Abstract
Metabolism impacts all cellular functions and plays a fundamental role in biology. In the last century, our knowledge of metabolic pathway architecture and the genomic landscape of disease have increased exponentially. Combined with these insights, advances in analytical methods for quantifying metabolites and systems approaches to analyze these data now provide powerful tools to study metabolic regulation. Here we review the diverse mechanisms cells use to adapt metabolism to specific physiological states and discuss how metabolic flux analyses can be applied to identify important regulatory nodes to understand normal and pathological cell physiology.
Beginning in the mid-19th century, biochemists began describing the network of reactions that comprise cell metabolism. With advances in molecular biology and genomics, the genes encoding metabolic enzymes have largely been defined. Recent links between cell signaling, metabolism, and the pathogenesis of disease have been discovered (Cairns et al., 2011; DeBerardinis and Thompson, 2012; Saltiel and Kahn, 2001), and rekindled an interest in understanding the metabolic network.
While the basic architecture of central carbon metabolism is known, the complexity of the network complicates identification of those nodes most amenable to therapeutic intervention. Detailed metabolic maps annotate the enzymes, substrates, products, and cofactors involved in cell biochemistry; however, metabolism is often considered at the level of individual reactions or pathways, and most studies interrogate metabolism by examining changes in enzyme expression or measuring relative changes in metabolite levels. While this offers insights, it is the flow of metabolites through metabolic networks that best characterizes the relationships between cell biology and biochemistry. Metabolite flux, or the rate of substrate conversion per cell, provides the energy for life. How cells control metabolism in some contexts is well described, but new mechanisms of regulation continue to be discovered. Indeed, components of central metabolism, such as the carrier responsible for transporting pyruvate into the mitochondria, have recently been identified (Bricker et al., 2012; Herzig et al., 2012). A growing appreciation for the role of metabolism in disease has accelerated research to understand metabolic control. Here we explore how cells regulate metabolism and discuss methods for quantifying metabolic processes.
The role of metabolism in cell physiology is context dependent
While the basic metabolic currencies remain the same across cells (e.g. ATP; acetyl coenzyme A, AcCoA; NADH; NADPH), the metabolic requirements of each cell type are determined by their tissue function and environment. For example, immune cells remain quiescent for extended periods but then rapidly proliferate upon stimulation. To accomplish this, cells shift from a state of low nutrient uptake that maintains basal functions to a state of increased nutrient uptake with activation of anabolic pathways that facilitates rapid growth and division (Wang et al., 2011). On the other hand, differentiated cardiac myocytes do not proliferate but have a high demand for ATP. As a result, these cells rely heavily on oxidative phosphorylation to efficiently generate ATP (Khairallah et al., 2004). Hepatocytes are tasked with controlling blood chemistry and thus need flexibility to perform energy-intensive processes such as the synthesis of glucose, amino acids, and macromolecules while also recycling byproducts of metabolism from other tissues into useful metabolites and excreting un-needed or toxic material as waste (Merritt et al., 2011). Each of these tissue physiologies requires different ratios of the metabolic currencies and employs unique regulatory circuits.
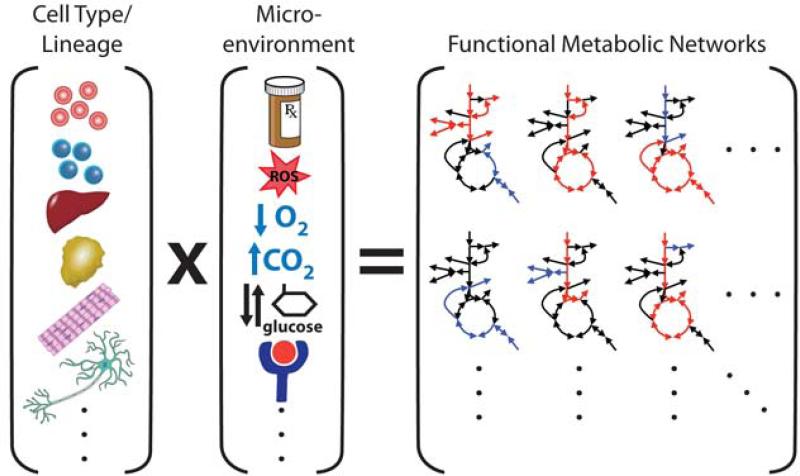
Nutrient availability can also vary for different cells. For instance, hepatocytes exist within a gradient of oxygen and nutrients imparted by the portal architecture of the liver (Puchowicz et al., 1999). For cells to contend with glucose limitation, one known adaptation is to decrease glucose oxidation and utilize amino acids, fatty acids, or their breakdown products to fuel mitochondrial respiration (Cahill et al., 1972; Krebs, 1966; Ruderman, 1975). Consumption of available macromolecules can also serve as a source of fuel, and catabolic pathways are activated during the process of macroautophagy to maintain energy homeostasis when other nutrients are limited (Lum et al., 2005). Therefore, both the tissue and microenvironment will determine the metabolic phenotype of cells and impact how regulatory events affect normal and disease states (Figure 1). While the core metabolic pathways used to adapt to these conditions (e.g. glycolysis, the pentose phosphate pathway (PPP), the tricarboxylic acid (TCA) cycle) are fairly constant, the means through which cells detect and respond to endogenous and exogenous signals are diverse. As such, alterations in cell metabolism have to be considered in relevant contexts.
Figure 1. Cellular and environmental heterogeneity promote metabolic diversity.
Cells express lineage-specific networks of metabolic enzymes and regulatory factors that support appropriate tissue functions. Microenvironments, hormonal cues, and/or pharmacological perturbations will elicit adaptive metabolic responses that are unique to the metabolic networks of individual cells. Thus, a diverse set of metabolic phenotypes is observed in normal and disease states.
Cells employ diverse mechanisms to regulate metabolism
Organisms and cells have evolved systems to modulate metabolic flux over short and long time scales. Though not discussed in detail here, hormones and other extracellular factors communicate signals between tissues to regulate metabolic function (Randle, 1963; Saltiel and Kahn, 2001). At the cellular level, genes encoding enzyme isoforms and regulatory factors allow tissue- and context-specific responses, and the abundance of these proteins is controlled by mRNA transcription, splicing, stability, and translation (Cairns et al., 2011). Another means of regulating enzyme function is post translational modifications (PTM), which provide a feedback mechanism for metabolites that act as substrates for PTM reactions (Zhao et al., 2010). Finally, small molecules can affect metabolic flux by allosteric effects on enzymes (Carminatti et al., 1971). The goal of these processes is to parse metabolites through pathways in the proportions needed to match the requirements of individual cells.
