Abstract
The hippocampus is a cortical region thought to play an important role in learning and memory. Most of our knowledge about the detailed organization of hippocampal circuitry responsible for these functions is derived from anatomical studies. These studies present an incomplete picture, however, because the functional character and importance of connections are often not revealed by anatomy. Here, we used a physiological method (photostimulation with caged glutamate) to probe the fine pattern of functional connectivity between the CA3 and CA1 subfields in the mouse hippocampal slice preparation. We recorded intracellularly from CA1 and CA3 pyramidal neurons while scanning with photostimulation across the entire CA3 subfield with high spatial resolution. Our results show that, at a given septotemporal level, nearby CA1 neurons receive synaptic inputs from neighboring CA3 neurons. Thus, the CA3 to CA1 mapping preserves neighbor relations.
The concept of cortical maps plays a central role in our understanding of cortical function. Many, if not all, cortical areas are organized according to some sort of map, and this fact, together with the idea that the hippocampus contains a “cognitive map” (1), naturally leads to the idea that the hippocampus might have a map-like organization. The existence of such an hippocampal map would be important for understanding the function of this important structure (2) because knowing what is represented in the map, and how this representation is arranged, would place constraints on what computations the hippocampus could carry out. Whatever arrangement a hippocampal map might have, the very idea of a map involves a knowledge of the basic intracortical circuitry, and of the topological arrangement of that circuitry. Our goal here is to investigate the topology of hippocampal CA3 to CA1 mapping. That is, we wish to determine the extent to which neighbor relations in CA3 are preserved in the projections to CA1.
To place our goals in context, we need to review briefly the current understanding of hippocampal organization as revealed by anatomical and physiological methods.
Most of our knowledge about the detailed organization and connectivity of circuits within the hippocampus is derived from extensive anatomical studies (3–11). The pioneering works of Ramon y Cajal and Lorente de No established that the fields of the hippocampal formation are linked by a sequence of unique and largely unidirectional connections, the fundamental “trisynaptic circuit”: The dentate gyrus receives its major input from the entorhinal cortex via the so-called perforant path. The granule cells of the dentate gyrus project via the mossy fibers to the CA3 field of the hippocampus. Pyramidal cells of the CA3 field in turn give rise to collateralized axons, the Schaffer collaterals, that terminate within CA3 as associational connections and also provide a major input to the CA1 field.
Andersen and colleagues proposed an important extension of this basic tri-synaptic circuit concept: according to their view, the hippocampus is organized in parallel “lamellae,” or small strips or slices (4); this is the so-called “lamellar hypothesis.” The lamellar hypothesis formed a conceptual basis for the use of the in vitro hippocampal slice preparation as a means of studying synaptic physiology because all components of the tri-synaptic hippocampal circuit would be contained in an easily isolated slice amenable to manipulation and study. The lamellar hypothesis was, however, revisited by Amaral and colleagues (5, 8) and found to be an incomplete description of hippocampal connectivity because Schaffer collateral projections from CA3 to CA1 were shown to provide a wide dispersion of information from a particular level in the hippocampus to much of its septotemporal extent.
Anatomical studies have shown that the projections from CA3 to CA1 are, however, far from random. Rather, the general target area in CA1 for a given CA3 neuron depends on the that neuron's septotemporal and transverse position within the CA3 cortical sheet (7, 8). Most relevant for the present work is the observation that the CA1 map is “flipped” in the transverse direction relative to the CA3 map so that CA3 neurons nearest CA1 tend to project to the closest CA1 neurons, and CA3 neurons farthest from CA1 tend to project to the most transversely distant CA1 cells. Here, we confirm and extend this anatomical observation by showing, with a functional method, that the flipped mapping is topological in the sense that neighbors in CA3 project to neighbors in CA1.
