Abstract
To identify genes important for hair-cell function, we conducted a mutagenic screen in zebrafish. Larvae from one mutant line, ru848, were unresponsive to acoustic stimuli and unable to balance. The mutation results in a 90% reduction in hair-cell number and partial retinal degeneration by 5 days postfertilization. We localized the recessive ru848 mutation by positional cloning to the zebrafish homolog of the human Choroideremia gene, which encodes Rab escort protein 1. This protein is essential for the normal prenylation of Rabs. Mutations in the human gene induce choroideremia, a disease marked by slow-onset degeneration of rod photoreceptors and retinal pigment epithelial cells. The degenerative phenotype resulting from a null mutation in the zebrafish gene indicates that hair cells and retinal cells require Rab escort protein 1 for survival.
The internal ear, one of the most complex organs of a vertebrate organism, converts mechanical stimuli into neuronal responses. The task of mechanotransduction is accomplished by the ear's sensory receptor, the hair cell. The lateralline organs of fishes and aquatic amphibians also use this receptor to detect water movements. Despite our detailed knowledge of the hair cell's mechanical and electrical properties (1), our progress in understanding its development and operation has been impeded by our lack of knowledge of the relevant molecular components.
One strategy used to identify genes important for hair-cell function is the investigation of their loss-of-function phenotypes, deafness and disequilibrium. The principal advantage of such a genetic approach is that detailed characterization of the mutant phenotype provides explicit clues about the role of the gene involved. Large-scale mutagenesis screens for mechanosensory mutants in Drosophila melanogaster and Caenorhabditis elegans have identified molecules critical for vibration and touch sensitivity (2–4). The absence of hair cells in invertebrates, however, makes screens specific to hair-cell defects impossible in these organisms.
To identify important proteins in the hair cell, we have created point mutations randomly throughout the zebrafish genome and screened for impairment of ear and lateral-line-organ development and function. Several of the identified mutations have been subjected to further phenotypic analysis and genetic mapping and cloning. We report here the characterization of a mutation that identifies an important process in hair cells and serves as a model of degenerative disease in the human retina.
Materials and Methods
ru848 Identification and Breeding. Zebrafish were maintained in aquaria (Marine Biotech, Beverly, MA) according to published protocols (5). Embryos were raised in a 28.5°C incubator in facility water containing 1 μg/ml methylene blue and staged as described (6). Male fish of the TL strain were mutagenized with ethylnitrosourea and bred for three generations to create homozygous F3 larvae (7) that were screened for hearing defects by observing their response to a sharp tap on the side of their container. Heterozygous carrier fish of the ru848 mutant line were selected from the F2 generation and outcrossed to WT animals of the WIK strain for mapping.
Morphological Analysis. Fish were mounted on coverslips in 2% methylcellulose in facility water and imaged with a dissecting microscope. To label neuromasts, larvae were placed for 5 min in a 200-μM solution of 4-(4-(diethylamino)styr yl)-N-methylpyridinium iodide (4-Di-2-ASP; Molecular Probes) with 0.1 mg/ml BSA and observed under epifluorescence with a fluorescein filter block.
Measurement of Inner-Ear Microphonic Potentials. Microphonic potentials in response to vibratory stimuli were measured from the otic vesicles of WT and ru848 mutant larvae at 5–7 days postfertilization (dpf). An individual larva was transferred to a recording chamber, visualized under a dissecting microscope, and immobilized in three drops of mounting solution containing 1.2% agarose in anuran standard saline solution. A vibratory stimulus was applied to the larval surface at the posterior edge of the otic vesicle by a glass rod pulled to a tip ≈30 μm in diameter and driven by a calibrated piezoelectric actuator.
Responses were recorded with glass microelectrodes whose resistances were 10–20 MΩ when filled with standard saline solution and 500 mM gentamicin. An electrode was advanced until its tip visibly penetrated the labyrinth. Responses measured with a directly coupled amplifier were amplified ×104, filtered at 3–2,000 Hz, and recorded at a sampling frequency of 5 kHz.
Phalloidin Labeling. Larvae were fixed overnight at 4°C in 4% paraformaldehyde in PBS, rinsed in PBS containing 10 mg/ml BSA and 1% DMSO, permeabilized overnight at 4°C in 2% Triton X-100, and labeled for 3 h with Alexa 488 phalloidin (2.5 units/ml; Molecular Probes). Mounted on coverslips, the larvae were imaged with a ×40 water-immersion objective lens on a laser-scanning confocal microscope (MRC1024ES, Bio-Rad).