Because metabolic needs can fluctuate on the order of seconds or persist for prolonged periods, no single molecule or system can effectively control fluxes in a way that is adaptive for every situation. For example, ATP is rapidly metabolized and in the heart the ATP pool can turnover more than 6 times per minute (Jacobus, 1985); at such rates if consumption remains constant, a 10% decrease in ATP production would halve ATP levels in less than one minute. Cells must therefore rapidly sense and respond to perturbations in energy state. However, a per-cell quantity of a single metabolite is not always informative of physiological state. Individual reaction rates are influenced by the abundance of both the substrate and product, and cell control systems often detect metabolite ratios. For example, the concentration of ATP in cells is far above the Km for most enzymes that use ATP as a substrate (Albe et al., 1990). When coupled to other reactions, ATP hydrolysis to ADP supplies energy to make those reactions favorable, but also makes the ratio of ATP to ADP the relevant parameter in determining whether cells have sufficient energy. The ATP/ADP ratio is itself buffered by both creatine kinase and adenylate kinases. When demand for ATP is low, the high ATP/ADP ratio is used to produce creatine-phosphate which can then regenerate ATP from ADP when the ATP/ADP ratio falls (Bessman and Geiger, 1981). This latter system is important in muscle to support contractile function during periods of prolonged ATP demand. Adenylate kinases can convert two ADP to produce an ATP and AMP to maintain energy charge in periods of nutrient stress (Dzeja et al., 1998). This promotes a high ATP/AMP ratio and represents the energy charge of a cell. Indeed, AMP allosterically regulates key metabolic enzymes that control flux into glycolysis and oxidative phosphorylation to increase ATP production.
At the signal transduction level, AMP-activated protein kinase (AMPK) is a sensor that responds to changes in the ratio of ATP to AMP (and ADP) and coordinates diverse metabolic responses (Hardie, 2011; Oakhill et al., 2011). AMPK is a heterotrimeric complex with each subunit encoded by more than one gene, and tissue-specific expression of different isoforms provides a genetically encoded means of mediating heterogeneous regulatory responses (Viollet et al., 2009). AMPK affords both short- and long-term feedback control for cells by controlling the activity of numerous proteins through phosphorylation such as glucose transporters, 3-hydroxy-3-methyl-glutaryl-CoA reductase, and AcCoA carboxylase (Hardie, 2011). By increasing glucose uptake and decreasing cholesterol and lipid synthesis, this regulation provides a means of rapidly modulating ATP production and consumption, respectively. AMPK also regulates the mammalian target of rapamycin complex-1 (mTORC1) (Inoki et al., 2003). Inhibition of mTORC1 is crucial for cell survival under conditions of stress, as rapamycin-mediated inhibition can decrease biosynthetic processes that consume ATP and prevent bioenergetic catastrophe (Choo et al., 2010). Additionally, activation of autophagy through phosphorylation of ULK1 can provide additional fuel to support ATP production in mitochondria (Egan et al., 2011). Metabolic control over longer time scales is accomplished by AMPK through control of gene expression. SREBP1 is involved in lipid and carbohydrate metabolism, and AMPK is thought to inactivate this transcription factor through phosphorylation (Li et al., 2011). Another important target of AMPK is PGC1α, which acts as a transcriptional co-activator of PPARγ to drive mitochondrial biogenesis (Jager et al., 2007). In this manner, AMPK induces changes in the cell that facilitate ATP generation. Together, these short- and long-term controllers mediate a coordinated response to stabilize the energy state of cells.
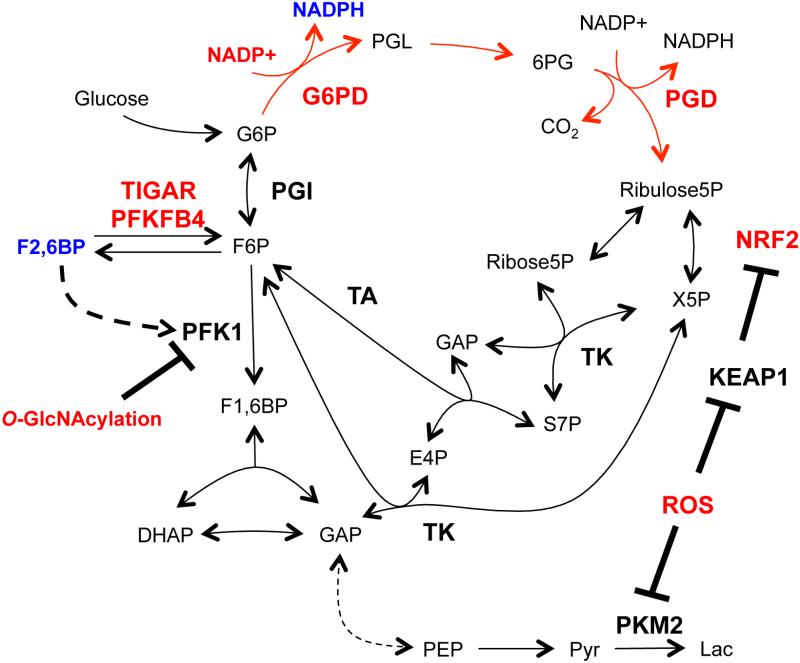
Cells also have both fast-acting and long-term control solutions for adapting to fluctuations in cellular redox state. The oxidative PPP helps supply cells with NADPH to maintain reduced glutathione and mitigate damage caused by reactive oxygen species (ROS). Glucose-6-phosphate dehydrogenase (G6PD) catalyzes the first committed step in the oxidative PPP and directly converts NADP+ to NADPH. Because these cofactors are a product and substrate of the enzyme, a falling NADPH/NADP+ ratio stimulates flux through G6PD (Holten et al., 1976). However, additional regulatory events can also influence flux through this pathway that branches from glycolysis just above the reaction catalyzed by phosphofructokinase (PFK1) (Figure 2). PFK1 activity is responsive to levels of the metabolite fructose-2,6-bisphosphate (F2,6BP), and F2,6BP abundance is controlled by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphate-2-phosphatase (PFK2), a bifunctional enzyme system designed to regulate F2,6BP levels and PFK1 in response to signaling inputs (Pilkis et al., 1995). While this system forms a key circuit in the liver to control glycolysis and gluconeogenesis, selective expression of PFK2 isoforms with differential kinase and phosphatase activity can also impact PFK1 activity in other cells. In fact, expression of the PFKFB4 isoform of PFK2 is critical for redox homeostasis and cell survival in some prostate cancers (Ros et al., 2012). Another enzyme that influences F2,6BP abundance is the fructose-2,6-bisphosphate-2-phosphatase TIGAR, which is a target of p53 in response to stress. Expression of TIGAR leads to decreased F2,6BP levels and PFK1 activity, which shunts more glucose carbon into the oxidative PPP (Bensaad et al., 2006). Additionally, p53 can interact with G6PD and modulate oxidative PPP activity (Jiang et al., 2011). Glycosylation of PFK1 itself also affects enzyme activity and regulates glucose flux to the oxidative PPP, potentially allowing a more rapid stress response than new enzyme synthesis (Yi et al., 2012). More distal regulatory events in glycolysis can also influence PPP flux, as ROS-mediated oxidation of cysteine residues in the pyruvate kinase M2 isoform (PKM2) can acutely divert glycolytic flux toward the PPP (Anastasiou et al., 2011). Finally, antioxidant response pathways such as NRF2 are triggered to activate transcriptional programs that increase expression of PPP and other enzymes that scavenge ROS (Mitsuishi et al., 2012). It is likely that different combinations of the above strategies facilitate oxidative stress responses for cells in different situations.
