6(tm2416) single mutants (Paired Student’s t-test, n ≥ 5 assays).
In all, ***P < 0.001, *P < 0.05, n.s.: not significant; mean +/− SEM.
Next, we examined where DAF-2 acts to antagonize INS-7within the learning circuit that we have previously mapped (Ha et al., 2010). The interneuron RIA is the main postsynaptic neuron of URX (White et al., 1986), which is the release site of INS-7 in negatively regulating learning. Intriguingly, we have previously shown that RIA plays an essential role in the neural circuit underlying aversive olfactory learning. Laser ablation of RIA did not impair olfaction, but completely abolished the ability to generate learned olfactory aversion to the training pathogen PA14 (Ha et al., 2010). Similarly, genetic ablation of RIA, via ectopic expression of a cell-death molecule selectively in RIA (Pglr-3::caspase), led to a complete loss of learning ability (Figure 6B), which further confirms a key role of RIA in aversive olfactory learning. Thus, we hypothesized that RIA is the target neuron for the “INS-6 ⊣INS-7”pathway and URX-generated INS-7 inhibits learning by antagonizing DAF-2 function in RIA.
To test this possibility, we asked whether the activity of DAF-2 in RIA could counteract the inhibitory effect of URX-generated INS-7 on learning (Figure 5G). We selectively increased daf-2 expression in RIA by using the FLP/FRT approach (Davis et al., 2008). We used a transgene that specifically expressed FLP recombinase in RIA (Pglr-3::flp), and a second transgene, Pdpy-30-FRT-mcherry-terminator-FRT-gfp-SL2-daf-2, which was ubiquitously expressed (Brockie et al., 2001; Seydoux and Fire, 1994). In wild-type animals that overexpressed ins-7 in URX (Pgcy-36::ins-7), we either (1) co-expressed the above transgenes, which led to RIA-specific FLP-mediated excision of mcherry and the transcription terminator and subsequent increased expression of daf-2 only in RIA (Figure 6C); or (2) separately expressed these two transgenes (Figure 6D). We found that the negative effect on learning by Pgcy-36::ins-7 was completely reversed by specifically increasing daf-2 expression in RIA (Figure 6C), but not in control animals (Figure 6D). Thus, URX-produced INS-7 plays a negative role in learning by antagonizing DAF-2 in RIA.
To further assess the antagonistic effect of INS-7 on DAF-2 in RIA, we also examined the nuclear accumulation of DAF-16, the FOXO transcription factor known to act downstream of DAF-2 in many physiological processes (Kenyon et al., 1993; Lin et al., 1997; Lin et al., 2001b; Ogg et al., 1997). DAF-2 prevents the nuclear accumulation of DAF-16, where decreased DAF-2 pathway activity promoted the nuclear localization of a DAF-16::GFP fusion (Lin et al., 2001b). Here, we first confirmed the expression of daf-16::gfp (Lin et al., 2001b) in RIA, among many other cells, through its co-localization with an mcherry reporter expressed in RIA (Pglr-3::mcherry) (Figure 6E). Next, we measured the nuclear and cytoplasmic levels of DAF-16::GFP in the RIA neurons of transgenic animals that overexpressed ins-7 in URX (Pgcy-36::ins-7) and of their non-transgenic wild-type siblings (Figure 6F). We found that elevated expression of ins-7 in URX increased nuclear accumulation of DAF-16::GFP in RIA (Figure 6F), which further substantiates that URX-generated INS-7 antagonizes the DAF-2 signal transduction in RIA.