Transmission Electron Microscopy. Animals were fixed for 18 h at room temperature in 200 mM glutaraldehyde, 400 mM formaldehyde, 4 mM CaCl2, and 100 mM sodium cacodylate at pH 7.4. After a wash, specimens were postfixed for 2 h at 4°C in 50 mM OsO4, 4 mM CaCl2, and 80 mM sodium cacodylate at pH 7. After decalcification in 100 mM EGTA and en bloc staining with uranyl acetate during dehydration in graded ethanol concentrations, larvae were embedded in epoxy plastic. Sections were cut at a thickness of 70 nm, stained with lead citrate, and examined at an accelerating voltage of 80 kV.
Scanning Electron Microscopy. To remove the gelatinous cupulae from neuromasts, larvae were exposed for 30 min at room temperature to 80 μg/ml subtilopeptidase A (protease XXIV, Sigma) in anuran standard saline solution. Specimens were then treated by a modification of the TAO technique (8). After dehydration in graded ethanol concentrations and critical-point drying from liquid CO2, the uncoated specimens were examined at an accelerating voltage of 3 kV in a scanning electron microscope (1550, LEO Electron Microscopy, Cambridge, U.K.) with a 10-μm aperture.
Genetic Mapping. DNA isolation and PCRs were performed as described (9). Bulked segregant analysis was conducted by using primers (Research Genetics, Huntsville, AL) flanking each of 215 simple sequence-length polymorphisms (10). Bacterial artificial chromosome (BAC) fingerprinting data were obtained from the Wellcome Trust Sanger Institute (Hinxton, U.K.), which was also the source of genomic sequence information from the shotgun assembly. Information on ESTs was found at The Institute for Genomic Research Zebrafish Gene Index (www.tigr.org/tdb/tgi/zgi) and GenBank (www.ncbi.nlm.nih.gov/Genbank). BACs were obtained from the CHORI-211 library (Children's Hospital Oakland Research Institute, Oakland, CA). The protein sequence alignments were constructed by using the clustalw algorithm (macvector, Accelrys, San Diego).
RNA Isolation. Total RNA was isolated according to the manufacturer's directions by homogenizing tissue in TRIzol reagent (Invitrogen). cDNA was prepared by reverse-transcribing 500–1,000 ng of total RNA with the SuperScript First-Strand Synthesis System for RT-PCR by using the oligo(dT) primer (Invitrogen). For expression analysis, a short fragment of RNA was amplified by using the PCR; all primer sequences are available on request.
RNA Injection. The coding region of the gene of interest was amplified by the PCR from WT TL cDNA. After the product had been cloned into the CS2++ vector, SP6 RNA polymerase was used to transcribe REP1 according to protocol (mMessage mMachine Kit, Ambion). Approximately 100 pg of RNA was injected into each embryo before the four-cell stage (11). The phenotype was assessed by counting numbers of neuromasts labeled with 4-Di-2-ASP at 5 dpf. A day later, the larvae were fixed in methanol and genotyped by PCR amplification and sequencing of the genomic region containing the mutation.
Morpholino Injection. A morpholino was synthesized that was designed to bind to the region spanning the start codon of the relevant RNA (5′-GATCCTCCGCAGCCATCTTGAAACC; GeneTools, Philomath, OR). As a negative control, a second oligomer was synthesized with five mismatches (5′-GATgCTaCGCAGCgATCTTcAAtCC). Each morpholino was resuspended at a concentration of 100 μM in Danieau solution [58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, and 5 mM Hepes at pH 7.2] supplemented with 0.025% phenol red. Developing blastulae with one to four cells were injected with ≈5 ng per embryo.
Results
Phenotypic Analysis. Zebrafish larvae at 5 dpf normally display a characteristic startle response to acoustic stimulation: the fish curl into a C-shaped bend, then flip in the opposite direction to move away from the stimulus (12). The ru848 mutant line was initially identified in a behavioral screen by the absence of this startle response and by defective swimming behavior. Mutant fish did not respond to an acoustic stimulus, but remained lying on their sides at the bottom of the dish throughout the assay. In response to a touch stimulus at the tip of the tail, however, mutant fish consistently displayed avoidance behavior beginning with the stereotyped C-bend. We therefore concluded that the absence of a response to sound did not reflect an inability to initiate and execute a startle response. In contradistinction to WT animals, mutant fish did not swim directly away from the touch stimulus, but instead looped back toward the probe and continued circling on the spot or eventually swam away with a corkscrew motion. This abnormal swimming behavior is indicative of a balance defect.