Figure 2. Multiple regulatory events converge on the oxidative pentose phosphate pathway.
The oxidative pentose phosphate pathway (PPP; red arrows) generates NADPH to maintain reduced glutathione, combat reactive oxygen species (ROS), and maintain redox homeostasis. Transcriptional events and direct enzyme regulation via metabolic intermediates and PTMs mediate both long-lasting and rapid responses to oxidative stress by modulating oxidative PPP flux. Factors that increase oxidative PPP flux are shown in red, while those that reduce flux are shown in blue.
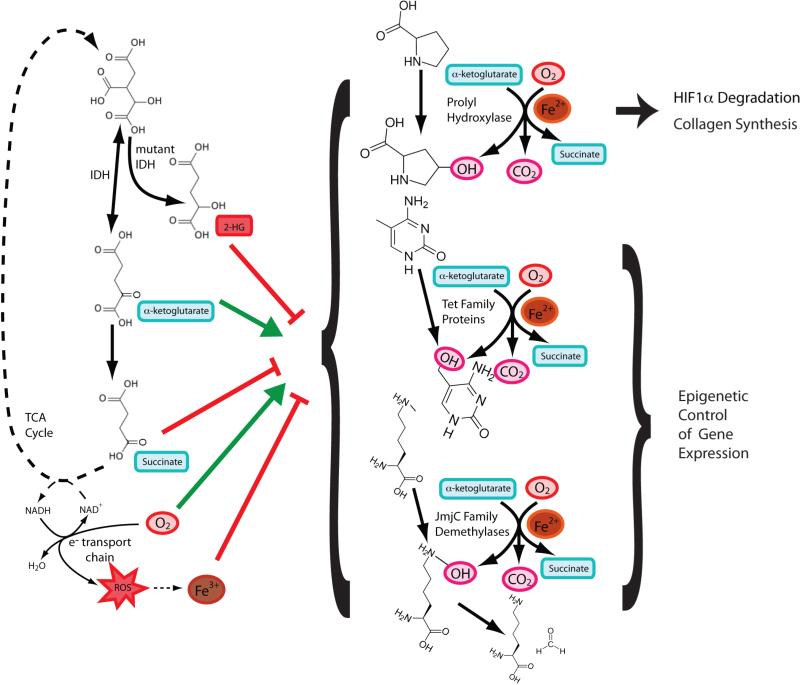
Cells must sense and respond to environmental changes. A signaling response to low oxygen is mediated by the EGLN-family of alpha-ketoglutarate (αKG)-dependent dioxygenases. The half-life of the hypoxia inducible factor (HIF) transcription factor complex α subunit is short when oxygen levels are high because the protein is hydroxylated by the EGLN proteins, recognized by the von Hippel Lindau E3 ubiquitin ligase, and targeted for degradation (Ivan et al., 2001; Jaakkola et al., 2001). In hypoxia, oxygen becomes limiting for protein hydroxylation and active HIF transcription factor accumulates to induce an adaptive gene expression program (Kaelin and Ratcliffe, 2008). In addition, because the enzymatic mechanism of HIFα-modification by EGLN proteins requires O2, αKG, and Fe2+ while producing both succinate and the hydroxylated species (Figure 3), O2 is not the only factor that influences enzyme activity and HIFα stabilization (Bruick and McKnight, 2001; Epstein et al., 2001). Changes in cellular redox state can increase HIFα levels because increased ROS oxidizes Fe2+ and limits EGLN activity, and elevated ROS levels during hypoxia can activate HIF-dependent transcription (Chandel et al., 1998). Emerging evidence suggests that other αKG-dependent dioxygenases in the TET and JmjC protein families can hydroxylate DNA and histones to influence the epigenetic state of cells (Tahiliani et al., 2009; Tsukada et al., 2006) (Figure 3). In turn, αKG-dependent dioxygenase activity can be directly modulated by metabolite levels. Succinate and fumarate abundances increase with oncogenic loss-of-function mutations in the TCA cycle enzymes SDH and FH, and both metabolites can affect dioxygenase activity (Xiao et al., 2012). For the same reason, changes in nutrient levels or regulatory events that lead to alterations in αKG/succinate ratios might also influence HIF-dependent transcription (Tennant et al., 2009). (D)-2-hydroxyglutarate (2HG), an oncometabolite that accumulates to high levels in cells as a direct product of cancer-associated mutant IDH enzymes, can inhibit αKG-dependent dioxygenase activity (Chowdhury et al., 2011; Figueroa et al., 2010; Xu et al., 2011). This effect of 2HG can block differentiation of cells by inhibition of TET2 and other αKG-dependent dioxygenases that regulate chromatin structure and collagen maturation and has been implicated in the pathogenesis of IDH-mutant tumors (Figueroa et al., 2010; Lu et al., 2012; Sasaki et al., 2012a; Sasaki et al., 2012b). Increasing the complexity of this metabolic regulation, 2HG can also replace αKG as co-substrate for a subset of these dioxygenases (Koivunen et al., 2012). These findings illustrate how multiple aspects of metabolism can influence αKG-dependent dioxygenase enzyme activity and have effects on cell physiology, including promotion of cancer.
Figure 3. αKG-dependent dioxygenases integrate numerous metabolic inputs and elicit pleiotropic effects within cells.
The prolyl hydroxylases, TET proteins, and JmjC domain-containing proteins all carry out similar enzymatic reactions. These dioxygenases split molecular oxygen in an Fe2+-dependent reaction where one oxygen is transferred as a hydroxyl group and the other oxygen is used in a decarboxylation reaction the converts αKG to succinate. Each family of proteins has different hydroxylation substrates. All of these enzymes are influenced by events affecting the redox state of Fe or any of the substrates and products, as well as by levels of (D)-2-hydroxyglutarate, succinate, or fumarate that accumulate as a result of cancer-associated mutations in TCA cycle enzymes.