Yet, the possibility remains that URX-produced INS-7 could act on RIA directly or indirectly via secondary signals. We hypothesized that if INS-7 directly regulated RIA, expressing ins-7 in RIA should produce the INS-7 peptide in the local environment of RIA and allow direct interaction between INS-7 and RIA to rescue the learning phenotype of the ins-7(tm2001) mutation. Indeed, we found that RIA-specific expression of ins-7 (Pglr-3::ins-7) in ins-6(tm2416); ins-7(tm2001) double mutants fully rescued the suppressive effect of ins-7(tm2001) on the learning defect of ins-6(tm2416) (Figure 6G), similar to ins-7 expression in URX (Figures 4H–4J). These results support the possibility that URX-generated INS-7 directly regulates RIA by antagonizing DAF-2, which together with the above data, reveal the cellular and circuit mechanisms underlying the role of the INS-6 and INS-7 pathway in regulating learning (“INS-6 in ASI ⊣INS-7 in URX ⊣DAF-2 in RIA → learning”; Figure 7E).
ILP signaling regulates RIA neuronal property
Having mapped RIA, the key interneuron in the learning circuit, as the target neuron of the ILP pathway in aversive olfactory learning, we then asked whether ILP signaling regulates RIA neuronal properties. Using intracellular calcium imaging on transgenic animals that selectively expressed the calcium-sensitive fluorescent protein GCaMP3 (Tian et al., 2009) in RIA, we have previously shown that the single neurite of RIA is compartmentalized into different functional domains that exhibit independent calcium dynamics (Hendricks et al., 2012). At the same time, these axonal domains also display synchronized calcium responses that are evoked by sensory inputs detected by upstream neurons (Hendricks et al., 2012). We measure these synchronized responses by scoring synchronized calcium influx or efflux events, which are time points when all axonal domains display a positive or negative rate of change in GCaMP3 signals, respectively. We compute the average rate of calcium flux in all axonal domains to quantify these synchronized events (Hendricks et al., 2012). In this study, we subjected the animals to alternating OP50-conditioned versus PA14-conditioned media and switched the stimuli every 2 seconds. We averaged four trials of synchronized RIA calcium responses for each animal and generated the time histograms in Figures 7A and 7C, based on the results of multiple animals. Previously, we have shown that switching between OP50-conditioned and PA14-conditioned media evokes calcium responses of AWC, a sensory neuron upstream of RIA (Ha et al., 2010). Here we found that switching from OP50 to PA14 suppressed the synchronized RIA activity in wild type, as demonstrated by the negative rates of calcium flux, whereas the reverse switch from PA14 to OP50 activated RIA by generating positive rates of calcium flux (Figures 7A and 7C). These temporal responses reveal the pattern of RIA synchronized activity that is evoked by alternating stimuli of OP50 and PA14.
Next, we asked whether the INS-6 and INS-7 pathway regulates the sensory-evoked RIA calcium signals. We first examined the learning-defective ins-6(tm2416) mutants (Figure 1C). Strikingly, the RIA calcium activity evoked by switching between OP50 and PA14 was significantly disrupted in ins-6(tm2416) mutants. The most striking change in these mutants was an ectopic increase in RIA calcium flux rate evoked by the switch from OP50 to PA14 (Figures 7A and 7B). Because INS-6 positively regulates learning by repressing ins-7 expression in URX and increasing URX expression of ins-7 (Pgcy-36::ins-7) in wild-type animals disrupts learning (Figure 5), we hypothesized that Pgcy-36::ins-7 should generate a defect in RIA calcium responses similar to that of ins-6(tm2416). Indeed, we observed that expressing Pgcy-36::ins-7 in wild type also generated a strong ectopic increase in RIA calcium flux rate in response to the switch from OP50 to PA14, significantly disrupting the pattern of RIA activity (Figures 7C and 7D). Because RIA is critically required for learning [(Ha et al., 2010) and Figure 6B], our results together demonstrate that the INS-6 and INS-7 pathway modulates learning ability by regulating RIA neuronal activity (Figure 7E).