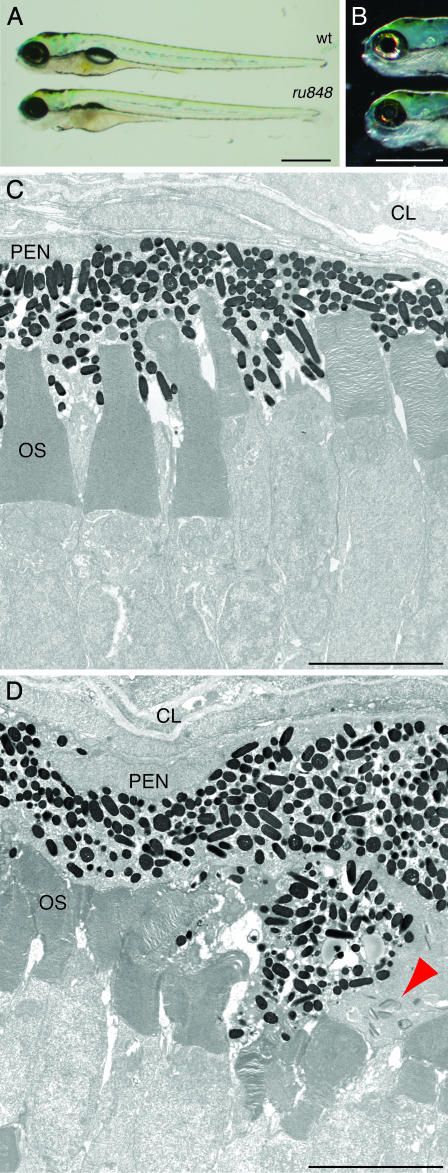
Despite these behavioral defects, the overall appearance of 5-dpf mutant larvae was similar to that of WT fish (Fig. 1A). The most prominent defect was an uninflated swim bladder. In addition, mutant larvae had noticeably smaller eyes and irregular eye pigmentation (Fig. 1B): mutants displayed a patchy, discontinuous distribution of iridophore pigment cells. Transmission electron microscopy showed that the retinal layers were disrupted in mutants (Fig. 1 C and D). The pigment epithelium was particularly disordered, with some cells invading the outer nuclear layer. There were also small, dense, presumably pyknotic cells present throughout the neural retina. Additional degeneration occurred on the posterior surface of the developing lens. The cells of mutant animals were marked by discrete cytoplasmic inclusions that were not observed in WT fish. Discoidal and comprising ≈10 well organized laminae, each structure was surrounded by a lightly stained, amorphous mass; no bounding membrane was present. The inclusions were prominent in pigment epithelial cells of the retina (Fig. 1D) and occurred as well in putative Schwann cells of the developing otic ganglion.
Fig. 1.
Morphology of ru848 mutant and WT larvae at 5 dpf. (A) A transilluminated mutant larva (Lower) lacks an inflated swim bladder and has a slightly smaller eye than the WT (Upper). (B) Incident illumination shows reflective iridiphores that cover almost the entire eye in the WT larva (Upper) but are sparse in the mutant (Lower). (C) In a transmission electron micrograph, the photoreceptor outer segments (OS) in the WT retina are neatly arrayed and interdigitated with processes of the pigment-epithelial cells. (D) In the mutant retina, the outer segments are disheveled and the pigment-epithelial cells are hypertrophic and disorganized. A cluster of cytoplasmic inclusions in a mutant pigment epithelial cell is marked by a red arrowhead. CL, choroid layer; PEN, pigment epithelial nucleus. (Scale bars: 500 μm, A and B;5 μm, C and D.)
Beginning on the sixth day, edema was observed around the heart and abdomen, the eyes continued to degenerate, and the mutant larvae began to die. Proper feeding was precluded by the absence of a functional swim bladder and perhaps by disequilibrium. The lack of nutrition was not likely to have been the sole cause of the widespread degeneration seen in mutants, however, for unfed WT larvae survived until at least 10 dpf.
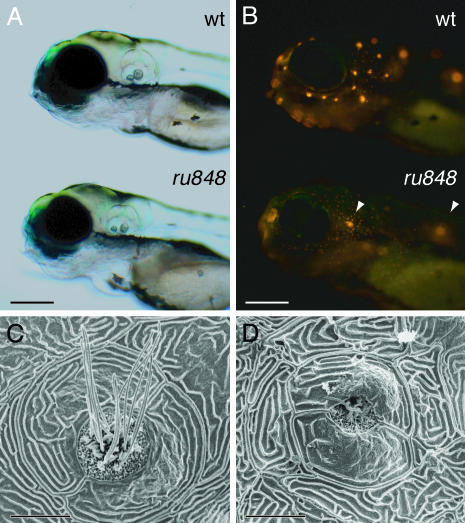
Each neuromast on one side of a WT larva can be uniquely identified as a member of one of eight lateral lines (13). As assayed by observation under differential interference contrast optics, the primordia from which neuromasts originate migrate normally in mutant fish (data not shown). Neuromast hair cells can be labeled in the living organism with the fluorescent compound 4-Di-2-ASP, which is thought to traverse the mechanically sensitive channels near the stereociliary tips (14). At 5 dpf, mutant neuromasts were labeled poorly save for two: the second neuromast of the otic line, situated above the otic vesicle, and the first neuromast of the ventral posterior line, situated dorsal to the yolk (Fig. 2 A and B). Similar results were observed when 4-Di-2-ASP was iontophoresed directly into a mutant's otic vesicle, where it was taken up by only a few hair cells directly beneath the otoliths (data not shown).