Complex regulation underlies the Warburg effect
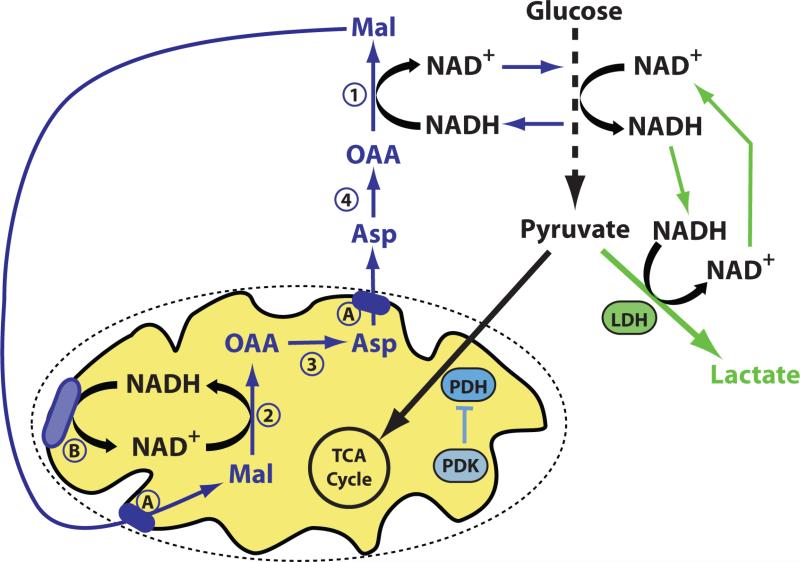
The increased uptake of glucose and conversion to lactate in the presence of oxygen, also known as the “Warburg Effect”, or aerobic glycolysis, is a characteristic feature of many cancers (Cairns et al., 2011; Vander Heiden et al., 2009). Multiple regulatory events, including HIF-dependent transcription, have been implicated in driving glucose to lactate conversion. However, instead of one event causing the Warburg effect, numerous factors play a role in determining the fate of glucose in cancer cells (Figure 4). HIF and other signaling pathways control flux through glycolysis, which ends with the production of pyruvate. At the center of pyruvate metabolism lies the pyruvate dehydrogenase (PDH) complex, a multi-subunit complex that catalyzes the oxidative decarboxylation of pyruvate to AcCoA and conversion of NAD+ to NADH in the mitochondria. This complex is subject to regulation by PDH kinases (PDKs) and phosphatases that control phosphorylation of the E1α subunit of the PDH complex to control its activity. Different PDK isoforms respond to various signaling and metabolic inputs to control carbon entry into the TCA cycle, including hypoxia and nutrient levels (Kim et al., 2006; Papandreou et al., 2006). Apart from phosphorylation, PDH complex activity is influenced by the mitochondrial NAD+/NADH ratio and AcCoA levels (Roche and Hiromasa, 2007). The complex regulation of PDH activity illustrates how diverse metabolic inputs can affect pyruvate fate.
Figure 4. Enzymes controlling pyruvate fate as well as metabolism of NAD+/NADH can influence the Warburg effect.
Increased LDH activity and/or decreased PDH activity can shunt glucose-derived pyruvate to lactate. However, metabolism of glucose to pyruvate requires conversion of NAD+ to NADH, and NAD+ must be regenerated from NADH for glycolysis to continue. Pyruvate to lactate conversion efficiently recycles NADH back to NAD+, while oxidation of NADH to NAD+ involving the mitochondria requires four separate enzymatic reactions (numbered 1-4), metabolite transport across the mitochondrial membranes (A), and coupling to the electron transport chain (B) (or other routes of mitochondrial NADH oxidation).
Although subject to less complex regulation than PDH, flux through the enzyme lactate dehydrogenase (LDH) to convert pyruvate to lactate is also not dependent on only the amount of enzyme. Different LDH isoforms can bias the directionality of the reaction to either promote lactate excretion as a waste product or consumption of lactate as a fuel, but in all cases the interconversion of lactate and pyruvate involves NAD+ and NADH as co-substrates (Bittar et al., 1996; Cahn et al., 1962). Thus, flux through LDH is affected by the redox state of these cofactors. In fact, in some contexts the pyruvate/lactate ratio has been used as a surrogate for the NAD+/NADH ratio (Brindle et al., 2011; Newsholme and Crabtree, 1979). Importantly, this relationship between pyruvate/lactate and NAD+/NADH will only hold true if the reaction is at equilibrium; however metabolic reactions in living cells are not necessarily close to equilibrium so caution must be used in broadly applying metabolite ratios to extrapolate information about a specific node in metabolism. The specific isoform of pyruvate kinase can also influence pyruvate to lactate conversion in cells. The PKM2 isoform promotes anabolic metabolism with glucose to lactate conversion, while high pyruvate kinase activity from the PKM1 isoform promotes efficient pyruvate oxidation in the TCA cycle (Anastasiou et al., 2012). While this phenomenon is not completely understood, it may reflect the influence of pyruvate kinase activity on feedback controls within the metabolic network such as the NAD+/NADH ratio rather than channeling of pyruvate to a specific compartment.
Lower pyruvate kinase activity associated with PKM2 can decouple glycolytic flux from ATP synthesis (Vander Heiden et al., 2010) and may account for the low NAD+/NADH ratio observed in cancer cells (Hung et al., 2011). Even though the cytosolic and mitochondrial pools of NAD(H) are separate, metabolic shuttle systems allow for the transfer of reducing equivalents between compartments, and these are subject to regulation at the level of mitochondrial membrane transport, activity of dehydrogenase enzymes in each compartment, and flux through other enzymes that shuttle electrons between dehydrogenase reactions (Figure 4). LDH may therefore offer a more efficient means of regenerating cytosolic NAD+ to support the high glycolytic rates of cancer cells. Proliferating cells may favor the production of lactate based on kinetic considerations, which might explain the propensity of PKM2-expressing cells to make lactate. In addition, metabolites that participate in the glycerol-phosphate and malate-aspartate shuttle systems responsible for moving reducing equivalents across the mitochondrial membranes are also required for biosynthesis. Glycerol-3-phosphate is a side product of glycolysis and is involved in lipid synthesis, while aspartate is used to synthesize proteins and nucleotides. This illustrates that no single event can explain a metabolic phenotype such as the Warburg effect. Instead, complex regulation allows metabolism to be tuned by a variety of inputs to match cellular needs. In the context of understanding disease mechanisms, changes in any single component of the system may or may not be important, and the rate-limiting or rate-controlling steps may also vary, making it challenging to identify the mechanism of metabolic dysfunction in disease models based on gene expression changes alone. Therefore, efforts to quantify metabolic flux in complex systems are important to understand the contribution of altered cell metabolism to disease.
