Abstract
Essential tremor (ET), one of the most common adult-onset movement disorders, has been associated with cerebellar Purkinje cell degeneration and formation of brainstem Lewy bodies. Recent findings suggest that genetic variants of the leucine-rich repeat and Ig domain containing 1 (LINGO-1) gene could be risk factors for ET. The LINGO-1 protein contains both leucine-rich repeat (LRR) and immunoglobulin (Ig)-like domains in its extracellular region, as well as a transmembrane domain and a short cytoplasmic tail. LINGO-1 can form a ternary complex with Nogo-66 receptor (NgR1) and p75. Binding of LINGO-1 with NgR1 can activate the NgR1 signaling pathway, leading to inhibition of oligodendrocyte differentiation and myelination in the central nervous system. LINGO-1 has also been found to bind with epidermal growth factor receptor (EGFR) and induce downregulation of the activity of EGFR–PI3K–Akt signaling, which might decrease Purkinje cell survival. Therefore, it is possible that genetic variants of LINGO-1, either alone or in combination with other genetic or environmental factors, act to increase LINGO-1 expression levels in Purkinje cells and confer a risk to Purkinje cell survival in the cerebellum.
Here, we provide a concise summary of the link between LINGO-1 and neurodegeneration and discuss various hypotheses as to how this could be potentially relevant to ET pathogenesis.
Keywords: Epidermal growth factor receptor, essential tremor, LINGO-1, Nogo-66, oligodendrocyte, Purkinje cells
Introduction
Essential tremor (ET), one of the most common adult-onset movement disorders, is characterized by action and postural tremor. The prevalence is approximately 3–6% in people over the age of 65 years.1,2 ET patients usually have an 8–12-Hz kinetic tremor of the arms, which is often accompanied with head and voice tremor.1 This disorder is common across different races worldwide.2 ET patients may also have other non-tremor features, including cognitive and psychiatric symptoms.3,4 Based on pathological findings, some authors have categorized ET into two groups: patients with cerebellar Purkinje cell degenerative changes and patients with brainstem Lewy bodies.5–9 Family and twin studies have provided evidence for a genetic contribution to ET, and segregation of ET within families containing multiple affected members has been reported.10,11 The sequence variant (rs9652490 G allele) of the leucine-rich repeat and Ig domain containing 1 (LINGO-1) gene was recently identified to be a risk factor for ET in American and European populations in a genome-wide association study.12 Subsequently, most studies were able to replicate this finding.12–21 Vilarino-Guell et al.15 showed that LINGO-1 variants, including rs9652490 and variants of LINGO2, a paralog of LINGO-1, are associated with ET and PD. The LINGO-1 rs9652490 variant has been found to associate with ET in Europeans,14 North Americans,16 and Singaporeans.17 In summary, six studies comprising 2834 ET patients and 16,638 healthy controls support a positive link between the LINGO-1 variant rs9652490 and ET. However, four studies comprising 711 ET patients and 2,186 healthy controls reported negative results.18–21 Some authors have shown that ET is also associated with additional LINGO-1 variants (rs488687, rs3144, rs8028808, and rs12905478).15 The association of ET with other LINGO-1 variants suggests possible allelic heterogeneity of the LINGO-1 gene. Environmental factors may also play a role in ET.22 Here, we provide a concise summary of the link between LINGO-1 and neurodegeneration, and hypothesize its possible relevance to ET pathogenesis.