Fig. 2.
Disruption of lateral-line hair cells by the ru848 mutation. (A) Transmitted-light illumination shows the grossly normal structure of the otocysts in WT (Upper) and mutant (Lower) larvae at 5 dpf. (B) Under fluorescent illumination, the neuromasts form a characteristic pattern on the head of a WT larva (Upper) labeled with 4-Di-2-ASP. The mutant larva (Lower) bears only two brightly labeled neuromasts (arrowheads). (C) In a scanning electron micrograph, the center of a WT neuromast at 6 dpf displays hair bundles with short stereocilia and long kinocilia. The neuromast is enclosed by two periderm cells. (D) In a mutant, the periderm cells appear relatively normal, but in the center of the neuromast stand only two kinocilia that are much shorter than those seen in the WT. (Scale bars: 200 μm, A and B; 10 μm, C and D.)
To examine the structure of the neuromasts in more detail, we fixed larvae at 6 dpf for scanning electron microscopy. The perimeter of each neuromast is delineated by two semilunar periderm cells that surround the centrally located hair bundles. In WT larvae, every neuromast displayed multiple hair bundles, each comprising short stereocilia and a single long kinocilium (Fig. 2C). The general shape of the neuromasts was preserved in the mutants, but the hair bundles were degenerate or completely absent (Fig. 2D).
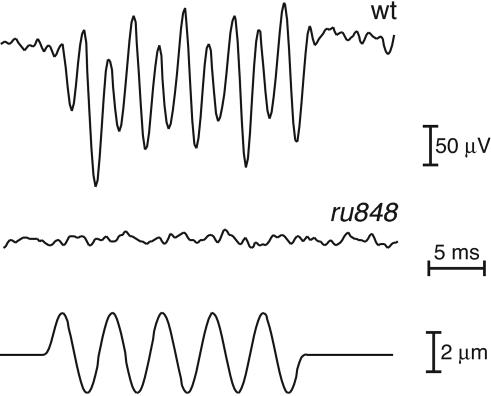
We recorded extracellular microphonic potentials from developing ears in response to vibratory stimulation of WT zebrafish larvae at 5–7 dpf (Fig. 3). Iontophoretic application of gentamicin, an aminoglycoside blocker of mechanoelectrical transduction, abolished the microphonic response within 10 min, confirming the origin of the recorded responses in the mechanoelectrical transduction process. We were unable to record similar electrical responses from the inner ears of 10 mutants, indicating that these fish had impaired mechanoelectrical transduction.
Fig. 3.
Analysis of hair-cell function in WT and mutant fish at 7 dpf. While a vibratory stimulus (200 Hz, ±2 μm; Bottom) was provided by a glass probe, extracellular microphonic potentials were recorded and averaged over 50–100 stimulus epochs. The WT larvae displayed a robust electrical response at twice the stimulus frequency (Top), reflecting the activation of two populations of hair cells with opposing orientations. No response to the vibratory stimulus was seen in the 10 mutant larvae examined (Middle).