Physiological studies of hippocampal activity and the functional correlates of neural firing (1, 12, 13) have provided further important clues to the organization of hippocampal circuits. These studies have shown that the activity of hippocampal pyramidal neurons in rodents exploring the environment is characterized by “place fields,” areas of the environment in which these cells are reliably activated. Interestingly, no systematic relationship has been observed between places in the environment and anatomical locations in the hippocampus (14). Thus, in terms of global topography, no obvious relationship has been found between the region of hippocampus in which a particular neuron is located and the region of space spanned by its place field, suggesting the possibility that the hippocampus might be organized as a parallel distributed network without an orderly underlying map-like organization. Some studies have shown, however, that factors other than the animal's absolute spatial position, including movement direction, speed, and even non-spatial cues stemming from olfactory and auditory systems, also influence hippocampal neuronal activity (15, 16). It is therefore possible that neighboring cells within the hippocampus might be arranged in the manner of the primary sensory cortices, with nearby cells sharing little overlap in place field location but significant overlap in other (as yet uncharacterized) functional variables (17). Consistent with this idea, Hampson et al. (18) showed that neurons encoding task-relevant features of a delayed-non-match-to-sample task were distributed within defined segments of the hippocampus, providing evidence of a functional topography, at least along the septotemporal axis (18).
In this study, we used photostimulation with caged glutamate (19, 20) in the mouse hippocampal slice preparation to probe functional connectivity between the CA3 and CA1 subfields. We find that the CA3 to CA1 mapping is an orderly one that preserves neighbor relations.
Methods
General Electrophysiology. Hippocampal coronal (acute) slices (350 μm) were prepared from 3- to 5-week-old mice. Slices were taken with a DSK Microslicer (Ted Pella, Inc., Redding, CA) while immersed in ice-cold solution composed of 200 mM Sucrose, 1.9 mM KCl, 1.2 mM NaH2PO4, 33.0 mM NaHCO3, 10.0 mM d-glucose, 4.0 mM MgCl2, 0.7 mM CaCl2, and 0.4 mM ascorbic acid. Slices were then stored submerged in a room-temperature solution composed of 120 mM NaCl, 3.5 mM KCl, 1.25 mM NaH2PO4, 26.0 mM NaHCO3, 10.0 mM d-glucose, 4.0 mM MgCl2, 0.7 mM CaCl2, and 0.4 mM ascorbic acid for over 1.5 h before recording. During the experiment, slices were placed in the recording chamber and perfused with the recording solution (120 mM NaCl/3.5 mM KCl/1.25 mM NaH2PO4/26.0 mM NaHCO3/10.0 mM d-glucose/1.3 mM MgCl2/2.5 mM CaCl2/0.4 mM ascorbic acid). Fifty micromolar “caged” glutamate [γ-(α-carboxy-2-nitrobenzyl) ester, trifluoroacetate, l-glutamic acid; CNB-glutamate, Molecular Probes] was included for photostimulation. An infrared Olympus (Melville, NY) differential interference contrast microscope with a ×40, 0.8 numerical aperture (N.A.) water-immersion lens was used to visualize and target CA1 and CA3 pyramidal neurons for whole-cell recordings. Glass microelectrodes (4–6 MΩ resistance) were filled with an intracellular solution (170 mM K-gluconate/10 mM Hepes/10 mM NaCl/2 mM MgCl2/0.5 mM EGTA/0.1 mM CaCl2/3.5 mM Mg-ATP/1 mM Li-GTP) containing 0.5% biocytin for cell labeling.
Photostimulation. Neurons were stimulated by a brief flash (5 ms) of UV light from an argon-ion laser focused to a small spot (≈15 μm) in the plane of the brain slice through the same ×40 microscope objective used for differential interference contrast imaging. The intensity of the laser light was adjusted so as to elicit spikes from CA3 neurons only when stimulating near the recording site (see Results). Whole-cell voltage clamp recordings (at -60 mV) were made from one or two postsynaptic CA1 neurons and inward excitatory postsynaptic currents (EPSCs) resulting from photostimulation of presynaptic CA3 neurons were measured by using two Axon Instruments (Foster City, CA) amplifiers (Axopatch 1A and 1D). In some experiments, CA3 spiking activity resulting from direct dendritic photostimulation was monitored under current-clamp conditions. Cells with initial membrane potentials more depolarized than -55 mV were not included in the data sets.
To map the locations of input to single cells, sites 25 or 50 μm apart spanning all of CA3 were stimulated sequentially in a pseudorandom pattern that covered the entire subfield (Fig. 1 A). In addition, photostimulation trials were interleaved with control trials (no stimulation) to obtain spontaneous postsynaptic currents (sEPSCs). In most cases, two neurons were recorded simultaneously and/or sequential recordings were made from multiple cells while photostimulation was repeated. After completion of photostimulation and recordings, the laser (3-s pulse) was used to burn alignment sites (<10 μm) into the slice so that photostimulation coordinates could be assigned to their corresponding position in the tissue (21).