The INS-6 and INS-7 peptides display distinct signaling properties
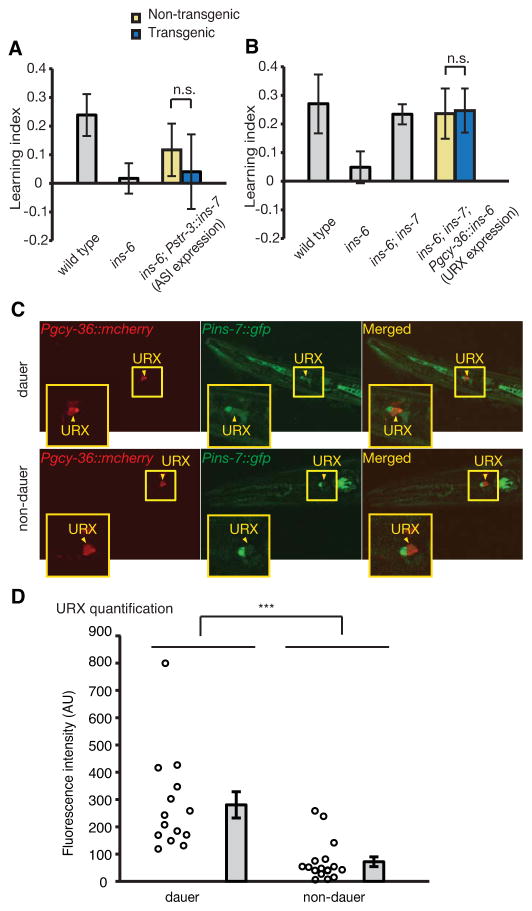
INS-6 and INS-7 belong to the type-β class of the C. elegans ILP superfamily and are predicted to share some similarities in protein structures (Pierce et al., 2001). However, these two ILPs play distinct roles in regulating aversive olfactory learning: ASI-generated INS-6 enables learning, while URX-generated INS-7 prevents it. To further understand these differences, we examined whether INS-6 and INS-7 can functionally substitute for each other. First, we found that, unlike INS-6 from ASI (Pstr-3::ins-6 in Figure 2B), INS-7 from ASI (Pstr-3::ins-7) did not significantly improve the learning ability of ins-6(tm2416) mutants (Figure 8A), suggesting INS-7 function in ASI is distinct from that of INS-6. Conversely, we found that INS-6 expressed from URX (Pgcy-36::ins-6) did not obviously alter the learning ability of ins-6; ins-7 double mutants (Figure 8B), which is again different from the potent effect of INS-7 from URX (Pgcy-36::ins-7) on these animals (Figures 4H and 4I). This difference between Pgcy-36::ins-6 and Pgcy-36::ins-7 is not likely due to an inability of URX to produce INS-6 properly, since Pgcy-36::ins-6 rescues the learning defect of ins-6(tm2416) mutants (Figure 5F). Thus, our results indicate that the functional differences between INS-6 and INS-7 in aversive olfactory learning reside in their peptide structures.
Figure 8. INS-6 and INS-7 have distinct signal properties that respond to environmental cues.
(A, B) Expression of ins-7 in the ASI neurons (Pstr-3::ins-7) does not rescue the learning defect of ins-6(tm2416) mutants (A); conversely, expression of ins-6 in URX (Pgcy-36::ins-6) does not rescue the learning phenotype of ins-7(tm2001) in ins-6(tm2416); ins-7(tm2001) double mutants (B) (Paired Student’s t-test, n.s.; not significant, n ≥ 5 assays, mean +/− SEM).
(C, D) Representative images (C) and quantification (D) of yxIs13[Pins-7::gfp] expression in URX in dauers and age-matched non-dauers. In D, each circle indicates a measurement of yxIs13[Pins-7::gfp] expression in URX from one animal (Student’s t-test, ***P < 0.001; n ≥ 15 animals; mean +/− SEM; AU, artificial unit).