LINGO-1: Gene, Structure and Roles
The LINGO-1 gene, encoding a transmembrane glycoprotein, was mapped to chromosome 15q24.23 The LINGO-1 protein belongs to the larger leucine-rich repeat Ig protein family (also referred to as the leucine-rich repeat transmembrane neuronal family (LRRTM) ).24,25 The LRRTM protein family was first described in 2003 and has been linked with psychiatric diseases.26 This protein family possesses three other human paralogs besides LINGO-1: Lingo-2, Lingo-3, and Lingo-4.27,28 The LINGO-1 protein has a predominant expression profile in the central nervous system (CNS), including the brain and spinal cord, with no detectable level of expression in non-neural tissues.29 Though little is known about Lingo-2, Lingo-3, and Lingo-4, studies have revealed that they are present in far lower levels in the CNS than LINGO-1.27,28 LINGO-1 is the co-receptor of Nogo-66 receptor (NgR1) and lack of LINGO-1 will significantly decrease the activity of the NgR1 signaling pathway. The involvement of Lingo-2, 3, and 4 in this signaling pathway is minimal.30
Human LINGO-1 has 614 amino acids.24,25 It contains both leucine-rich repeat (LRR) and Ig-like domains in their extracellular region followed by a transmembrane domain as well as a short cytoplasmic tail (about 70 amino acids).25,29–31 The extracellular LRR domain comprises 12 LRR flanked by an N terminal and a C terminal capping domain.30,31 This extracellular LRR domain of LINGO-1 binds with the LRR domain of NgR1 and significantly influences the activity of the NgR1 signaling pathway in the CNS.30,31 The LRR domains of proteins have been implicated to have a role in protein–protein interaction and their unique structure may help in cell signaling and/or cell adhesion in specific regions of the brain.25,29–31 The C-terminal part of LINGO-1 has a short cytoplasmic tail with about 70 amino acids.25,29–31 The cytoplasmic tail consists of a canonical epidermal growth factor receptor (EGFR)-like tyrosine phosphorylation site, which binds with EGFR and influences cellular activity of the EGFR–PI3K–Akt signaling pathway in the CNS.32,33 Recent studies suggest that LINGO-1 could undergo oligomerization, which may also be related to the physiological role of LINGO-1 in the CNS.25
In human brain, higher levels of LINGO-1 expression are seen in the hippocampus, thalamus and neocortex and lower levels of expression are seen in the cerebellum and spinal cord.29 In situ hybridization studies demonstrated that the mRNA of LINGO-1 could be predominantly detected during embryonic and postnatal stages.29 The expression of LINGO-1 increases after CNS injury.34 All of these findings suggest that LINGO-1 has some important unknown neuronal functions in the CNS. LINGO-1 could be involved with the inhibition of axon growth, disturbance of oligodendrocyte differentiation and myelination, as well as influence neuron survival via upregulation of the NgR1 signaling pathway and downregulation of the EGFR-PI3K-Akt signaling pathway.32,35–37 The possible pathological roles of LINGO-1 in ET as well as other neurodegenerative diseases will be discussed.
LINGO-1 and Nogo-66 Receptor
LINGO-1 is a functional component of the NgR1 (a ligand-binding subunit)/p75 or TROY (signal transducing subunits) signaling complex in the CNS.30 In this complex, LINGO-1 binds with NgR1 and p75 or TROY proteins to form a ternary complex and functions to facilitate the NgR1 signal pathway.30,35 However LINGO-1 has a very low level of expression in non-neuronal cells. Lack of LINGO-1 in non-neuronal cells contributes to inactivation of NgR1 even in the presence of its binding ligands (inhibitory molecules such as MAG, OMGp, and Nogo-66).30,35 In the CNS, activation of NgR1 by its ligands can result in the displacement of Rho from the dissociation inhibitor, thus activating RhoA.38,39 RhoA activation is a key event and can activate its downstream Rho-associated protein kinase (ROCK), leading to blockage of neurite outgrowth, disruption of myelination of axons and inhibition of the maturation of oligodendrocytes.38–40 Activation of the NgR1/P75 or TROY signaling pathway could inhibit neuron proliferation and might abrogate neuron regeneration after injury.38 In contrast, targeted disruption of endogenous LINGO-1 function can downregulate the activity of the NgR1/P75 or TROY signaling pathway, block RhoA activation, and may facilitate neurite outgrowth even in the presence of inhibitory molecules.30,41–44
LINGO-1 and EGF Receptor
The PI3K-Akt signaling pathway is an important signaling pathway downstream of EGFR.45,46 After binding with its ligands, EGFR forms dimers, and is phosphorylated and activated.45,46 The activated EGFR sends a signal to PI3K, which in turn propagates the signal and leads to the phosphorylation of Akt (formation of p-Akt).45,46 The p-Akt is an activated form of Akt that promotes cell survival and proliferation.45,46 The EGFR signaling pathway is related to survival and proliferation of neurons and also is linked with neurodegenerative diseases.47–52 The EGFR signaling pathway promotes proliferation, survival, and migration of neuronal stem cells47,48 and protects post-mitotic neuronal cells in vivo and in vitro against various stress-induced injuries.49 The lack of the EGFR can induce neurodegeneration in transgenic animals.51,52
Inhibition of LINGO-1 in glaucoma animal models could upregulate p-Akt and rescue retinal ganglion cells (RGCs) from cell death.53 Neuroprotection in LINGO-1 knockout mice is associated with increased p-Akt.32 LINGO-1 could interact with EGFR directly, while overexpression of LINGO-1 could induce dosage-dependent downregulation of EFGR levels in cells.32 Therefore, LINGO-1 might negatively regulate EGFR signaling pathways via accelerating EGFR internalization and degradation, and therefore downregulates the downstream PI3K-Akt signaling activity.32 This mechanism is related to negative regulation of neuronal survival by LINGO-1. Hence LINGO-1 antagonists (LINGO-1-Fc, dominant negative LINGO-1, and anti-LINGO-1 antibody) might disrupt LINGO-1's binding with EGFR and increase EGFR levels in cells. This could contribute to activation of the EGFR-PI3K-Akt signaling pathway and improvement of Purkinje cell survival in cerebellum of ET.