Integrative approaches to quantify metabolic flux
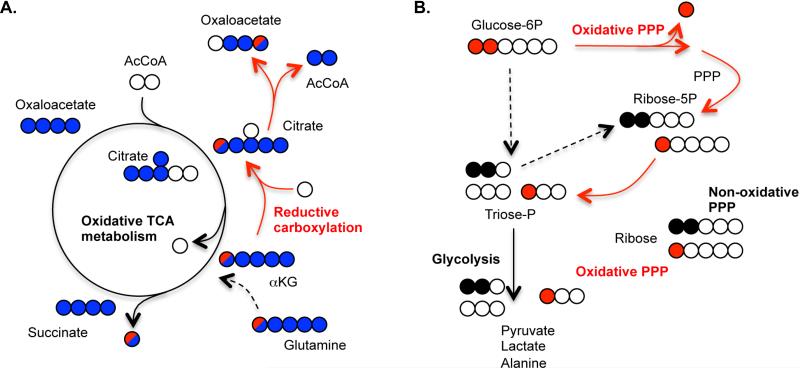
Aided by technological developments in chromatography, mass spectrometry (MS), and nuclear magnetic resonance (NMR), the metabolome (i.e., static abundance of metabolites) can be observed in increasing detail. The use of isotope-labeled tracers can add contrast to such datasets and allow visualization of how a specific nutrient contributes to metabolic processes (Hellerstein, 2004). As tracers are metabolized within cells, labeled atoms become enriched in various compounds, and this incorporation is a function of label flux into and out of the metabolite pools. Steady state isotopic labeling alone can provide information on relative fluxes within cells, in particular within pathways that consume multiple substrates. A priori design is important, as the specific tracer used influences what information can be ascertained from a given experiment (Metallo et al., 2009). For example, citrate labeling from uniformly 13C-labeled glutamine ([U-13C5]glutamine; labeled on all 5 carbons) is often used to estimate oxidative and reductive TCA metabolism in cultured cells (Figure 5A). When labeled glutamine is converted to 5-carbon labeled αKG, metabolism in one pass of the oxidative TCA cycle leads to 4-carbon labeled citrate because one of the labeled carbons is lost as 13CO2. In contrast, if the labeled αKG is reductively metabolized by IDH, then all 5 labels are retained in citrate. However, this labeling pattern can also be generated through reversible exchange reactions or via oxidative TCA cycling through malic enzyme and pyruvate carboxylase (Le et al., 2012), and additional measurements are needed to obtain a definitive readout of reductive flux. Singly labeled [1-13C]glutamine and [5-13C]glutamine tracers can provide a measure of net reductive flux when quantified on downstream metabolites such as aspartate, fumarate, or lipids because these carbons have different fates when metabolized by reductive or oxidative TCA pathways (Holleran et al., 1995; Metallo et al., 2012).
Figure 5. Probing metabolic pathways with stable isotope tracers.
A) Uniformly labeled [U-13C5]glutamine and singly labeled [1-13C]glutamine provide independent means of distinguishing reductive carboxylation from oxidative TCA metabolism when measuring 13C enrichment in various intermediates. Five carbon labeled citrate derived from uniformly labeled glutamine suggests reductive carboxylation while four carbon labeled citrate suggests oxidative TCA metabolism (blue circles). Five carbon labeled citrate can also be obtained from glutamine via pyruvate cycling (not shown), so label incorporation into citrate, oxaloacetate, and downstream metabolites from [1-13C]glutamine provides an independent assessment of reductive TCA flux (red circle). B) Glucose labeled on only the first and second carbons ([1,2-13C2]glucose) is often used to determine relative flux through the oxidative and non-oxidative PPP by assessment of singly versus doubly-labeled downstream intermediates.
Isotope tracers are also commonly used to make flux estimates at the branch-point between glycolysis and the oxidative PPP. Measuring 14CO2 loss from the first carbon of glucose can directly measure oxidative PPP flux provided that appropriate controls are included to account for oxidation of this carbon to 14CO2 at later steps in glucose metabolism (Anastasiou et al., 2011; Beatty et al., 1966; Ying et al., 2012). When using [1,2-13C2]glucose (labeled on only the 1 and 2 carbons) the carbon from the 1 position is lost as 13CO2 along the oxidative pathway, and the labeled carbon from the 2 position is retained on metabolites that re-enter glycolysis. Both labeled carbons remain on glycolytic intermediates when this tracer is not metabolized by the oxidative PPP (Figure 5B). Therefore, the quantity of singly versus doubly 13C-labeled metabolites provides a relative measure of oxidative PPP metabolism (Lee et al., 1998). This approach has been used to show that ribose for nucleotide synthesis is derived via the non-oxidative rather than the oxidative PPP in many cancer cells (Boros et al., 1998; Ying et al., 2012). These analyses are dependent upon specific assumptions regarding network operation (e.g., minimal pyruvate and pentose recycling). In some cases, the absence of a unique labeling pattern distinguishing two possibilities makes direct assessment of a particular flux impossible. Compartmentalization of pathways further complicates analysis. Whether reductive carboxylation of αKG is catalyzed in the cytosol (IDH1), the mitochondria (IDH2), or both cannot be definitively answered with the aforementioned experiments (Metallo et al., 2012; Mullen et al., 2012; Scott et al., 2011; Wise et al., 2011). More work is needed to develop better experimental and mathematical tools to tackle these questions, though approaches to model labeling data in the context of a network can provide insights.