Meanwhile, we also showed that the functions of INS-6 and INS-7 in learning depend on their expression from ASI and URX, respectively. These two sensory neurons detect a variety of internal and external environmental cues, such as population density and oxygen levels (Bargmann and Horvitz, 1991; Beverly et al., 2011; Busch et al., 2012; Cornils et al., 2011; Gray et al., 2004; McGrath et al., 2009; Persson et al., 2009; Zimmer et al., 2009), suggesting that the “INS-6 in ASI ⊣INS-7 in URX” pathway is likely differentially regulated under certain environmental and physiological contexts. Previously, we have found that worms that arrest as dauers under harsh conditions switch off ins-6 expression in ASI neurons (Cornils et al., 2011). Intriguingly, we found that these same animals also dramatically upregulated the expression of ins-7 in URX, despite the general reduction in yxIs13(Pins-7::gfp) expression in other cells (Figures 8C and 8D). Thus, these findings highlight the importance of the INS-6 and INS-7 signaling pathway in response to external and internal sensory and physiological cues.
DISCUSSION
Many animals, including humans, have multiple ILPs in their genomes, suggesting functional diversity and combinatorial activities within the ILP family. Here our study uncovers distinct roles for INS-6 and INS-7 in promoting versus inhibiting olfactory learning. We show that these two ILPs achieve antagonistic effects on learning through a conserved “ILP-to-ILP” signaling pattern, where INS-6 from ASI neurons suppresses ins-7 expression in URX (Figure 7E). Mechanistically, this ILP pathway regulates learning by antagonizing the DAF-2 insulin receptor in RIA, the key interneuron in the neural network underlying aversive olfactory learning. Moreover, we show that changes in signaling by INS-6 and INS-7 alter the neuronal property of the target neuron RIA (Figure 7E), which further links the ILP pathway to the learning network.
INS-6 and INS-7 exert opposite roles in olfactory learning via an “ILP-to-ILP” signaling pattern
Previous studies have suggested the complexity of the mechanisms behind the role of ILP signaling in learning and memory (Chen et al., 2011; Kauffman et al., 2010; Kern et al., 2001; Kodama et al., 2006; Lin et al., 2001a; Man et al., 2000; Tomioka et al., 2006). For example, insulin treatment of human subjects can be associated with memory improvement (Kern et al., 2001); whereas in C. elegans, the signaling of the DAF-2 insulin-like receptor has been shown to promote the association of salt with starvation, but inhibit the association of food signals with certain odorants (Kauffman et al., 2010; Tomioka et al., 2006). Now using deletion mutations that are specific for individual C. elegans ILPs and combinations of these mutations, our study reveals that the diverse regulatory activities of different ILPs can generate various behavioral outputs. We identify a positive role for INS-6 in learning and show that INS-6 represses the learning-inhibitory effect of another ILP, INS-7, via transcriptional repression of ins-7 in the URX neurons. URX-generated INS-7, in turn, negatively regulates learning by antagonizing the positive effect of the DAF-2 receptor in the RIA interneurons (Figure 7E). Our results suggest that ILPs can either enable or prevent learning, depending on the nature of the signaling cascade and the neural circuits through which each ILP is acting. Thus, the large number of ILPs in many animals, such as the 40 C. elegans ILPs, many of which are expressed in neurons (Brogiolo et al., 2001; Ikeya et al., 2002; Li et al., 2003; Liu and Lovenberg, 2008; Pierce et al., 2001; Rulifson et al., 2002), could provide a rich repertoire of neuromodulatory signals.
Studies in worms, flies and mice have shown that insulin and ILPs can affect metabolism and life span through an “ILP-to-ILP” pathway. In mice, inactivation of the insulin receptor in the pancreatic β-cells results in defective insulin secretion (Kulkarni et al., 1999). In flies, the activity of dFOXO in the head fat body non-autonomously modulates the neuronal transcription of one of the fly ILPs, leading to systemic effects on metabolism and life span (Hwangbo et al., 2004; Ikeya et al., 2002). In C. elegans, the FOXO transcription factor in the intestine regulates the expression of ins-7, which non-cell autonomously modulates the ILP pathway in distant tissues (Murphy et al., 2007). Here we demonstrate that a similar “ILP-to-ILP” signaling strategy, INS-6-to-INS-7, is employed to regulate aversive olfactory learning in C. elegans. Importantly, we have also identified the target neuron of the ILP pathway, thereby linking the activities of these neuromodulators to a neural network that underlies experience-dependent behavioral plasticity (Figure 7E).