LINGO-1 Negatively Regulates Neuron Survival
Glaucoma (increased intraocular pressure) results in slow degeneration of RGCs and their axons.55,56 In one study it was demonstrated that blocking of LINGO-1 with a soluble version of the extracellular domain of LINGO-1, LINGO-1-Fc fragment or primary antibody against LINGO-1 in glaucoma animal models could up-regulate Akt phosphorylation and rescue RGCs from cell death after ocular hypertension and optic nerve transaction.53 Another study revealed that LINGO-1 antagonism promotes functional recovery and axonal sprouting after spinal cord injury.57 Furthermore LINGO-1 expression is present in midbrain dopaminergic neurons in human and rodent brains.32 The expression level of LINGO-1 is elevated in the substantia nigra (SN) of patients with Parkinson's disease (PD) compared with age-matched controls32 and in animal PD models after neurotoxic damage.32 However in LINGO-1 knockout mice, dopaminergic neuron survival is higher.32 This neuroprotection in LINGO-1 knockout mice is accompanied by increased Akt phosphorylation (p-Akt).32 Both in vitro and in vivo data suggest that blockage of LINGO-1 protects and enhances neurite growth of midbrain neurons after blockage of LINGO-1.32 Thus there is a negative link between LINGO-1 and dopaminergic neuron survival, which might be related to pathological roles of LINGO-1 in Purkinje cell degeneration in ET.
LINGO-1 Nnegatively Regulates Oligodendrocytes Differentiation
LINGO-1 expression can be detected in neurons and in oligodendrocytes.54 Inhibition of LINGO-1 activity in vitro and in vivo promotes outgrowth of oligodendrocyte processes and leads to highly developed myelinated axons.41,42,58 In contrast, overexpression of LINGO-1 could contribute to delays in the onset of myelination.40 The maturation of oligodendocytes is vital to the myelination of neuron axons and the neurotrophic support to neurons.59–61 These functions are dependent on maturation of oligodendocytes and are related to neuronal functions and even neuron survival in the brain.59–61 The RhoA signaling pathway is related to oligodendrocyte differentiation and myelination.62,63 LINGO-1 activates the RhoA pathway and is a negative regulator of oligodendrocyte differentiation and myelination.40,57 However, detailed molecular mechanisms related to LINGO-1 induced inhibition of oligodendrocyte maturation remains to be elucidated. These findings have stimulated much interest in this molecule as a promising therapeutic target, in particular for the treatment of neurodegenerative diseases and diseases associated with myelin deficiencies, such as multiple sclerosis and leukodystrophies.
Possible Mechanisms of LINGO-1 Induced Neurodegeneration in ET
Several reports of association, driven largely by different rs9652490 alleles, suggest that these LINGO-1 variants when combined with other epigenetic and genetic factors possibly function to influence the LINGO-1 expression profile.64 The expression level of LINGO-1 is higher in dopaminergic neurons of PD patients.32 The expression level of LINGO-1 also increases in PD animal models after neurotoxin challenge32 and after CNS injury.54 Therefore, it would be interesting to determine whether the expression level of LINGO-1 in the brains of individuals carrying genetic variants of LINGO-1 is higher than that of non-carriers. The identified risk variants of LINGO-1 are all located in the introns of the LINGO-1 gene.65 The non-coding regions of genes including introns can often function as regulatory sequences that affect gene splicing, transcription, and translation.66 Intron variants could also significantly influence gene expression levels and even contribute to human disorders.67–70 Multiple variants of dopamine transporter (DAT1) gene have been shown to influence DAT1 gene expression and modulate susceptibility to bipolar disorder.67,68 An intronic polymorphism is associated with increased XRCC1 gene expression and leads to reduced apoptosis and familial breast cancer. Furthermore the rs9939609 A variation of the fat mass and obesity-associated (FTO) gene increases the mRNA level of the FTO gene.69 The intronic variants of the MEIS1 gene decreases expression of this gene and is associated with restless legs syndrome.70 Therefore it is possible that the intronic variants of LINGO-1 gene can lead to increased expression level of LINGO-1 and contribute to ET in the presence of other factors including oxidative stress and environmental neurotoxins.