Fig. 1.
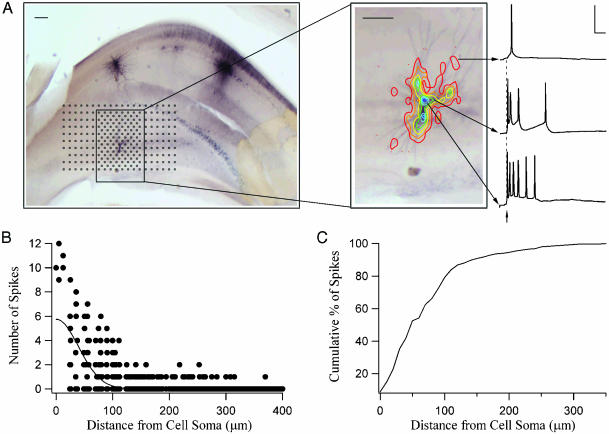
(A) A representative mouse hippocampal slice with biocytin-stained CA3 pyramidal cell. The black markers represent sites over CA3 that were photostimulated (25 or 50 μm apart). (Inset) Contour plots depicting the spiking response profile for this CA3 cell. The contours were generated by counting the number of spikes; each contour line represents from one spike (red = 1) to seven spikes (dark blue = 7). (Scale bar, 100 μm.) Sample traces are shown for some contours on the right. (Scale, 50 ms and 50 mV.) (B) Number of spikes generated by three CA3 neurons as a function of photostimulation distance from each cell's soma. (C) Cumulative percentage of spikes generated as a function of distance between the photostimulation site and the cell soma for all recorded CA3 cells (n = 8).
We recorded from 8 CA3 pyramidal cells and 26 CA1 pyramidal cells from a total of nine slices. An additional four slices were discarded because we were not able to elicit postsynaptic EPSCs in CA1 cells while photostimulating over CA3, presumably because of the angle at which the slice was cut. We estimated an upper bound to the number of neurons stimulated by each photostimulation flash over the pyramidal cell layer by measuring the neuronal cell body diameters in thin hippocampal CA3 sections, then determining the number of cells whose dendritic arbors would be expected to lie in a 15-μm diameter and 50-μm-deep cylinder (15-μm spot focused on the surface of the slice; 50-μm effective thickness due to light scatter).
Data Analysis. Twenty-five milliseconds preceding each laser flash as well as the subsequent 375 ms was recorded at 10 kHz and analyzed by using igor pro (WaveMetrics, Lake Oswego, OR). For each cell at each photostimulation site, the number of EPSCs (CA1 neuron) or action potentials (CA3 neuron) detected within 375 ms after photostimulation was used to generate contour plots (Figs. 1, 2, and 5) by using Delauney Triangulation (22) (igor pro). For CA1 cells, background rates of sEPSCs were low (0.32 ± 0.11 Hz, 0.12 ± 0.04 per 375-ms trace, n = 26), and values from each cell were subtracted from the response counts from the same cell before contour generation. To determine whether the responses generated were statistically significant, a Mann–Whitney test was performed comparing the measured responses to spontaneous activity traces. For each CA1 neuron, the location over CA3 yielding the maximal number of responses was determined (most prominent peak in Fig. 5A). As a measure of excitability overlap for a pair of cells, the area of intersection between the broadest contour lines was divided by the total contour area outlined by the two cells (normalized area of contour overlap) and plotted in Fig. 5B.
Fig. 2.