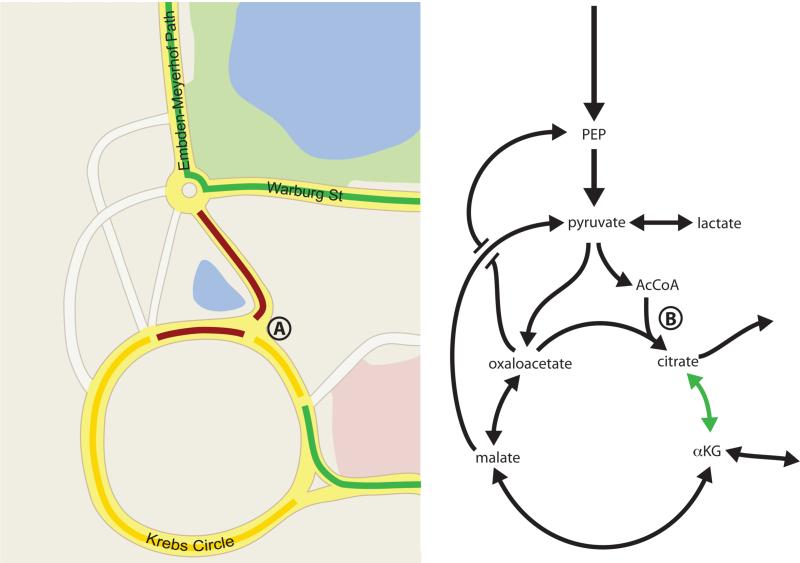
Much like traffic patterns on a highway, change in flow caused by an accident may affect traffic speeds at various locations on a map (Figure 6). In the context of metabolism, model-based approaches provide a means of analyzing these networks as a system. The hallmarks of these methods are 1) the interpretation of data within interconnected pathways and 2) the application of mass conservation principles. Such approaches first involve construction of a stoichiometric matrix, S, from metabolic pathway architecture, which defines the system to be analyzed and creates a mathematical framework that describes all possible metabolic interconversions within a proposed network (Quek et al., 2010; Zamboni, 2011). This matrix enables one to map the linear relationship between a flux vector, ν, and the rate of change of metabolite concentrations, .
Figure 6. Variables affecting traffic flow are analogous to the regulation of metabolic flux.
The available routes as well as congestion on each road will affect how quickly cars can navigate between two points. High traffic volume into a point where two roads merge (such as point A on the map) will cause a slowing of traffic as depicted in yellow or red, and result in fewer cars moving via that route per unit time. Road closures or traffic signals can further affect flux though the network of roads as depicted here in the style of Google Maps. Similar principles affect metabolite flow through reaction networks. Decreased flux from AcCoA into the TCA cycle at point B leads to a drop in citrate levels, allowing increased flux from αKG to citrate (green arrow). This example illustrates how a large change in enzyme level may have little effect if metabolite traffic is not constrained at that node, while small changes in enzyme activity can have dramatic effects on flux if a step is limiting. Thus, analysis of individual metabolites, enzyme levels, or maximal enzymatic capacities for single steps may or may not be informative of overall flux.
At steady state (i.e. metabolite concentrations are constant), the right hand side of this equation is zero. This simplifies estimation of fluxes and facilitates interpretation of metabolite labeling data. When combining steady state isotope distribution data with absolute measurements of major nutrient uptake and metabolite secretion, these network models can provide estimates of absolute fluxes throughout the pathways included in the model. By iteratively adjusting flux values to minimize the difference between simulated and measured labeling data one can estimate metabolic fluxes from this information (Sauer, 2006). This approach, referred to as metabolic flux analysis (MFA), depends on a near complete understanding of the network structure. In the case of “linear” pathways (those fueled by a single nutrient such as glucose) dynamic measurements of metabolite labeling are required. In this way, nonstationary MFA or kinetic flux profiling approaches can more effectively estimate fluxes using dynamic labeling information (Maier et al., 2008; Munger et al., 2008; Murphy et al., 2012). Importantly, metabolite pool size will influence labeling kinetics; therefore, reliable flux measurements can only be made when interpreting these data in the context of absolute metabolite concentrations. Finally, a sensitivity analysis to determine confidence intervals informs the significance of flux estimates (Antoniewicz et al., 2006). These approaches allow an analysis of metabolism that accounts for the systems-level character of biochemical networks.
Challenges and limitations with existing approaches to determine flux
While these approaches offer powerful tools for studying metabolism, results from such experiments must be interpreted in the context of specific limitations and assumptions. Much of our current knowledge of metabolic pathways was garnered through the use of radioactive tracers, typically through quantification of 3H or 14C loss (Consoli et al., 1987; Landau et al., 1995; Liang et al., 1997). These tracers have the advantage of exquisite sensitivity, but results can be misleading if a limited subset of metabolites are measured, or if label cannot be assigned to specific metabolites. Metabolomics approaches can quantify static metabolite abundances within cells, tissues, and plasma at relatively high-throughput, and this information can serve as a biomarker and/or suggest pathways that are deregulated in disease. However, it is often difficult to elucidate mechanisms from these data since metabolite levels are a function of both production and consumption. Due to reaction reversibility (i.e. metabolic exchange), cyclic pathways, and competition among enzymes/pathways, the abundances of individual compounds are not always informative of flux, or even the activity of a proximal enzyme.
Even small changes in reaction rates integrated over time can have a major impact on metabolite levels and cells. Although mutant IDH1 activity is significantly lower than that of the wild-type enzyme, the lack of appreciable enzyme capacity to eliminate 2HG causes this metabolite to accumulate to millimolar levels within tumors (Dang et al., 2009; Gross et al., 2010; Ward et al., 2010). Available enzyme capacity for many steps in metabolism is in excess, and changing enzyme levels with RNA interference or other genetic means might have minimal effects on substrate or product levels. When activity does change, distal points of the metabolic network may be indirectly affected. Activation of pyruvate kinase causes a larger decrease in serine levels than it does an increase in pyruvate levels (Anastasiou et al., 2012), perhaps because the pyruvate pool has multiple inputs other than pyruvate kinase while relative depletion of upstream glycolytic intermediates limits their shunting into serine biosynthesis.
In vitro studies of cells in isolation are informative with respect to cell autonomous metabolism; however, experiments with mammalian cells in culture do not necessarily mimic physiological microenvironments. Indeed, as metabolism is evaluated under more relevant conditions striking changes in pathway function are observed (Metallo et al., 2012; Wise et al., 2011). Isotope tracers are increasingly being applied to in vivo systems ranging from C. elegans to human patients (Fan et al., 2009; Marin-Valencia et al., 2012; Perez and Van Gilst, 2008; Sunny et al., 2011). Although the experimental considerations may differ in these contexts, the fundamental aspects of metabolic flux, isotope enrichment, and steady state assumptions remain important. While studying metabolism in animals eliminates potential artifacts related to cell culture, in vivo experiments present challenges. Both the strengths and limitations of these experiments arise from the complexity of animal physiology as well as the intrinsic heterogeneity of cells within tissues, and results therefore depend on the route of administration, tissue and method of sampling, nutritional state, and stress level of the animal (Ayala et al., 2010). In addition, labeling of some tracers during the course of a study can be affected by metabolic pathways such as the Cori cycle, where excreted products of glucose metabolism in peripheral tissues are recycled by the liver to regenerate partially labeled glucose (Katz et al., 1993). Many recent studies also suggest that metabolic symbiosis exists between cells in both normal and diseased states. Stroma can reduce cysteine to protect leukemia cells from drug treatment (Zhang et al., 2012), and mesenchymal stem cells can mediate chemoresistance of tumor cells through lipid metabolism (Roodhart et al., 2011). Subpopulations of breast epithelial cells may provide glutamine to neighboring cells via glutamine synthetase activity (Kung et al., 2011), and astrocytes metabolize glucose to provide lactate as fuel for neurons (Bittar et al., 1996). Therefore, mechanistic conclusions should be measured by the fact that in vivo findings represent the complete result of both organismal and cellular metabolism.