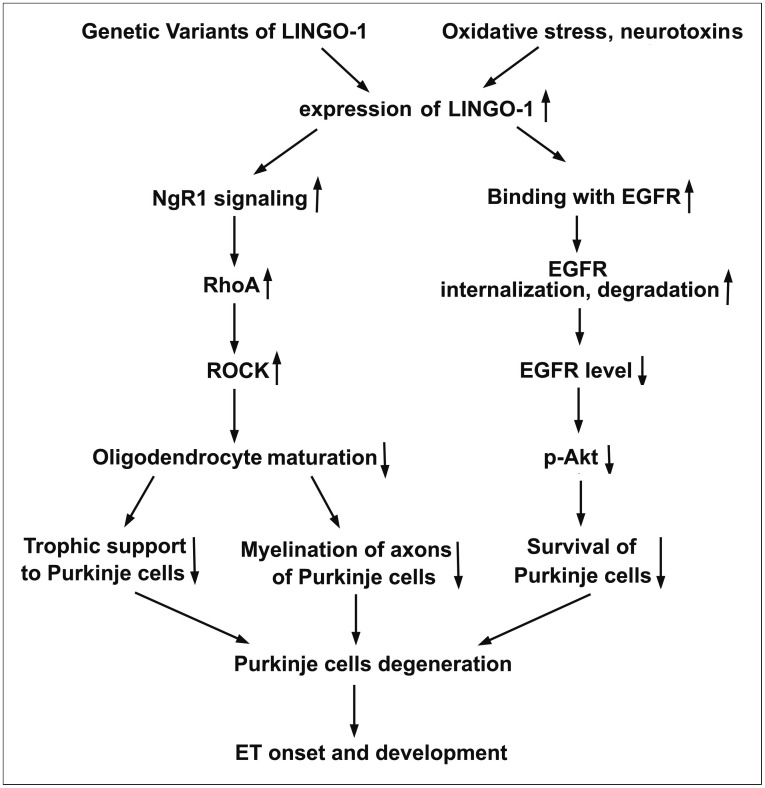
Increase in LINGO-1 expression might negatively regulate cell viability of Purkinje neurons due to downregulation of EGFR-PI3K-Akt signaling. The increased expression level of LINGO-1 can lead to inhibition of oligodendrocyte differentiation via activation of NgR1 signaling. The impaired oligodendocyte maturation will impair the neurotrophic support of oligodendocytes to Purkinje cells and disrupt the myelination of axons of Purkinje neurons in the cerebellum. All of these mechanisms might converge and contribute to Purkinje cell degeneration. Furthermore, LINGO-1 can activate the NgR1 signaling pathway, while activation of the NgR1 signaling pathway is found to prohibit neurogenesis in brain.71,72 This could contribute to decreased numbers of Purkinje cells in cerebellum to replace degenerated neurons. The potential mechanisms of LINGO-1-induced Purkinje neuron degeneration are summarized in Figure 1.
Figure 1. Hypothetical mechanisms for LINGO-1 induced cerebellum Purkinje cell degeneration in ET. LINGO-1 might contribute to Purkinje cell degeneration in the cerebellum via regulation of the activity of the NgR1–p75 signaling pathway and EGFR–PI3K–Akt signaling pathway:
Discussion
LINGO-1, the key negative regulator of the oligodendrocyte differentiation, has been considered to be a potential therapeutic target for CNS disorders, including degenerative diseases and CNS injury. We have discussed various hypotheses as to how LINGO-1 could be involved in the pathophysiology of ET. However, additional clinical and in vivo studies will be required to further decipher the role LINGO-1 in neurodegeneration. Future post-mortem studies to determine the expression levels of LINGO-1 in Purkinje cells in the cerebellum of LINGO-1 variants carriers with or without ET compared with age-matched healthy control subjects might be helpful to answer the question whether genetic variants of LINGO-1 lead to an increased expression of LINGO-1 in Purkinje cells. Transgenic LINGO-1 animal models overexpressing the LINGO-1 risk variants will also be useful. This is based on the presumption that the identified risk variants are the actual causative variants. However, all the identified risk variants are located in introns and it is not clear if these risk variants could influence the splicing of the LINGO-1 gene or have an effect on LINGO-1 protein translation. This will be the challenge for developing LINGO-1 genetic variant animal models. Long-term longitudinal clinical, imaging, and neurophysiologic studies to evaluate asymptomatic LINGO-1 variant carriers to assess their symptom onset and clinical progression will be useful to determine the penetrance and pathogenicity of these LINGO-1 risk variants. Potential neuroprotective drug trials targeting suitable molecular targets, such as LINGO-1, may offer hope to ET patients.
Footnotes
Funding: None.
Competing Interests: The authors report no conflict of interest.