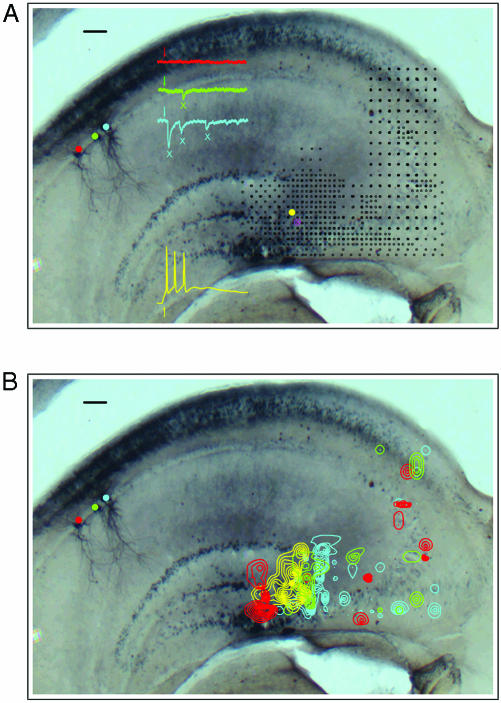
(A) A mouse hippocampal slice with recordings from three CA1 cells (red, green, and blue) and one CA3 cell (yellow). The black markers represent sites over CA3 that were photostimulated (25 μm or 50 μm apart). Sample responses from each cell to photostimulation at a single site (yellow marker) are also shown. (B) Contour plots depicting the response profiles for each cell shown in A. The contours were generated from responses to stimulation of CA3 cell (shown in yellow). (Scale bar, 100 μm.)
Fig. 5.
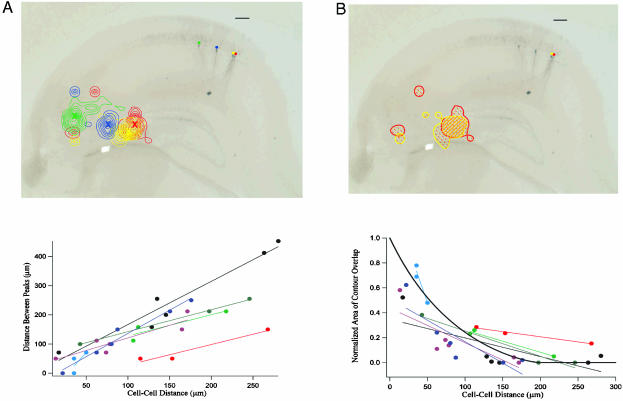
(A) Distance between the location of the most prominent peaks of the CA3 input source regions for CA1 neurons plotted vs. intercellular distance in CA1 for each cell pair (see Methods). Neuronal pairs recorded from the same slice are shown in the same color. The best-fit lines for each slice data set are also included to show the trends. (B) Normalized area of contour overlap in the CA3 response profiles plotted versus intercellular distance in CA1 for each cell pair (see Methods). Neuronal pairs recorded from the same slice are shown in the same color. The best-fit lines for each slice data set are also included to show the trend. The thick black line represents the same overlap measure for two circles of diameter 200 μm as a function of the distance between their centers. (Scale bar, 100 μm.)
Tissue Processing. Slices were fixed by immersion in a 0.1 M phosphate buffer for 12–14 h. Slices were then double-stained for cytochrome oxidase and biocytin as described (23).
Results
Although our main concern is the topological properties of the CA3 to CA1 mapping, we have first carried out experiments to characterize the photostimulation technique in the context of the hippocampal circuits. Specifically, we have determined the spatial resolution of the technique and the specificity of the connections detected, and have estimated the reliability of the method.
Spatial Resolution of Photostimulation. Focal release of caged glutamate (scanning laser photostimulation) is a physiological method that can permit stimulation of neurons with high spatial resolution (19–21). Here, we use photostimulation to create an input map of source regions located in CA3 for individual pyramidal neurons located in CA1.
In addition to their major projections to CA1 via the Schaffer collaterals, neurons in CA3 have recurrent axons contributing extensive associational projections that terminate within CA3 itself (11, 24, 25). These associational projections could confound connectivity data because one might not know whether the stimulated area in CA3 was directly connected to a particular recorded CA1 neuron, or whether that area made projections to other CA3 neurons that in turn made synapses onto the CA1 neuron. To address this issue and to maximize the spatial resolution of stimulation, we sought experimental conditions which would activate CA3 neurons minimally, only when focal glutamate release occurred directly over their dendritic arbor. We did this by reducing the concentration of caged glutamate in the perfusion solution, reducing the intensity and duration of the uncaging laser pulse, and not blocking inhibitory inputs (see Methods).
