Abstract
The emergence of pandemic H1N1/2009 influenza demonstrated that pandemic viruses could be generated in swine. Subsequent re-introduction of H1N1/2009 to swine has occurred in multiple countries. Through systematic surveillance of influenza viruses in swine from a Hong Kong abattoir, we characterize a reassortant progeny of H1N1/2009 with swine viruses. Continued reassortment of H1N1/2009 with swine influenza viruses could produce variants with transmissibility and altered virulence for humans. Global systematic surveillance of influenza viruses in swine is warranted.
Pandemic H1N1/2009 influenza virus emerged from swine in Mexico to infect humans and has rapidly spread to over 200 countries (1). This virus was generated by multiple reassortment events, and each of its precursor gene segments has circulated in swine for more than 10 years (2, 3). Infection of swine with H1N1/2009 virus has been observed in multiple countries (4), But because of a paucity of systematic surveillance of swine influenza worldwide, questions remain whether H1N1/2009 will become established in swine and become a reservoir of reassortment that may produce novel viruses of potential threat to public health.
Over the past 10 years, systematic virological surveillance of influenza viruses in swine has been ongoing in a Hong Kong abattoir wherein over 80% of swine tested originate from adjacent provinces in China (2, 5). A total of 32 H1N1 and H1N2 viruses were isolated in fortnightly surveys from June 2009 to February 2010 (table S1) (6). Since 22 October 2009, 10 H1N1/2009 viruses were isolated from 4 of 8 sampling occasions. Phylogenetic analysis shows that all eight genes of these viruses belonged to the H1N1/2009 lineage (fig. S1). Pandemic H1N1/2009 viruses isolated on the same sampling occasion were genetically identical, suggesting transmission of viruses occurred within swine herds. But viruses from different sampling dates were genetically distinct from each other and also from H1N1/2009-like swine viruses isolated in other countries, indicating multiple independent introductions of these viruses from humans to swine. The H1N1/2009 viruses had not been detected in our surveys until October 2009, supporting the contention that this virus lineage did not arise from China (2).
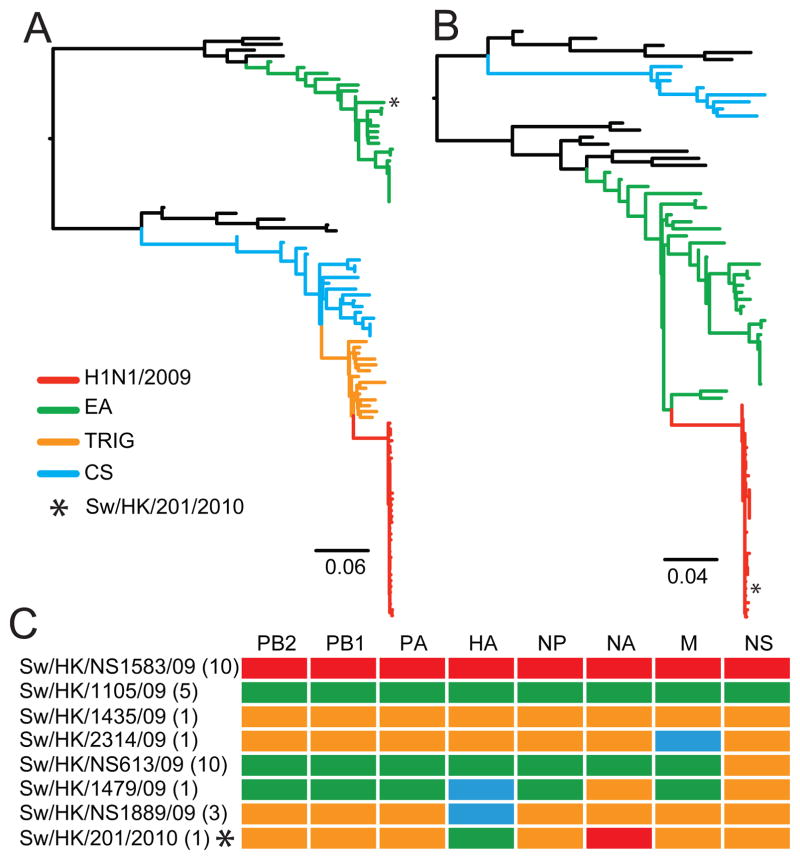
Three major lineages of swine H1 influenza viruses have been prevalent in swine in our surveys in the past 10 years: classical swine H1N1 (CS), European ‘avian-like’ H1N1 (EA) and triple-reassortant H1N2 viruses (TRIG) (Fig. 1A, B) (2). The remaining 22 viruses described here include five EA H1N1, one TRIG H1N2 and 16 reassortant viruses belonging to five different genotypes (Fig. 1C).