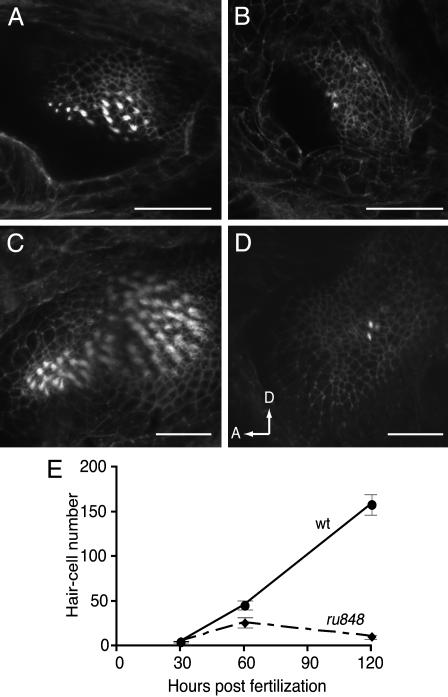
Signs of sensory-epithelial degeneration were visible in the inner ears of mutant embryos from 30 h postfertilization (hpf). In mutant embryos at this stage, the full complement of hair cells and hair bundles was visible, but the edges of the epithelium were marked by cellular debris. In fixed tissue, fully mature hair cells can be identified by the distinctive appearance of hair bundles labeled with fluorescent phalloidin. The number of mature hair cells at each stage was assessed by counting the hair bundles visible in serial confocal microscopic sections. At 30 hpf, the numbers of hair bundles present in mutant and WT embryos were indistinguishable. A difference in hair-cell count was apparent from 60 hpf and became more pronounced at 120 hpf owing to reduction of total hair-cell number in the mutant (Fig. 4). These results suggest that the gene mutated in ru848 is required for hair-cell survival.
Fig. 4.
Reduction in the number of inner-ear hair cells in mutant larvae. The total number of hair cells was assessed at three developmental times by counting the number of Alexa 488-phalloidin-labeled hair bundles in all sensory epithelia. A reduction in number became noticeable from 60 hpf, as shown here in single confocal sections through WT (A) and mutant (B) posterior maculae. By 120 hpf, many more hair cells had been added to the posterior macula in the WT (C), but little change was observed in the mutant (D), as seen in projections of Z-series through the posterior maculae. In D, arrows show anterior (A) and dorsal (D) directions. (Scale bars: 50 μm.) (E) Similar results were observed when hair cells from all five sensory epithelia in the inner ear were counted at three developmental stages. At least eight larvae were examined for each data point.
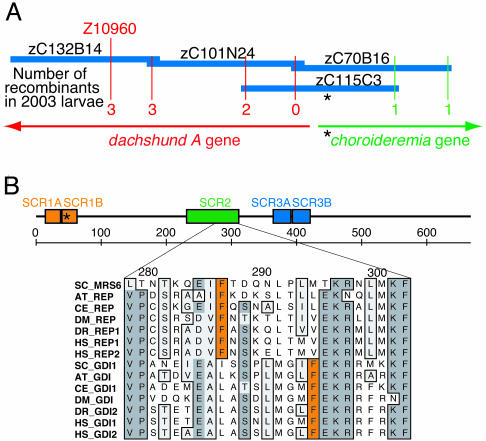
Positional Cloning. The mutant phenotype identified in the ru848 line was fully penetrant and varied little either between clutches or between larvae within a clutch. We used genetic mapping techniques to determine the underlying causative mutation. Bulked segregant analysis with simple sequence length polymorphisms (10, 15) revealed that the polymorphic marker z4425 in linkage group 21 cosegregated with the mutant phenotype. Further characterization of markers in the same region identified a second simple sequence length polymorphism, z10960, that was 7.2 cM distant on the opposite side of the mutation. Just over 2,000 mutant larvae were genotyped at these two polymorphic markers to identify 225 recombination events between z4425 and the mutation, and only three between z10960 and the mutation (Fig. 5A).
Fig. 5.
Mapping of the ru848 mutation. (A) Sequence and BAC fingerprint information was used to create a contig spanning the mutant locus and anchored to the genetic map at marker z10960. The number of recombinations observed at polymorphic loci on opposite sides of the mutation are indicated (red and green). This contig contained exons from two genes, dachshund A and chm. The asterisk indicates the location of the mutation. (B) In a diagram of the REP1 protein sequence, the boxed areas outline stretches of sequence similarity [structurally conserved regions (SCRs)] found in REPs and the closely related Rab GDIs. The ruler indicates amino acid numbers. The alignment below shows a segment of SCR2; the positions of the phenylalanine residues indicated in orange clearly distinguish the REP and GDI subfamilies. The species abbreviations and accession numbers for the alignment are: for S. cerevisiae (SC) MRS6, NP_015015; GDI1, NP_011062; for Arabidopsis thaliana (AT) REP, NP_187306; GDI, T49943; for C. elegans (CE) REP, NP_497423; GDI, NP_502788; for D. melanogaster (DM) REP, NP_477420; GDI, NP_523524; for Homo sapiens (HS) REP1, NP_000381; REP2, NP_001812; GDI1, NP_001484; GDI2, NP_001485; and for Danio rerio (DR) REP1, AY500891; GDI2, AY391428.
The sequence amplified by the z10960 primers was used to search the zebrafish genome shotgun assembly. A number of nearby BAC clones were identified that were used in turn to query the shotgun assembly. BAC inserts were ordered on the basis of sequence overlap at their 5′ and 3′ ends. Through examination of the BAC clone fingerprinting database, a single BAC (zC115C3) was identified that defined the critical interval known to contain the ru848 mutation.