Fig. 1A shows the response profile for a representative CA3 neuron (recorded in current-clamp) to photostimulation as the stimulation site was scanned (25-μm steps near the vicinity of the cell body; 50-μm steps elsewhere) over the entire CA3 subfield. Note that photostimulation near the soma of the cell generates more action potentials than more distant photostimulation (Fig. 1 A Inset), probably reflecting a greater functional glutamate receptor density and less electrotonic attenuation near the soma. Spiking responses of the type shown by the representative traces in Fig. 1 A were accumulated at each CA3 stimulation site to yield a response profile for the recorded neuron. We generated a contour map for the CA3 cell to represent this spatial information (Fig. 1 A Inset; see Methods). Note that the contours lines roughly match the dendritic arborization of this neuron; no spikes were generated when the flashed site was not directly over the cell's dendritic arbor.
Fig. 1B shows the number of action potentials generated as a function of stimulation-site distance from cell soma for three CA3 cells. The percentage of all spikes generated for all of the recorded CA3 cells (n = 8) as a function of distance from each cell's soma is plotted in Fig. 1C: ≈80% of all spikes are elicited when the photostimulation site is within 100 μm of the cell's soma, and roughly 90% of all spikes are generated when the stimulation site is within 150 μm. We therefore conclude that, under our stimulation conditions, recurrent excitation within CA3 due to associational projections from distant neurons will not affect functional connectivity results. Thus, if photostimulation at a particular site in CA3 results in postsynaptic EPSCs in a CA1 neuron, we may be confident that the presynaptic CA3 cell somas are located close to the photostimulation site.
CA3–CA1 Functional Connectivity. Fig. 2A shows a hippocampal slice in which recordings were made from three CA1 neurons (voltage-clamp, blue, green, and red) and one CA3 neuron (current-clamp, yellow) while the photostimulation site was scanned over the entire CA3 subfield (black markers). Typical sample responses of each cell to stimulation at a single site are also shown in this figure. Note that whereas photostimulation at this single site elicits a strong synaptic response in the blue cell, indicating that the blue cell receives inputs from one or more neurons in the photostimulated region, the red cell does not respond at all. Photostimulation allows the exploration of source regions (regions that elicit responses in recorded CA1 neurons) at higher spatial resolution (areas with greater black marker density). Also note that the CA3 cell is again activated directly from photostimulation at the depicted site and thus responds by firing action potentials.
Responses of the type shown by the representative traces in Fig. 2 A were accumulated at each CA3 stimulation site to yield a spatial response profile for each individual recorded neuron, and we again generated contour maps for each cell to define quantitatively the CA1 neuron's input source regions in CA3. Contours representing synaptic input maps, or source regions, were generated for CA1 cells by counting the number of elicited EPSCs (shown by green and blue Xs in Fig. 2 A; see Methods for a detailed description). The average sEPSC activity was subtracted before contour map generation. Fig. 2B shows the response profiles for each of the recorded cells in this slice. The area encompassed by the contours, or source region, is an indication of the location of the CA3 cell or cells that connect to the CA1 neurons from which we were recording. Because the isolated source spots evident in Fig. 2B are not significantly greater than what is expected by chance (given the background activity; see Methods), and because their spatial distribution does not conform to the outlines of a neuronal dendritic tree, we conclude that, in our slice preparation, cells within CA1 receive inputs from only circumscribed and specific regions of CA3. We cannot, of course, exclude the possibility that some particular isolated source spots reflect actual circuit connections.
We recorded from 8 CA3 pyramidal cells and 26 CA1 pyramidal cells in 9 mouse hippocampal slices. The average area over which spiking activity could be elicited for individual CA3 neurons was 51,612 ± 1,459 μm2, and the spatial extent of the main source region connecting to each of the CA1 neurons was 35,229 ± 4,487 μm2 (the area encompassed by the broadest contour lines). These values correspond to circular areas with diameters of ≈250 μm and 200 μm, respectively; the former is in rough accordance with the spatial extent of dendritic arborization for hippocampal CA3 pyramidal neurons in the slice preparation (26, 27).