Fig. 1.
Maximum likelihood phylogenies of the influenza (A) hemagglutinin and (B) neuraminidase genes showing major swine H1N1/H1N2 and H1N1/2009 lineages. * denotes phylogenetic position of the novel reassortant virus. Identical phylogenies with virus names shown are provided in fig. S1A, B. Bar represents nucleotide substitutions per site. (C) Lineages of reassortant swine viruses identified through phylogenetic analyses, with the name of a representative virus and number of each variant isolated listed to the left.
One was a novel reassortant (A/swine/Hong Kong/201/2010 [H1N1]) with a H1N1/2009-like neuraminidase (NA) gene, an EA-like hemagglutinin (HA) gene and the six internal genes derived from TRIG lineage viruses (Fig. 1, fig. S1). The TRIG internal gene cassette (with its new EA derived M gene) therefore continues to be adept at acquiring novel HA and NA genes (7). The identity of the novel virus has been confirmed by direct polymerase chain reaction detection of the eight gene segments in the original swab specimen (6). The HA gene of A/swine/Hong Kong/201/2010 grouped within the EA swine lineage, in a basal phylogenetic position to the H1N1 and H1N2 swine viruses isolated during the study period (Fig. 1A, fig. S1A). Hemagglutination inhibition indicated neither H1N1/2009 vaccine nor natural infection reliably elicits cross-protective antibody to A/swine/Hong Kong/201/2010 (table S2) (6). The NA gene sequence grouped within the H1N1/2009 NA clade (100% boostrap support) indicating it was derived from H1N1/2009 (Fig. 1B, fig. S1B). Comparison with the consensus of all available H1N1/2009 NA genes showed a single silent nucleotide substitution in A/swine/Hong Kong/201/2010. The amino-acid sequences of A/swine/Hong Kong/201/2010 showed predicted resistance to the adamantanes but not to oseltamivir, similar to recently described Hong Kong swine viruses (2).
The H1N1/2009 virus has remained antigenically and genetically stable and of relatively low virulence for humans since its detection in humans in April 2009 (1). Our results show that the introduction of H1N1/2009 virus to swine has provided it with opportunities for reassortment. Furthermore, H5N1 and H9N2 viruses have been occasionally isolated from swine in Asia (5), providing the possibility for the incorporation of avian virus genes into mammalian adapted viruses. Phylogenetic analyses on the emergence of the 1918, 1957 and 1968 pandemics suggests that all three of these pandemics evolved undetected in an intermediate mammalian host for some years before they were recognized in humans (8). The 2009 pandemic, though mild and apparently contained at present, could undergo further reassortment in swine and gain virulence. It is therefore important that surveillance in swine is greatly heightened and that all eight gene segments are genetically characterized so that such reassortment events are rapidly identified.
Supplementary Material
Acknowledgments
This work was supported in part by the NIAID contract HHSN266200700005C and the Area of Excellence Scheme of the University Grants Committee (grant AoE/M-12/06) of the Hong Kong SAR Government. We acknowledge the Food and Environmental Hygiene Department of Hong Kong for facilitating this study.
Footnotes
Authors declare no conflict of interest. Sequence data generated from this study were deposited in GenBank with accession no. XXXXXX–XXXXXX.
References and Notes
- 1.World Health Organization. Pandemic (H1N1) 2009 (update 89, 2010; http://www.who.int/csr/don/2010_02_26/en/index.html)
- 2.Smith GJD, et al. Nature. 2009;459:1122. [Google Scholar]
- 3.Garten RJ, et al. Science. 2009;325:197. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasma T, Joseph T. Emerg Infect Dis. 2010 doi: 10.3201/eid1604.091636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris JSM, et al. J Virol. 2001;75:9679. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Materials and methods are available as supporting material on Science Online.
- 7.Bastien N, et al. J Infect Dis. 2010;201:1178. doi: 10.1086/651507. [DOI] [PubMed] [Google Scholar]
- 8.Smith GJD, et al. Proc Natl Acad Sci USA. 2009;106:11709. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.