Two unique, nonrepetitive coding regions were present within this BAC (Fig. 5A). One represented a portion of a previously cloned zebrafish gene, dachshund A, of which only the first exon lay within the critical interval. Sequencing of this exon disclosed no mutations in ru848 animals.
The second gene identified in BAC zC115C3 was a close homolog of the human gene Choroideremia, which encodes Rab escort protein 1 (REP1). We accordingly term the zebrafish gene choroideremia (chm). Sequencing of chm exons in both mutant and WT larvae identified a nonsense mutation in the second exon resulting from the transition of a cytosine to a thymine. The mutation converts a glutamine codon (CAA) to a stop codon (UAA) 32 aa after the start of the coding region.
REPs are required for the posttranslational modification of Rab GTPases (16), which coordinate vesicle trafficking in eukaryotic cells (17). REPs contain highly conserved stretches of amino acids, the structurally conserved regions (18), which also occur in Rab guanine-nucleotide dissociation inhibitors (GDIs). Examination of a diagnostic region that clearly differentiates between the two families confirmed that chm encodes a REP and not a Rab GDI (Fig. 5B).
RNA Expression. In adult animals, we detected chm RNA in skeletal muscle and in all of the organs tested, including the heart, liver, ovary, testis, and eye. Because the survival of a limited number of neuromast and inner-ear hair cells might be attributable to the presence of maternally derived chm RNA, we conducted PCR amplification of reverse-transcribed cDNA derived from RNA isolated from WT zebrafish embryos at 2 hpf. We observed the presence of chm RNA at that time, before embryos begin to transcribe their own genes (19) (data not shown).
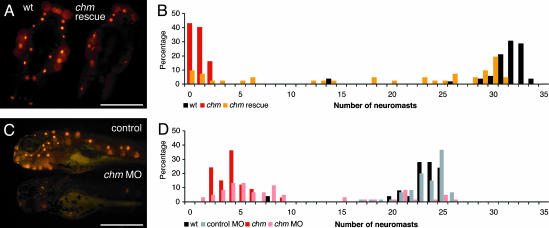
Gene Confirmation. To confirm the phenotypic impact of the mutation in chm, we transcribed in vitro RNA representing the full ORF for WT zebrafish REP1 for injection into embryos that were derived from heterozygous chm parents. As assessed at 5 dpf by counting the number of neuromasts labeled with 4-Di-2-ASP on one side of each injected animal, ≈50% of the injected mutant larvae displayed a fully rescued lateral-line organ phenotype (Fig. 6 A and B). An occasional animal showed a unilaterally rescued phenotype resulting from the localization of the injected RNA in only one cell at the two-cell stage, and thus to only one side of the larva. About 25% of the injected mutant larvae showed no signs of phenotypic rescue, probably because they did not receive a full dose of RNA owing to variability in the injection procedure.
Fig. 6.
Confirmation of the mutation in chm as responsible for the ru848 phenotype. (A) REP1 mRNA transcribed in vitro rescues the mutant phenotype. Dorsal views of WT (Left) and mutant (Right) larvae at 5 dpf after injection with in vitro-transcribed REP1 mRNA. The mutant larva has been partially rescued by the exogenous transcript. On the left side of the mutant fish, an almost normal number of neuromasts can be observed, but on the right side, the only labeled neuromast is that typically seen above the ear in uninjected mutant larvae. The WT larva was also injected with the exogenous transcript but showed no sign of impairment caused by overexpression of this gene. (B) A histogram of the number of labeled neuromasts at 5 dpf in mutant larvae after injection with chm mRNA (orange) documents a distribution distinct from those for uninjected WT (black) and mutant (red) controls. The injected mutant larvae display a broad distribution in neuromast number, with the majority of the mutants displaying at least a partially rescued phenotype. (C)Ina “knockdown” of REP1 translation, the morpholino (MO)-injected larva (Lower) has only one labeled neuromast above the ear and one in the posterior lateral line. The control-injected larva (Upper) has the normal complement of neuromasts. (D) In a histogram of neuromast numbers at 3 dpf, WT larvae injected with a control morpholino (MO) (gray) have a distribution in neuromast number similar to that seen in uninjected WT larvae (black). In contrast, the majority of WT larvae injected with the chm-targeted morpholino (pink) display a strikingly reduced number of labeled neuromasts, similar to that in uninjected mutant larvae (red). (Scale bars: 500μm.)