The INS-6-to-INS-7 pathway regulates the activity of the neural network underlying olfactory learning
Neuropeptides, including ILPs, can regulate experience-dependent plasticity in chemosensation (Chalasani et al., 2010; Kauffman et al., 2010; Kodama et al., 2006; Root et al., 2011; Tomioka et al., 2006; Yamada et al., 2010). In several cases, the target cells are mapped to sensory neurons. In Drosophila, the short neuropeptide F and insulin act on specific olfactory receptor neurons to mediate starvation-dependent modulation of food-search behavior (Root et al., 2011). In C. elegans, INS-1, an ILP released from the interneuron AIA, limits the activity of AWC olfactory sensory neurons, which contributes to adaptation upon prolonged odor exposure (Chalasani et al., 2010). Here we demonstrate that the ILP pathway of “INS-6-to-INS-7” antagonizes the DAF-2 receptor in the interneuron RIA (Figure 6), which is critically required for aversive olfactory learning. Increasing URX-generated INS-7, which can result from loss of INS-6, disrupts learning (Figure 5G), presumably by downregulating DAF-2 activity in RIA. Consistent with this idea, increasing INS-7 signal in URX increases nuclear localization of DAF-16 in RIA (Figures 6E and 6F) and a further increase in DAF-2 activity in RIA suppresses the inhibitory effect of increased URX-generated INS-7 on learning (Figures 6C and 6D). Strikingly, the same increase in URX-INS-7 signal or a decrease in INS-6 signal disrupts RIA activity patterns (Figures 7A–7D). Thus, these results demonstrate that the signal strength of INS-6 and INS-7 are critical for RIA activity, which can contribute to an appropriate state of the learning circuit that is receptive to experience-dependent changes (Figure 7E).
The regulation of a learning network by this ILP pathway is particularly intriguing given that the function of INS-6 and INS-7 in learning requires their specific expression in ASI and URX, respectively, two sensory neurons that detect environmental cues. These cues include population density and oxygen levels, all of which regulate neuronal activity and behavior (Bargmann and Horvitz, 1991; Beverly et al., 2011; Busch et al., 2012; Cornils et al., 2011; Gray et al., 2004; McGrath et al., 2009; Persson et al., 2009; Zimmer et al., 2009). Interestingly, we find that some of these cues, such as dauer-inducing cues, switch off INS-6 expression in ASI (Cornils et al., 2011), and likewise increase INS-7 expression in URX (Figures 8C and 8D). Thus, we propose that INS-6 and INS-7 monitor environmental conditions and convey such information to the nervous system, thereby tuning neural activity to generate appropriate behaviors. Considering that ILPs and their receptors not only regulate multiple physiological and behavioral events but are also expressed in the nervous systems of many organisms, including humans [(Cornils et al., 2011) and references therein], we propose that the ILP function in monitoring both environment and physiology is conserved and this conservation will likely extend to modulation of neuronal activity to promote coordinated responses.