The value of systems-based approaches is inherently limited by the accuracy of the molecular networks. Reactions that are not accounted for will affect MFA. We continue to identify metabolites and pathways in various biological systems where activity only arises in certain cell types or conditions (Strelko et al., 2011; Vander Heiden et al., 2010). Accounting for such activity within networks is not always possible, but the consequences of their presence are important to understand pathway function in disease. In this way, metabolic modeling can be useful, as a failure to adequately fit labeling data to a network may indicate that specific reaction(s) are missing. Nevertheless, the application of MFA to disease models provides an important means of investigating metabolic regulation by specific genes and pathways. No single methodology can completely characterize metabolism in all settings, highlighting the value of applying interdisciplinary approaches to understand metabolic regulation and its contribution to disease.
Acknowledgements
We thank members of the Metallo and Vander Heiden laboratories for comments and Brooke Bevis for help generating the figures. CMM is supported by the American Cancer Society Institutional Research Grant 70-002. MVH is supported by the Burroughs Wellcome Fund, the Damon Runyon Cancer Research Foundation, the Smith Family, and the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albe KR, Butler MH, Wright BE. Cellular concentrations of enzymes and their substrates. J Theor Biol. 1990;143:163–195. doi: 10.1016/s0022-5193(05)80266-8. [DOI] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012 doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Ayala JE, Samuel VT, Morton GJ, Obici S, Croniger CM, Shulman GI, Wasserman DH, McGuinness OP. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech. 2010;3:525–534. doi: 10.1242/dmm.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty CH, Basinger GM, Bocek RM. Pentose cycle activity in muscle from fetal, neonatal and infant rhesus monkeys. Arch Biochem Biophys. 1966;117:275–281. doi: 10.1016/0003-9861(66)90413-9. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Bessman SP, Geiger PJ. Transport of energy in muscle: the phosphorylcreatine shuttle. Science. 1981;211:448–452. doi: 10.1126/science.6450446. [DOI] [PubMed] [Google Scholar]
- Bittar PG, Charnay Y, Pellerin L, Bouras C, Magistretti PJ. Selective distribution of lactate dehydrogenase isoenzymes in neurons and astrocytes of human brain. J Cereb Blood Flow Metab. 1996;16:1079–1089. doi: 10.1097/00004647-199611000-00001. [DOI] [PubMed] [Google Scholar]
- Boros LG, Lee PW, Brandes JL, Cascante M, Muscarella P, Schirmer WJ, Melvin WS, Ellison EC. Nonoxidative pentose phosphate pathways and their direct role in ribose synthesis in tumors: is cancer a disease of cellular glucose metabolism? Med Hypotheses. 1998;50:55–59. doi: 10.1016/s0306-9877(98)90178-5. [DOI] [PubMed] [Google Scholar]
- Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, et al. A Mitochondrial Pyruvate Carrier Required for Pyruvate Uptake in Yeast, Drosophila, and Humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle KM, Bohndiek SE, Gallagher FA, Kettunen MI. Tumor imaging using hyperpolarized 13C magnetic resonance spectroscopy. Magn Reson Med. 2011;66:505–519. doi: 10.1002/mrm.22999. [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Cahill GF, Jr., Aoki TT, Brennan MF, Muller WA. Insulin and muscle amino acid balance. Proc Nutr Soc. 1972;31:233–238. doi: 10.1079/pns19720042. [DOI] [PubMed] [Google Scholar]
- Cahn RD, Zwilling E, Kaplan NO, Levine L. Nature and Development of Lactic Dehydrogenases: The two major types of this enzyme form molecular hybrids which change in makeup during development. Science. 1962;136:962–969. doi: 10.1126/science.136.3520.962. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Carminatti H, Jimenez de Asua L, Leiderman B, Rozengurt E. Allosteric properties of skeletal muscle pyruvate kinase. J Biol Chem. 1971;246:7284–7288. [PubMed] [Google Scholar]
- Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, Cantley LC, Blenis J. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell. 2010;38:487–499. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consoli A, Kennedy F, Miles J, Gerich J. Determination of Krebs cycle metabolic carbon exchange in vivo and its use to estimate the individual contributions of gluconeogenesis and glycogenolysis to overall glucose output in man. J Clin Invest. 1987;80:1303–1310. doi: 10.1172/JCI113206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzeja PP, Zeleznikar RJ, Goldberg ND. Adenylate kinase: kinetic behavior in intact cells indicates it is integral to multiple cellular processes. Mol Cell Biochem. 1998;184:169–182. [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellerstein MK. New stable isotope-mass spectrometric techniques for measuring fluxes through intact metabolic pathways in mammalian systems: introduction of moving pictures into functional genomics and biochemical phenotyping. Metab Eng. 2004;6:85–100. doi: 10.1016/j.ymben.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, Kunji ERS, Martinou JC. Identification and Functional Expression of the Mitochondrial Pyruvate Carrier. Science. 2012;337:93–96. doi: 10.1126/science.1218530. [DOI] [PubMed] [Google Scholar]
- Holleran AL, Briscoe DA, Fiskum G, Kelleher JK. Glutamine metabolism in AS-30D hepatoma cells. Evidence for its conversion into lipids via reductive carboxylation. Mol Cell Biochem. 1995;152:95–101. doi: 10.1007/BF01076071. [DOI] [PubMed] [Google Scholar]
- Holten D, Procsal D, Chang HL. Regulation of pentose phosphate pathway dehydrogenases by NADP+/NADPH ratios. Biochem Biophys Res Commun. 1976;68:436–441. doi: 10.1016/0006-291x(76)91164-5. [DOI] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jacobus WE. Respiratory control and the integration of heart high-energy phosphate metabolism by mitochondrial creatine kinase. Annu Rev Physiol. 1985;47:707–725. doi: 10.1146/annurev.ph.47.030185.003423. [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Katz J, Wals P, Lee WN. Isotopomer studies of gluconeogenesis and the Krebs cycle with 13C-labeled lactate. J Biol Chem. 1993;268:25509–25521. [PubMed] [Google Scholar]
- Khairallah M, Labarthe F, Bouchard B, Danialou G, Petrof BJ, Des Rosiers C. Profiling substrate fluxes in the isolated working mouse heart using 13C-labeled substrates: focusing on the origin and fate of pyruvate and citrate carbons. Am J Physiol Heart Circ Physiol. 2004;286:H1461–1470. doi: 10.1152/ajpheart.00942.2003. [DOI] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs HA. The regulation of the release of ketone bodies by the liver. Adv Enzyme Regul. 1966;4:339–354. doi: 10.1016/0065-2571(66)90027-6. [DOI] [PubMed] [Google Scholar]