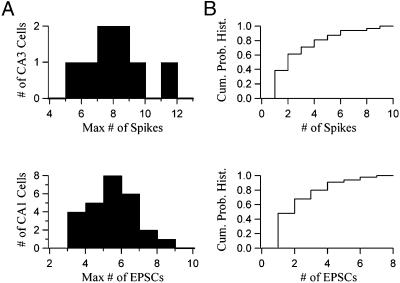
We sought to estimate the number of neurons in each CA3 source region that make functional connections to a CA1 neuron. Calculations based on the size of the photostimulation spot, the CA3 neuronal soma size, and density provide an upper bound estimate of 20 for the number of neurons exposed to glutamate for each uncaging light flash over the pyramidal cell layer (see Methods). Fig. 3 shows that only a small fraction of these potentially stimulated neurons make actual functional contacts to a recorded CA1 cell. Fig. 3A Upper presents a histogram of the maximal number of elicited spikes from each CA3 cell on photostimulation (n = 8, average = 7.6). This histogram measures the maximal response of each CA3 cell to photostimulation at its hot-spot, the peak location in its spatial response profile. Fig. 3A Lower shows a histogram of the maximal number of elicited EPSCs for each CA1 cell on photostimulation at its hot-spot, the peak location in its primary source region (n = 26, average = 5.0). The fact that the maximal number of elicited CA1 EPSCs is less than the maximal number of elicited CA3 spikes suggests again that very few CA3 neurons in each source region make functional contacts to the recorded CA1 cell. This possibility is further supported by Fig. 3B. Fig. 3B Upper shows a cumulative probability histogram of the number of spikes elicited over all photostimulation locations for a representative CA3 cell. Fig. 3B Lower exhibits a cumulative probability histogram of the number of elicited EPSCs over all stimulated locations for a representative CA1 cell. Note that the two distributions are similar, indicating that the shapes of CA3 spiking response profiles and CA1 source regions are similar; this observation is consistent with the possibility that very few CA3 cells in each region make functional connections to CA1 cells.
Fig. 3.
(A Upper) Histogram of the maximal number of elicited spikes from each CA3 cell on photostimulation at that cell's most responsive location (n = 8, average = 7.6). (A Lower) Histogram of the maximal number of measured EPSCs for each CA1 cell on photostimulation at that cell's best location in its primary source region (n = 26, average = 5.0). (B Upper) Cumulative probability histogram of the number of spikes elicited over all photostimulation locations for a representative CA3 cell. (B Lower) Cumulative probability histogram of the number of measured EPSCs over all photostimulation locations for a representative CA1 cell.
Reliability of Photostimulation Responses. How reproducible are the responses elicited by photostimulation? Fig. 4A shows the spiking responses of two CA3 cells to repeated photostimulation over the same site. The figure reveals that a burst of spikes is reliably elicited in response to each flash. Fig. 4B shows postsynaptic EPSCs from a CA1 neuron while the same site over CA3 was flashed repeatedly. We conclude that photostimulation can be reliably used to elicit spiking responses in CA3 neurons and EPSCs in CA1 neurons and can thus serve as a tool for functional mapping of circuitry in this preparation.
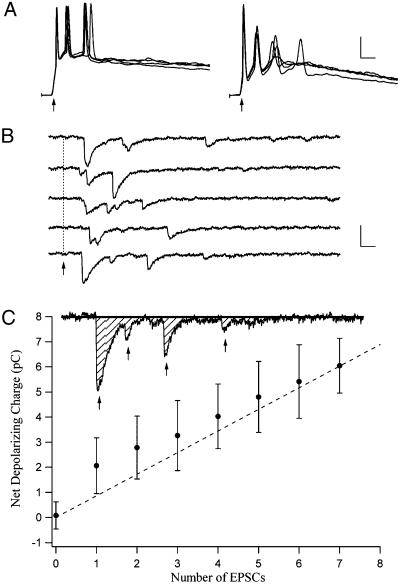
Fig. 4.
(A) Responses from two CA3 neurons in response to four photostimulations for each cell. (Scale, 50 ms and 20 mV.) (B) EPSCs from a CA1 neuron in response to multiple photostimulations at the same site in CA3. Arrow, onset of photostimulation. (Scale, 25 ms and 20 pA.) (C) Total charge transfer for a CA1 neuron as a function of the number of measured EPSCs for that cell. (Scale, 25 ms and 20 pA.)
Note however, the significant variability in the size of the elicited CA1 EPSCs in Fig. 4B; the initial EPSC tends to be much larger than subsequent depolarizing currents. This variability led us to investigate the implications of using EPSC number as the measure for generating the source region contours. We sought to link the number of elicited EPSCs to another measure of postsynaptic response, the net inward charge generated in the cell as a result of photostimulation. Fig. 4C shows that although there is some variability in the net depolarizing charge measured in a CA1 cell for each elicited EPSC, a clear relationship between the two measures is apparent. We therefore conclude that the number of EPSCs provides a good estimate of the “strength of connection” from the stimulation point in CA3 to the recorded CA1 neuron.