Neuron-specific expression and intrinsic signaling properties confer functional diversity among ILPs
Tissue-specific activities have been reported for DAF-2 and some ILPs, like INS-6, in generating systemic effects on physiology, like dauer formation and longevity (Apfeld and Kenyon, 1998; Cornils et al., 2011; Wolkow et al., 2000). Here we demonstrate neuron-specific ILP function in regulating olfactory learning. Under normal conditions, ins-6 is only expressed in the ASI neurons and this expression switches to the ASJ neurons when animals have entered the dauer program (Cornils et al., 2011). However, only ins-6 expression in ASI, but not in ASJ, enables learning (Figures 2B and 2C). Similarly, despite being expressed in multiple neurons, only INS-7 in the URX neurons inhibits learning by antagonizing DAF-2 in the postsynaptic neuron RIA (Figures 4 and 6). We have also shown that INS-6 positively regulates learning by suppressing ins-7 expression specifically in URX (Figure 5). Similar to other neuropeptides (Edwards, 1998; Richmond and Broadie, 2002), this regulation is likely independent of synapse, since ASI does not form synapses or gap junctions with URX (White et al., 1986). Indeed, ectopically expressing INS-6 in the local environment of URX is sufficient to enable learning (Figure 5F), supporting the possibility of a direct effect of INS-6 on URX through a paracrine-dependent mechanism. Many secreted signaling molecules, including certain TGF-β ligands, WNT and Hedgehog, provide positional information to target tissues, based on the spatial expression pattern of the signals (Drossopoulou et al., 2000; Nellen et al., 1996; Zecca et al., 1996). Since the cell bodies of both URX and ASI are located on the dorsal side of the head and are only about one or two cells apart, while ASJ is located on the ventral side and is further away from URX (White et al., 1986), our findings suggest that INS-6 may act in a similar manner.
The cellular specificity in ILP function may also depend on differences in responses of specific neurons, or non-neuronal cells, to external or internal environmental cues. In addition, the intracellular presence or absence of signaling pathway(s) required to process the ILPs and/or the nature of the local extracellular environment may also regulate the cellular specificity of the ILP signal. Indeed, extracellular matrix properties and the cell surface attributes of signal-releasing cells, as well as the endocytotic activity of the neighboring cells, have been implicated in promoting specific spatial activity of secreted signaling molecules (Affolter and Basler, 2007). Thus, the mechanisms through which different ILP-producing cells interpret environment and experience to regulate diverse physiological events await further investigation.
Despite some similarities in protein structural elements, INS-6 and INS-7 cannot functionally substitute for each other (Figures 8A and 8B), revealing their distinct signaling properties. This functional diversity is consistent with their differences in amino acid composition (Pierce et al., 2001). Moreover, different ILPs may function either as homodimers or heterodimers that bind different receptors with different affinities (Adams et al., 2000; Alvino et al., 2011; Lawrence et al., 2007). Since both the invertebrate and vertebrate genomes encode large ILP families (Brogiolo et al., 2001; Ikeya et al., 2002; Li et al., 2003; Liu and Lovenberg, 2008; Pierce et al., 2001; Rulifson et al., 2002), our findings further highlight the importance of understanding the activities of individual ILPs, as well as their potential interactions and cross-regulations at a systems level, in the function of the nervous system.
EXPERIMENTAL PROCEDURES
C. elegans strains were cultivated at 20°C under standard conditions (Brenner, 1974). The aversive olfactory training and assays were performed as described in Zhang et al., 2005. Embryos were isolated by bleaching gravid adults. The naive animals were grown on a plate containing a lawn of E. coli OP50 and the trained animals were exposed to P. aeruginosa PA14 on a plate containing a PA14 lawn and an OP50 lawn. The choice assay was similar to standard chemotaxis assays, except that bacterial suspensions were used as odor sources (Figure 1A). Images were obtained either with an Olympus FV1000A confocal microscope or with a Nikon Eclipse TE2000-U. For DAF-16 subcellular localization, phase-contrast bright field images were also taken for nucleus identification. The fluorescence intensity of images was measured using FV1000 or ImageJ (NIH). Details on strains and experimental procedures are included in the Supplemental Information.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure (P40 OD010440), for strains; S. Mitani for tm2416, tm2001, tm1907, tm2308; and G. Ruvkun for GR1333, C. Kenyon for the daf-16::gfp strain, and I. Katic and J.-L. Bessereau for daf-2 cDNA constructs. This work was supported by funding from the Novartis Research Foundation and Wayne State University to J.A. and The Esther A. and Joseph Klingenstein Fund, March of Dimes Foundation, The Alfred P. Sloan Foundation, The John Merck Fund and NIH (R01 DC009852) to Y.Z.
Footnotes
SUPPLEMENTAL DATA accompany the paper.