We also performed the converse experiment, injecting a modified antisense oligonucleotide, or morpholino, into WT embryos to “knock down” the translation of chm mRNA. This strategy effectively reproduced the phenotype seen in ru848 mutant larvae, as evidenced by a reduction in the number of labeled neuromasts in injected larvae at 3–4 dpf to the level observed in the mutants (Fig. 6 C and D). The number of hair cells within each neuromast was also diminished, as was the ability of injected larvae to respond to an acoustic stimulus (data not shown). At 3 dpf, the gross appearance of the animals was similar to that of ru848 mutant larvae, with small eyes and irregular iridophore pigmentation. Injection of a control morpholino produced no observable phenotype.
Discussion
We have characterized a recessive mutation in the zebrafish that causes deafness, disequilibrium, and abnormality of the lateralline organ. The mutation additionally affects the eye, where it produces retinal degeneration. Our observations indicate that the protein product of the mutated gene, REP1, is necessary for the survival and perhaps development of hair cells in the inner ear and lateral-line organ, as well as of several cell types in the retina.
Some hair cells develop, but subsequently degenerate, in chm mutants; moreover, the mutation may affect the continued production and maturation of hair cells that normally occurs throughout early development. In the maculae of mutant internal ears, a few hair cells survive longer than the majority. These may include the tether cells, the first hair cells to differentiate within the inner ear (20). The two neuromasts that contain a few hair cells in mutants at 5–6 dpf are likewise among the first neuromasts to differentiate (13, 21). These observations indicate that precocious hair cells are the most stable, perhaps because they receive the greatest allotment of normal REP1 translated from maternal mRNA.
We have identified a point mutation in the second exon of chm. Because this mutation introduces a stop codon after only 32 aa in the 666-aa protein, it is very likely to represent a null mutation. The rescue of mutant embryos by injection of WT chm RNA and the ability of a chm-targeted morpholino to phenocopy the mutation provide strong evidence that the observed mutation causes the ru848 phenotype.
REP1 is required for a crucial step in the posttranslational processing of Rab GTPases, a subset of the Ras family of small G proteins with at least 60 members in the human genome (22). Because individual Rabs participate in specific transport steps of membrane-bounded organelles and localize to different subcellular compartments, they are thought to be the primary organizers of vesicular transport and membrane compartmentalization in eukaryotic cells. REP1 binds newly synthesized Rabs in their inactive form and presents them to the enzyme Rab geranylgeranyl transferase (16), which adds geranylgeranyl groups to cysteine residues at the carboxyl termini of Rabs (23).
Some Rab escort protein function is necessary for growth in all eukaryotic cells, for it provides a basis for prenylation and targeting of Rabs, without which vesicle transport would be severely impaired. In the yeast Saccharomyces cerevisiae, for example, the homolog of REP1 (MRS6) is an essential gene (18). In its absence, Rab GTPases remain unmodified and accumulate in the cytosol (24), precluding their conversion into active, GTP-bound forms and thus preventing the recruitment of their effectors to the membrane surface. The cytoplasmic inclusions in chm mutant cells might represent accumulations of unprenylated Rabs or other proteins whose processing is interrupted by the absence of prenylated Rabs.
Mutations in human Choroideremia cause the X-linked disorder choroideremia, which is characterized by slow degeneration of the rod photoreceptors, retinal pigment epithelium, and choroidal capillary network (25, 26). Choroideremia almost always occurs without other symptoms; the rare concurrence of choroideremia and deafness probably results from the codeletion of both Choroideremia and the nearby DFN3 locus (27, 28). There has been no animal model for this disease; deletion of the homologous gene in the mouse, for example, renders heterozygous females unable to carry either heterozygous female or hemizygous male embryos to term (29). The creation of a null mutation in the zebrafish therefore provides a much-needed avenue into the investigation of this disease.
In humans, the two orthologs REP1 and REP2 are both widely expressed. Although in vitro assays have shown that both REP1 and REP2 can present Rabs to Rab GGTase for prenylation, their efficacy varies depending on the specific Rab being assayed (30). Because of the restriction of the choroideremia phenotype to the eye, it is thought that the expression of REP2 permits cell survival in other organs. We have found no evidence for a compensatory REP in the zebrafish database of ESTs, the zebrafish genome database, or the published genome of the pufferfish Fugu rubripes; however, these databases all remain incomplete. Moreover, there is no evidence in the human or zebrafish for a splice isoform that skips the second exon, resulting in translation of a modified protein. The zebrafish phenotype, however, is more specific than what might be expected in view of the general cellular requirement for prenylated Rabs. This may be explained if hair cells are especially susceptible to reduced levels of this protein, whereas other cell types are able to function for a time with a limited amount of maternally derived transcript or protein. Prenylated Rabs are known to be very stable, and a transient supply of correctly folded MRS6 is enough to maintain normal growth rates in yeast (24).