Topography in the CA3–CA1 Functional Connection Map. Does the mapping from CA3 to CA1 preserve neighbor relations? Fig. 5A shows that there is a relation between the location of a CA1 cell and the region over CA3 that excites that cell maximally (location of the most prominent peak in the contours; colored Xs in Fig. 5A Upper). Pairs of cells (along with their best-fit lines) from the same slice are shown in the same color. Although between-slice variability is apparent, we see that, for each individual slice, as the distance between the recorded pair of CA1 neurons increases, so does the distance between the most prominent peaks in the source regions. Furthermore, cells in the proximal area of CA1 (closest to the dentate gyrus) are maximally responsive to photostimulation in the distal area of CA3, and vice versa. This global topographic arrangement (distal CA3 to proximal CA1) was present in all of our slices (n = 9), across all 32 cell pairs, and is consistent with tendencies in the anatomical data (5, 6, 8).
Another measure of this topographic arrangement is presented in Fig. 5B. For each CA1 cell pair, we measured the overlap in their CA3 input regions as the intersection of their broadest contour lines (Fig. 5B Upper; see Methods). Fig. 5B shows that nearby neurons in CA1 tend to have a significant amount of overlap in the regions over CA3 from which they receive their inputs. As the intercellular distance increases in CA1, this measure of overlap for the cell pair gradually drops. We again conclude that neighbor-to-neighbor relations are preserved in the connectivity pathway from CA3 to CA1.
Discussion
In this study, we used a physiological method (focal release of caged glutamate by scanning laser photostimulation combined with intracellular recordings of EPSCs) to probe functional circuitry between CA3 and CA1 in the hippocampal slice preparation to determine the extent of any underlying organization in the Schaffer collateral projections. We showed that photostimulation is an efficient and reliable technique for investigating functional connections in this preparation. Our results demonstrate that, at a specific septotemporal level, nearby CA1 neurons receive synaptic inputs from topographically organized source regions, indicating that neighbor-to-neighbor relations are preserved in CA3 to CA1 mapping.
Our observations suggest that only a few neurons in each CA3 primary source region make synaptic contacts to a given CA1 neuron in our slices. We stress, however, that CA3 axons may well (probably do, in fact) project outside the plane of the slice and then return back to that plane to make functional connections. The connections within a lamella, then, likely are denser than our analysis reveals. Experiments using iontophoretic injections of the anterograde tracer Phaseolus vulgaris leucoag-glutinin (PHA-L) showed that projections in the long (septotemporal) axis are generally even more prominent and well organized than those that run in the transverse axis (in the lamellae) (5–8). These “looping” axons that followed a less direct path to their target would be severed by the slicing procedure. Therefore, our experimental setup does not allow us to conclude that a particular region in CA3 is not connected to a given CA1 cell at the same septotemporal level in the intact animal because we do not elicit EPSCs on photostimulation in that region in the slice preparation.
Although there is evidence that a significant fraction of projections from CA3 cells are outside the slice (5), those that remain within the slice display a striking topographic arrangement. Thus, our data support a revised version of the lamellar hypothesis in that each slice (lamella) does indeed contain not only typical circuitry (the tri-synaptic circuit) but also at least a partial expression of an orderly map that preserves local relationships. We also show that at each septotemporal level, projections from CA3 to CA1 are arranged in a globally topographical manner with proximal CA3 projecting to distal CA1 and distal CA3 projecting to proximal CA1, consistent with the work of Amaral and colleagues (6) and Li et al. (7). The resolution of our method does not, however, permit us to determine whether the mapping is also orderly with respect to position of terminals on the CA1 neuron dendritic tree as suggested by the anatomical studies (7, 8). A most important extension of our experiments would involve the technically difficult evaluation of the extent to which neighbor relations are also preserved in the septotemporal direction.
Our finding that the CA3 to CA1 mapping appears to be topological, that is, that neighbor relations are preserved, naturally gives rise to the question of what the map represents. Answering this question will be essential to understanding what computations are performed by the hippocampus.
Abbreviation: EPSC, excitatory postsynaptic current.