AUTHOR CONTRIBUTIONS: Y.Z. conceived of the study. Z.C., M.H., J.A. and Y.Z. designed experiments, analyzed data and co-wrote the paper; all authors performed the experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams TE, Epa VC, Garrett TP, Ward CW. Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol Life Sci. 2000;57:1050–1093. doi: 10.1007/PL00000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- Alvino CL, Ong SC, McNeil KA, Delaine C, Booker GW, Wallace JC, Forbes BE. Understanding the mechanism of insulin and insulin-like growth factor (IGF) receptor activation by IGF-II. PLoS One. 2011;6:e27488. doi: 10.1371/journal.pone.0027488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- Beverly M, Anbil S, Sengupta P. Degeneracy and neuromodulation among thermosensory neurons contribute to robust thermosensory behaviors in Caenorhabditis elegans. J Neurosci. 2011;31:11718–11727. doi: 10.1523/JNEUROSCI.1098-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Busch KE, Laurent P, Soltesz Z, Murphy RJ, Faivre O, Hedwig B, Thomas M, Smith HL, de Bono M. Tonic signaling from O(2) sensors sets neural circuit activity and behavioral state. Nat Neurosci. 2012;15:581–591. doi: 10.1038/nn.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani SH, Kato S, Albrecht DR, Nakagawa T, Abbott LF, Bargmann CI. Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci. 2010;13:615–621. doi: 10.1038/nn.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DY, Stern SA, Garcia-Osta A, Saunier-Rebori B, Pollonini G, Bambah-Mukku D, Blitzer RD, Alberini CM. A critical role for IGF-II in memory consolidation and enhancement. Nature. 2011;469:491–497. doi: 10.1038/nature09667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornils A, Gloeck M, Chen Z, Zhang Y, Alcedo J. Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development. 2011;138:1183–1193. doi: 10.1242/dev.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MW, Morton JJ, Carroll D, Jorgensen EM. Gene activation using FLP recombinase in C. elegans. PLoS Genetics. 2008;4:e1000028. doi: 10.1371/journal.pgen.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drossopoulou G, Lewis KE, Sanz-Ezquerro JJ, Nikbakht N, McMahon AP, Hofmann C, Tickle C. A model for anteroposterior patterning of the vertebrate limb based on sequential long- and short-range Shh signalling and Bmp signalling. Development. 2000;127:1337–1348. doi: 10.1242/dev.127.7.1337. [DOI] [PubMed] [Google Scholar]
- Edwards RH. Neurotransmitter release: variations on a theme. Curr Biol. 1998;8:R883–885. doi: 10.1016/s0960-9822(07)00551-9. [DOI] [PubMed] [Google Scholar]
- Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- Gronke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genetics. 2010;6:e1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha HI, Hendricks M, Shen Y, Gabel CV, Fang-Yen C, Qin Y, Colon-Ramos D, Shen K, Samuel AD, Zhang Y. Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron. 2010;68:1173–1186. doi: 10.1016/j.neuron.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- Hendricks M, Ha H, Maffey N, Zhang Y. Compartmentalized calcium dynamics in a C. elegans interneuron encode head movement. Nature. 2012;487:99–103. doi: 10.1038/nature11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua QX, Nakagawa SH, Wilken J, Ramos RR, Jia W, Bass J, Weiss MA. A divergent INS protein in Caenorhabditis elegans structurally resembles human insulin and activates the human insulin receptor. Genes Dev. 2003;17:826–831. doi: 10.1101/gad.1058003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biology. 2010;8:e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kern W, Peters A, Fruehwald-Schultes B, Deininger E, Born J, Fehm HL. Improving influence of insulin on cognitive functions in humans. Neuroendocrinology. 2001;74:270–280. doi: 10.1159/000054694. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kodama E, Kuhara A, Mohri-Shiomi A, Kimura KD, Okumura M, Tomioka M, Iino Y, Mori I. Insulin-like signaling and the neural circuit for integrative behavior in C. elegans. Genes Dev. 2006;20:2955–2960. doi: 10.1101/gad.1479906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RN, Bruning JC, Winnay JN, Postic C, Magnuson MA, Kahn CR. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96:329–339. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, McKern NM, Ward CW. Insulin receptor structure and its implications for the IGF-1 receptor. Curr Opin Struct Biol. 2007;17:699–705. doi: 10.1016/j.sbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Yeh SH, Lin CH, Lu KT, Leu TH, Chang WC, Gean PW. A role for the PI-3 kinase signaling pathway in fear conditioning and synaptic plasticity in the amygdala. Neuron. 2001a;31:841–851. doi: 10.1016/s0896-6273(01)00433-0. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genetics. 2001b;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Liu C, Lovenberg TW. Relaxin-3, INSL5, and their receptors. Results and problems in cell differentiation. 2008;46:213–237. doi: 10.1007/400_2007_055. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man HY, Lin JW, Ju WH, Ahmadian G, Liu L, Becker LE, Sheng M, Wang YT. Regulation of AMPA receptor-mediated synaptic transmission by clathrin-dependent receptor internalization. Neuron. 2000;25:649–662. doi: 10.1016/s0896-6273(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Marder E. Neuromodulation of neuronal circuits: back to the future. Neuron. 2012;76:1–11. doi: 10.1016/j.neuron.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, Bargmann CI. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Vizuete A, Fierro Gonzalez JC, Gahmon G, Burghoorn J, Navas P, Swoboda P. Lifespan decrease in a Caenorhabditis elegans mutant lacking TRX-1, a thioredoxin expressed in ASJ sensory neurons. FEBS Lett. 2006;580:484–490. doi: 10.1016/j.febslet.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Murphy CT, Lee SJ, Kenyon C. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:19046–19050. doi: 10.1073/pnas.0709613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Peckol EL, Troemel ER, Bargmann CI. Sensory experience and sensory activity regulate chemosensory receptor gene expression in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2001;98:11032–11038. doi: 10.1073/pnas.191352498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson A, Gross E, Laurent P, Busch KE, Bretes H, de Bono M. Natural variation in a neural globin tunes oxygen sensing in wild Caenorhabditis elegans. Nature. 2009;458:1030–1033. doi: 10.1038/nature07820. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Broadie KS. The synaptic vesicle cycle: exocytosis and endocytosis in Drosophila and C. elegans. Curr Opin Neurobiol. 2002;12:499–507. doi: 10.1016/s0959-4388(02)00360-4. [DOI] [PubMed] [Google Scholar]
- Root CM, Ko KI, Jafari A, Wang JW. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell. 2011;145:133–144. doi: 10.1016/j.cell.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Sze JY, Victor M, Loer C, Shi Y, Ruvkun G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M, Adachi T, Suzuki H, Kunitomo H, Schafer WR, Iino Y. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 2006;51:613–625. doi: 10.1016/j.neuron.2006.07.024. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans Roy Soc London Ser B. 1986:314. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans lifespan by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Yamada K, Hirotsu T, Matsuki M, Butcher RA, Tomioka M, Ishihara T, Clardy J, Kunitomo H, Iino Y. Olfactory plasticity is regulated by pheromonal signaling in Caenorhabditis elegans. Science. 2010;329:1647–1650. doi: 10.1126/science.1192020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Direct and long-range action of a wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- Zhao W, Chen H, Xu H, Moore E, Meiri N, Quon MJ, Alkon DL. Brain insulin receptors and spatial memory. Correlated changes in gene expression, tyrosine phosphorylation, and signaling molecules in the hippocampus of water maze trained rats. J Biol Chem. 1999;274:34893–34902. doi: 10.1074/jbc.274.49.34893. [DOI] [PubMed] [Google Scholar]
- Zimmer M, Gray JM, Pokala N, Chang AJ, Karow DS, Marletta MA, Hudson ML, Morton DB, Chronis N, Bargmann CI. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron. 2009;61:865–879. doi: 10.1016/j.neuron.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.