- Kung HN, Marks JR, Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet. 2011;7:e1002229. doi: 10.1371/journal.pgen.1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau BR, Wahren J, Chandramouli V, Schumann WC, Ekberg K, Kalhan SC. Use of 2H2O for estimating rates of gluconeogenesis. Application to the fasted state. J Clin Invest. 1995;95:172–178. doi: 10.1172/JCI117635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WN, Boros LG, Puigjaner J, Bassilian S, Lim S, Cascante M. Mass isotopomer study of the nonoxidative pathways of the pentose cycle with [1,2-13C2]glucose. Am J Physiol. 1998;274:E843–851. doi: 10.1152/ajpendo.1998.274.5.E843. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Buettger C, Berner DK, Matschinsky FM. Chronic effect of fatty acids on insulin release is not through the alteration of glucose metabolism in a pancreatic beta-cell line (beta HC9). Diabetologia. 1997;40:1018–1027. doi: 10.1007/s001250050783. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Maier K, Hofmann U, Reuss M, Mauch K. Identification of metabolic fluxes in hepatic cells from transient 13C-labeling experiments: Part II. Flux estimation. Biotechnol Bioeng. 2008;100:355–370. doi: 10.1002/bit.21746. [DOI] [PubMed] [Google Scholar]
- Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt ME, Harrison C, Sherry AD, Malloy CR, Burgess SC. Flux through hepatic pyruvate carboxylase and phosphoenolpyruvate carboxykinase detected by hyperpolarized 13C magnetic resonance. Proc Natl Acad Sci U S A. 2011;108:19084–19089. doi: 10.1073/pnas.1111247108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Walther JL, Stephanopoulos G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol. 2009;144:167–174. doi: 10.1016/j.jbiotec.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TA, Dang CV, Young JD. Isotopically nonstationary (13)C flux analysis of Myc-induced metabolic reprogramming in B-cells. Metab Eng. 2012 doi: 10.1016/j.ymben.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme EA, Crabtree B. Theoretical principles in the approaches to control of metabolic pathways and their application to glycolysis in muscle. J Mol Cell Cardiol. 1979;11:839–856. doi: 10.1016/0022-2828(79)90480-2. [DOI] [PubMed] [Google Scholar]
- Oakhill JS, Steel R, Chen ZP, Scott JW, Ling N, Tam S, Kemp BE. AMPK is a direct adenylate charge-regulated protein kinase. Science. 2011;332:1433–1435. doi: 10.1126/science.1200094. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008;8:266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Pilkis SJ, Claus TH, Kurland IJ, Lange AJ. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase: a metabolic signaling enzyme. Annu Rev Biochem. 1995;64:799–835. doi: 10.1146/annurev.bi.64.070195.004055. [DOI] [PubMed] [Google Scholar]
- Puchowicz MA, Bederman IR, Comte B, Yang D, David F, Stone E, Jabbour K, Wasserman DH, Brunengraber H. Zonation of acetate labeling across the liver: implications for studies of lipogenesis by MIDA. Am J Physiol. 1999;277:E1022–1027. doi: 10.1152/ajpendo.1999.277.6.E1022. [DOI] [PubMed] [Google Scholar]
- Quek LE, Dietmair S, Kromer JO, Nielsen LK. Metabolic flux analysis in mammalian cell culture. Metab Eng. 2010;12:161–171. doi: 10.1016/j.ymben.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Randle PJ. Endocrine control of metabolism. Annu Rev Physiol. 1963;25:291–324. doi: 10.1146/annurev.ph.25.030163.001451. [DOI] [PubMed] [Google Scholar]
- Roche TE, Hiromasa Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell Mol Life Sci. 2007;64:830–849. doi: 10.1007/s00018-007-6380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodhart JM, Daenen LG, Stigter EC, Prins HJ, Gerrits J, Houthuijzen JM, Gerritsen MG, Schipper HS, Backer MJ, van Amersfoort M, et al. Mesenchymal stem cells induce resistance to chemotherapy through the release of platinum-induced fatty acids. Cancer Cell. 2011;20:370–383. doi: 10.1016/j.ccr.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M, Zamboni N, Schulze A. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov. 2012;2:328–343. doi: 10.1158/2159-8290.CD-11-0234. [DOI] [PubMed] [Google Scholar]
- Ruderman NB. Muscle amino acid metabolism and gluconeogenesis. Annu Rev Med. 1975;26:245–258. doi: 10.1146/annurev.me.26.020175.001333. [DOI] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio II, Cairns RA, McCracken S, Wakeham A, Haight J, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012a doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012b;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U. Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, Osterman AL, Smith JW. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J Biol Chem. 2011;286:42626–42634. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelko CL, Lu W, Dufort FJ, Seyfried TN, Chiles TC, Rabinowitz JD, Roberts MF. Itaconic acid is a mammalian metabolite induced during macrophage activation. J Am Chem Soc. 2011;133:16386–16389. doi: 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunny NE, Parks EJ, Browning JD, Burgess SC. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metab. 2011;14:804–810. doi: 10.1016/j.cmet.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant DA, Frezza C, MacKenzie ED, Nguyen QD, Zheng L, Selak MA, Roberts DL, Dive C, Watson DG, Aboagye EO, et al. Reactivating HIF prolyl hydroxylases under hypoxia results in metabolic catastrophe and cell death. Oncogene. 2009;28:4009–4021. doi: 10.1038/onc.2009.250. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollet B, Athea Y, Mounier R, Guigas B, Zarrinpashneh E, Horman S, Lantier L, Hebrard S, Devin-Leclerc J, Beauloye C, et al. AMPK: Lessons from transgenic and knockout animals. Front Biosci. 2009;14:19–44. doi: 10.2741/3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, 3rd, Peters EC, Driggers EM, Hsieh-Wilson LC. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N. (13)C metabolic flux analysis in complex systems. Curr Opin Biotechnol. 2011;22:103–108. doi: 10.1016/j.copbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Zhang W, Trachootham D, Liu J, Chen G, Pelicano H, Garcia-Prieto C, Lu W, Burger JA, Croce CM, Plunkett W, et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat Cell Biol. 2012;14:276–286. doi: 10.1038/ncb2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]