References
- 1.Dogu O, Sevim S, Camdeviren H, et al. Prevalence of essential tremor: door-to-door neurologic exams in Mersin Province, Turkey. Neurology. 2003;61:1804–1806. doi: 10.1212/01.wnl.0000099075.19951.8c. [DOI] [PubMed] [Google Scholar]
- 2.Louis ED, Ottman R, Hauser WA. How common is the most common adult movement disorder? estimates of the prevalence of essential tremor throughout the world. Mov Disord. 1998;13:5–10. doi: 10.1002/mds.870130105. [DOI] [PubMed] [Google Scholar]
- 3.Benito-Leon J, Louis ED. Essential tremor: emerging views of a common disorder. Nat Clin Pract Neurol. 2006;2:666–678. doi: 10.1038/ncpneuro0347. quiz 2p following 691. [DOI] [PubMed] [Google Scholar]
- 4.Louis ED, Benito-Leon J, Bermejo-Pareja F. Self-reported depression and anti-depressant medication use in essential tremor: cross-sectional and prospective analyses in a population-based study. Eur J Neurol. 2007;14:1138–1146. doi: 10.1111/j.1468-1331.2007.01923.x. [DOI] [PubMed] [Google Scholar]
- 5.Louis ED, Faust PL, Vonsattel JP, et al. Neuropathological changes in essential tremor: 33 cases compared with 21 controls. Brain. 2007;130:3297–307. doi: 10.1093/brain/awm266. [DOI] [PubMed] [Google Scholar]
- 6.Axelrad JE, Louis ED, Honig LS, et al. Reduced Purkinje cell number in essential tremor: a postmortem study. Arch Neurol. 2008;65:101–107. doi: 10.1001/archneurol.2007.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louis ED, Faust PL, Vonsattel JP. Purkinje cell loss is a characteristic of essential tremor. Parkinsonism Relat D. 2011;17:406–409. doi: 10.1016/j.parkreldis.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louis ED, Vonsattel JP. The emerging neuropathology of essential tremor. Mov Disord. 2008;23:174–182. doi: 10.1002/mds.21731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louis ED, Vonsattel JP, Honig LS, et al. Essential tremor associated with pathologic changes in the cerebellum. Arch Neurol. 2006;63:1189–1193. doi: 10.1001/archneur.63.8.1189. [DOI] [PubMed] [Google Scholar]
- 10.Shatunov A, Sambuughin N, Jankovic J, et al. Genomewide scans in North American families reveal genetic linkage of essential tremor to a region on chromosome 6p23. Brain. 2006;129:2318–2331. doi: 10.1093/brain/awl120. [DOI] [PubMed] [Google Scholar]
- 11.Illarioshkin SN, Rakhmonov RA, Ivanova-Smolenskaia IA, et al. [Molecular genetic analysis of essential tremor] Genetika. 2002;38:1704–1709. [PubMed] [Google Scholar]
- 12.Stefansson H, Steinberg S, Petursson H, et al. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nat Genet. 2009;41:277–279. doi: 10.1038/ng.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilarino-Guell C, Ross OA, Wider C, et al. LINGO1 rs9652490 is associated with essential tremor and Parkinson disease. Parkinsonism Relat D. 2010;16:109–111. doi: 10.1016/j.parkreldis.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thier S, Lorenz D, Nothnagel M, et al. LINGO1 polymorphisms are associated with essential tremor in Europeans. Mov Disord. 2010;25:709–715. doi: 10.1002/mds.22887. [DOI] [PubMed] [Google Scholar]
- 15.Vilarino-Guell C, Wider C, Ross OA, et al. LINGO1 and LINGO2 variants are associated with essential tremor and Parkinson disease. Neurogenetics. 2010;11:401–408. doi: 10.1007/s10048-010-0241-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark LN, Park N, Kisselev S, et al. Replication of the LINGO1 gene association with essential tremor in a North American population. Eur J Hum Genet. 2010;18:838–843. doi: 10.1038/ejhg.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan EK, Teo YY, Prakash KM, et al. Lingo1 variant increases risk of familial essential tremor. Neurology. 2009;73:1161–1162. doi: 10.1212/WNL.0b013e3181bacfc9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo X, Jiang H, Guo JF, et al. Screening for two SNPs of LINGO1 gene in patients with essential tremor or sporadic Parkinson's disease in Chinese population. Neurosci Lett. 2010;481:69–72. doi: 10.1016/j.neulet.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 19.Wu YW, Rong TY, Li HH, et al. Analysis of Lingo1 variant in sporadic and familial essential tremor among Asians. Acta Neurol Scand. 2011;124:264–268. doi: 10.1111/j.1600-0404.2010.01466.x. [DOI] [PubMed] [Google Scholar]