References
- 1.O'Keefe, J. & Dostrovsky, J. (1971) Brain Res. 34, 171-175. [DOI] [PubMed] [Google Scholar]
- 2.Scoville, W. B. & Milner, B. (1957) J. Neuropsychiatry Clin. Neurosci. 12, 103-113. [DOI] [PubMed] [Google Scholar]
- 3.Lorente de No, R. (1933) J. Psychol. Neurol. 45, 381-438. [Google Scholar]
- 4.Andersen, P., Bliss. T. V. P. & Skrede, K. K. (1971) Exp. Brain Res. 13, 222-238. [DOI] [PubMed] [Google Scholar]
- 5.Amaral, D. G. & Witter, M. P. (1989) Neuroscience 31, 571-591. [DOI] [PubMed] [Google Scholar]
- 6.Amaral, D. G. & Witter, M. P. (1993) Curr. Opin. Neurobiol. 3, 225-229. [DOI] [PubMed] [Google Scholar]
- 7.Li, X. G., Somogyi, P., Ylinen, A. & Buzsaki, G. (1994) J. Comp. Neurol. 339, 181-208. [DOI] [PubMed] [Google Scholar]
- 8.Ishizuka, N., Weber, J. & Amaral, D. G. (1990) J. Comp. Neurol. 295, 580-623. [DOI] [PubMed] [Google Scholar]
- 9.Ramon y Cajal, S. (1995) Histology of the Nervous System, trans. Swanson, N. & Swanson, L. W. (Oxford Univ. Press, Oxford).
- 10.Lorente de No, R. (1934) J. Psychol. Neurol. 46, 113-177. [Google Scholar]
- 11.Swanson, L. W., Kohler, C. & Bjorklund, A. (1987) in Handbook of Chemical Neuroanatomy, eds. Bjorklund, A., Hoekfelt, T. & Swanson, L. W. (Elsevier Science, Amsterdam), Vol. 5, pp. 125-277. [Google Scholar]
- 12.Wilson, M. A. & McNaughton, B. L. (1993) Science 271, 1055-1058. [DOI] [PubMed] [Google Scholar]
- 13.MacNaughton, B. L., Barnes, C. A. & O'Keefe, J. (1983) Exp. Brain Res. 52, 41-49. [DOI] [PubMed] [Google Scholar]
- 14.O'Keefe, J. & Nadel, L. (1978) The Hippocampus as a Cognitive Map (Clarendon, Oxford).
- 15.Eichenbaum, H., Fagan, A., Mathews, P. & Cohen, N. J. (1988) Behav. Neurosci. 103, 331-339. [DOI] [PubMed] [Google Scholar]
- 16.Wiener, S. I., Paul, C. A. & Eichenbaum, H. (1989) J. Neurosci. 9, 2737-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichenbaum, H., Wiener, S. I., Shapiro, M. L. & Cohen, N. J. (1989) J. Neurosci. 9, 2764-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampson, R. B., Simereal, J. D. & Deadwyler, S. A. (1999) Nature 402, 610-614. [DOI] [PubMed] [Google Scholar]
- 19.Katz, L. D. & Dalva, M. B. (1994) J. Neurosci. Methods 54, 205-218. [DOI] [PubMed] [Google Scholar]
- 20.Callaway, E. M. & Katz, L. C. (1993) Proc. Natl. Acad. Sci. USA 90, 7661-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danzker, J. L. & Callaway, E. M. (2000) Nat. Neurosci. 3, 701-707. [DOI] [PubMed] [Google Scholar]
- 22.Graham, R. & Yao, F. (1990) Am. Math. Mon. 97, 687-701. [Google Scholar]
- 23.Yabuta, N. H. & Callaway, E. M. (1998) J. Neurosci. 18, 9489-9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottlieb, D. I. & Cowan, W. M. (1973) J. Comp. Neurol. 149, 393-422. [DOI] [PubMed] [Google Scholar]
- 25.Swanson, L. W., Wyss, J. M. & Cowan, W. M. (1986) J. Comp. Neurol. 181, 681-716. [DOI] [PubMed] [Google Scholar]
- 26.Fitch, J. M., Juraska, J. M. & Washington, L. W. (1989) Brain Res. 479, 105-114. [DOI] [PubMed] [Google Scholar]
- 27.Turner, D. A., Li, X. G., Pyapali, G. K., Ylinen, A. & Buzsaki, G. (1995) J. Comp. Neurol. 356, 580-594. [DOI] [PubMed] [Google Scholar]