- 20.Lorenzo-Betancor O, Garcia-Martin E, Cervantes S, et al. Lack of association of LINGO1 rs9652490 and rs11856808 SNPs with familial essential tremor. Eur J Neurol. 2011;18:1085–1089. doi: 10.1111/j.1468-1331.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 21.Bourassa CV, Riviere JB, Dion PA, et al. LINGO1 Variants in the French-Canadian Population. Plos One. 2011;6:e16254. doi: 10.1371/journal.pone.0016254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis ED. Environmental Epidemiology of Essential Tremor. Neuroepidemiology. 2008;31:139–149. doi: 10.1159/000151523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carim-Todd L, Escarceller M, Estivill X, Sumoy L. LRRN6A/LERN1 (leucine-rich repeat neuronal protein 1), a novel gene with enriched expression in limbic system and neocortex. Eur J Neurosci. 2003;18:3167–3182. doi: 10.1111/j.1460-9568.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Aulia S, Li L, Tang BL. AMIGO and friends: an emerging family of brain-enriched, neuronal growth modulating, type I transmembrane proteins with leucine-rich repeats (LRR) and cell adhesion molecule motifs. Brain Res Rev. 2006;51:265–274. doi: 10.1016/j.brainresrev.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Mosyak L, Wood A, Dwyer B, et al. The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition. J Biol Chem. 2006;281:36378–36390. doi: 10.1074/jbc.M607314200. [DOI] [PubMed] [Google Scholar]
- 26.Lauren J, Airaksinen MS, Saarma M, Timmusk T. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–421. doi: 10.1016/S0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 27.Homma S, Shimada T, Hikake T, Yaginuma H. Expression pattern of LRR and Ig domain-containing protein (LRRIG protein) in the early mouse embryo. Gene Expr Patterns. 2009;9:1–26. doi: 10.1016/j.gep.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Haines BP, Rigby PW. Expression of the Lingo/LERN gene family during mouse embryogenesis. Gene Expr Patterns. 2008;8:79–86. doi: 10.1016/j.modgep.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Llorens F, Gil V, Iraola S, et al. Developmental analysis of Lingo-1/Lern1 protein expression in the mouse brain: interaction of its intracellular domain with Myt1l. Dev Neurobiol. 2008;68:521–541. doi: 10.1002/dneu.20607. [DOI] [PubMed] [Google Scholar]
- 30.Mi S, Lee X, Shao Z, et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7:221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Xu X, Zhang Y, et al. LINGO-1 interacts with WNK1 to regulate nogo-induced inhibition of neurite extension. J Biol Chem. 2009;284:15717–15728. doi: 10.1074/jbc.M808751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue H, Lin L, Lee X, et al. Inhibition of the leucine-rich repeat protein LINGO-1 enhances survival, structure, and function of dopaminergic neurons in Parkinson's disease models. Proc Natl Acad Sci U S A. 2007;104:14430–14435. doi: 10.1073/pnas.0700901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumaria A. LINGO-1, WNK1, and EGFR: a hypothesis. J Biol Chem. 2009;284:le9. doi: 10.1074/jbc.L808751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trifunovski A, Josephson A, Ringman A, et al. Neuronal activity-induced regulation of Lingo-1. Neuroreport. 2004;15:2397–2400. doi: 10.1097/00001756-200410250-00019. [DOI] [PubMed] [Google Scholar]
- 35.Shao Z, Browning JL, Lee X, et al. TAJ/TROY, an orphan TNF receptor family member, binds Nogo-66 receptor 1 and regulates axonal regeneration. Neuron. 2005;45:353–359. doi: 10.1016/j.neuron.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 36.Wang KC, Kim JA, Sivasankaran R, et al. P75 interacts with the Nogo receptor as a co-receptor for Nogo, MAG and OMgp. Nature. 2002;420:74–78. doi: 10.1038/nature01176. [DOI] [PubMed] [Google Scholar]
- 37.Yamashita T, Tohyama M. The p75 receptor acts as a displacement factor that releases Rho from Rho-GDI. Nat Neurosci. 2003;6:461–467. doi: 10.1038/nn1045. [DOI] [PubMed] [Google Scholar]
- 38.McGee AW, Strittmatter SM. The Nogo-66 receptor: focusing myelin inhibition of axon regeneration. Trends Neurosci. 2003;26:193–8. doi: 10.1016/S0166-2236(03)00062-6. [DOI] [PubMed] [Google Scholar]
- 39.Hunt D, Coffin RS, Anderson PN. The Nogo receptor, its ligands and axonal regeneration in the spinal cord; a review. J Neurocytol. 2002;31:93–120. doi: 10.1023/A:1023941421781. [DOI] [PubMed] [Google Scholar]
- 40.Mi S, Miller RH, Lee X, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- 41.Lee Z, Yang Z, Shao X, et al. NGF regulates the expression of axonal LINGO-1 to inhibit oligodendrocyte differentiation and myelination. J Neurosci. 2007;27:220–225. doi: 10.1523/JNEUROSCI.4175-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mi S, Hu B, Hahm K, et al. LINGO-1 antagonist promotes spinal cord remyelination and axonal integrity in MOG-induced experimental autoimmune encephalomyelitis. Nat Med. 2007;13:1228–1233. doi: 10.1038/nm1664. [DOI] [PubMed] [Google Scholar]
- 43.Zhao XH, Jin WL, Wu J, et al. Inactivation of glycogen synthase kinase-3beta and up-regulation of LINGO-1 are involved in LINGO-1 antagonist regulated survival of cerebellar granular neurons. Cell Mol Neurobiol. 2008;28:727–735. doi: 10.1007/s10571-007-9258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeon CY, Kim HJ, Morii H, et al. Neurite outgrowth from PC12 cells by basic fibroblast growth factor (bFGF) is mediated by RhoA inactivation through p190RhoGAP and ARAP3. J Cell Physiol. 224:786–94. doi: 10.1002/jcp.22184. [DOI] [PubMed] [Google Scholar]
- 45.Hakariya T, Shida Y, Sakai H, et al. EGFR signaling pathway negatively regulates PSA expression and secretion via the PI3K-Akt pathway in LNCaP prostate cancer cells. Biochem Biophys Res Commun. 2006;342:92–100. doi: 10.1016/j.bbrc.2006.01.106. [DOI] [PubMed] [Google Scholar]
- 46.Freudlsperger C, Burnett JR, Friedman JA, et al. EGFR-PI3K-AKT-mTOR signaling in head and neck squamous cell carcinomas: attractive targets for molecular-oriented therapy. Expert Opin Ther Targets. 15:63–74. doi: 10.1517/14728222.2011.541440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ayuso-Sacido A, Moliterno JA, Boockvar JA. Activated EGFR signaling increases proliferation, survival, and migration and blocks neuronal differentiation in neural stem cells. Neuro-Oncology. 2009;11:682–682. doi: 10.1007/s11060-009-0035-x. [DOI] [PubMed] [Google Scholar]
- 48.Ayuso-Sacido A, Moliterno JA, Kratovac S, et al. Activated EGFR signaling increases proliferation, survival, and migration and blocks neuronal differentiation in post-natal neural stem cells. J Neuro-Oncol. 2010;97:323–337. doi: 10.1007/s11060-009-0035-x. [DOI] [PubMed] [Google Scholar]
- 49.Sakanaka M, Peng H, Wen TC, et al. Epidermal growth factor protects neuronal cells in vivo and in vitro against transient forebrain ischemia- and free radical-induced injuries. J Cerebr Blood F Met. 1998;18:349–360. doi: 10.1097/00004647-199804000-00002. [DOI] [PubMed] [Google Scholar]
- 50.Chen-Plotkin A, Hu W, Siderowf A, et al. Plasma EGF Levels Predict Cognitive Decline in Parkinson's Disease. Neurology. 2011;76:A176–A176. [Google Scholar]
- 51.Wong RWC, Guillaud L. The role of epidermal growth factor and its receptors in mammalian CNS. Cytokine Growth F R. 2004;15:147–156. doi: 10.1016/j.cytogfr.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Kempermann G, Steiner B, Wolf SA. Adult neurogenesis and neurodegenerative disease. Regen Med. 2006;1:15–28. doi: 10.2217/17460751.1.1.15. [DOI] [PubMed] [Google Scholar]
- 53.Fu QL, Hu B, Wu W, et al. Blocking LINGO-1 function promotes retinal ganglion cell survival following ocular hypertension and optic nerve transection. Invest Ophthalmol Vis Sci. 2008;49:975–985. doi: 10.1167/iovs.07-1199. [DOI] [PubMed] [Google Scholar]
- 54.Mi S, Sandrock A, Miller RH. LINGO-1 and its role in CNS repair. Int J Biochem Cell Biol. 2008;40:1971–1978. doi: 10.1016/j.biocel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 55.Tataru LC, Bogdanici CM. [Glaucoma–neurodegenerative disease] Rev Med Chir Soc Med Nat Iasi. 2009;113:1120–1125. [PubMed] [Google Scholar]
- 56.Gupta N, Yucel YH. Glaucoma as a neurodegenerative disease. Curr Opin Ophthalmol. 2007;18:110–114. doi: 10.1097/ICU.0b013e3280895aea. [DOI] [PubMed] [Google Scholar]
- 57.Ji B, Sandrock A, Miller RH, et al. LINGO-1 antagonist promotes functional recovery and axonal sprouting after spinal cord injury. Mol Cell Neurosci. 2006;33:311–320. doi: 10.1016/j.mcn.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Rudick RA, Mi S, Sandrock AW., Jr LINGO-1 antagonists as therapy for multiple sclerosis: in vitro and in vivo evidence. Expert Opin Biol Ther. 2008;8:1561–1570. doi: 10.1517/14712598.8.10.1561. [DOI] [PubMed] [Google Scholar]
- 59.Wilkins A, Majed H, Layfield R, et al. Oligodendrocytes promote neuronal survival and axonal length by distinct intracellular mechanisms: a novel role for oligodendrocyte-derived glial cell line-derived neurotrophic factor. J Neurosci. 2003;23:4967–4974. doi: 10.1523/JNEUROSCI.23-12-04967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao Q, He Q, Wang Y, et al. Transplantation of ciliary neurotrophic factor-expressing adult oligodendrocyte precursor cells promotes remyelination and functional recovery after spinal cord injury. J Neurosci. 30:2989–3001. doi: 10.1523/JNEUROSCI.3174-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jakovcevski I, Filipovic R, Mo Z, et al. Oligodendrocyte development and the onset of myelination in the human fetal brain. Front Neuroanat. 2009;3:5. doi: 10.3389/neuro.05.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baer AS, Syed YA, Kang SU, et al. Myelin-mediated inhibition of oligodendrocyte precursor differentiation can be overcome by pharmacological modulation of Fyn-RhoA and protein kinase C signalling. Brain. 2009;132:465–481. doi: 10.1093/brain/awn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajasekharan S, Baker KA, Horn KE, et al. Netrin 1 and Dcc regulate oligodendrocyte process branching and membrane extension via Fyn and RhoA. Development. 2009;136:415–426. doi: 10.1242/dev.018234. [DOI] [PubMed] [Google Scholar]
- 64.Vilarino-Guell C, Ross OA, Wider C, et al. LINGO1 rs9652490 is associated with essential tremor and Parkinson disease. Parkinsonism Relat D. 16:109–111. doi: 10.1016/j.parkreldis.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng H, Gu S, Jankovic J. LINGO1 variants in essential tremor and Parkinson's disease. Acta Neurol Scand. 2011;125:1–7. doi: 10.1111/j.1600-0404.2011.01516.x.2011. [DOI] [PubMed] [Google Scholar]
- 66.Wang GJ, Yang P, Xie HG. Gene variants in noncoding regions and their possible consequences. Pharmacogenomics. 2006;7:203–209. doi: 10.2217/14622416.7.2.203. [DOI] [PubMed] [Google Scholar]
- 67.Kelsoe JR, Greenwood TA, Schork NJ, Eskin E. Identification of additional variants within the human dopamine transporter gene provides further evidence for an association with bipolar disorder in two independent samples. Mol Psychiatr. 2006;11:125–133. doi: 10.1038/sj.mp.4001764. [DOI] [PubMed] [Google Scholar]
- 68.Kelsoe JR, Greenwood TA. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics. 2003;82:511–520. doi: 10.1016/S0888-7543(03)00142-3. [DOI] [PubMed] [Google Scholar]
- 69.Hoffstedt J, Wahlen K, Sjolin E. The common rs9939609 gene variant of the fat mass- and obesity-associated gene FTO is related to fat cell lipolysis. J Lipid Res. 2008;49:607–611. doi: 10.1194/jlr.M700448-JLR200. [DOI] [PubMed] [Google Scholar]
- 70.Rouleau GA, Xiong L, Catoire H, et al. MEIS1 intronic risk haplotype associated with restless legs syndrome affects its mRNA and protein expression levels. Hum Mol Genet. 2009;18:1065–1074. doi: 10.1093/hmg/ddn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ng CE, Tang BL. Nogos and the Nogo-66 receptor: factors inhibiting CNS neuron regeneration. J Neurosci Res. 2002;67:559–565. doi: 10.1002/jnr.10134. [DOI] [PubMed] [Google Scholar]
- 72.Funahashi S, Hasegawa T, Nagano A, Sato K. Differential expression patterns of messenger RNAs encoding Nogo receptors and their ligands in the rat central nervous system. J Comp Neurol. 2008;506:141–160. doi: 10.1002/cne.21541. [DOI] [PubMed] [Google Scholar]