Our data suggest that one or more Rabs sensitive to the loss of REP1 are uniquely required in hair cells and particular cell types of the retina. The isolation of the chm mutant in the zebrafish provides an avenue for the identification of the proteins that mediate the effects of REP1 in these cells. The mutant also affords a novel opportunity for investigation of the pathogenesis of the human X-linked disease choroideremia.
Acknowledgments
We thank I. Muñoz-Sanjuán for providing the CS2++ vector; A. Afolalu, D. Chosid, P. Espitia, R. Fendrick, and J. McDonald for zebrafish husbandry; and Y. Asai, W. Liedtke, H. Lopez-Schier, B. M. McDermott, Jr., F. Pataky, and N. Rosenkranz for assistance with the maintenance and analysis of mutant lines. T. B. Friedman and the members of our research group provided valuable comments on the manuscript. This research was supported by National Institutes of Health Grant DC00241. A.J.H. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: BAC, bacterial artificial chromosome; dpf, days postfertilization; 4-Di-2-ASP, 4-(4-(diethylamino)styryl)-N-methylpyridinium iodide; GDI, guanine-nucleotide dissociation inhibitor; hpf, hours postfertilization; REP, Rab escort protein.
References
- 1.Hudspeth, A. J. (1989) Nature 341, 397-404. [DOI] [PubMed] [Google Scholar]
- 2.Kernan, M., Cowan, D. & Zuker, C. (1994) Neuron 12, 1195-1206. [DOI] [PubMed] [Google Scholar]
- 3.Herman, R. K. (1996) BioEssays 18, 199-206. [DOI] [PubMed] [Google Scholar]
- 4.Walker, R. G., Willingham, A. T. & Zuker, C. S. (2000) Science 287, 2229-2234. [DOI] [PubMed] [Google Scholar]
- 5.Westerfield, M. (1995) The Zebrafish Book (Univ. of Oregon Press, Eugene).
- 6.Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. (1995) Dev. Dyn. 203, 253-310. [DOI] [PubMed] [Google Scholar]
- 7.Haffter, P., Granato, M., Brand, M., Mullins, M. C., Hammerschmidt, M., Kane, D. A., Odenthal, J., van Eeden, F. J., Jiang, Y. J., Heisenberg, C. P., et al. (1996) Development (Cambridge, U.K.) 123, 1-36. [DOI] [PubMed] [Google Scholar]
- 8.Dunnebier, E. A., Segenhout, J. M., Kalicharan, D., Jongebloed, W. L., Wit, H. P. & Albers, F. W. (1995) Hear. Res. 90, 139-148. [DOI] [PubMed] [Google Scholar]
- 9.Zhang, J., Talbot, W. S. & Schier, A. F. (1998) Cell 92, 241-251. [DOI] [PubMed] [Google Scholar]
- 10.Shimoda, N., Knapik, E. W., Ziniti, J., Sim, C., Yamada, E., Kaplan, S., Jackson, D., de Sauvage, F., Jacob, H. & Fishman, M. C. (1999) Genomics 58, 219-232. [DOI] [PubMed] [Google Scholar]
- 11.Hyatt, T. M. & Ekker, S. C. (1999) Methods Cell Biol. 59, 117-126. [DOI] [PubMed] [Google Scholar]
- 12.Kimmel, C. B., Patterson, J. & Kimmel, R. O. (1974) Dev. Psychobiol. 7, 47-60. [DOI] [PubMed] [Google Scholar]
- 13.Raible, D. W. & Kruse, G. J. (2000) J. Comp. Neurol. 421, 189-198. [PubMed] [Google Scholar]
- 14.Nishikawa, S. & Sasaki, F. (1996) J. Histochem. Cytochem. 44, 733-741. [DOI] [PubMed] [Google Scholar]
- 15.Michelmore, R. W., Paran, I. & Kesseli, R. V. (1991) Proc. Natl. Acad. Sci. USA 88, 9828-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andres, D. A., Seabra, M. C., Brown, M. S., Armstrong, S. A., Smeland, T. E., Cremers, F. P. & Goldstein, J. L. (1993) Cell 73, 1091-1099. [DOI] [PubMed] [Google Scholar]
- 17.Zerial, M. & McBride, H. (2001) Nat. Rev. Mol. Cell. Biol. 2, 107-117. [DOI] [PubMed] [Google Scholar]
- 18.Waldherr, M., Ragnini, A., Schweyer, R. J. & Boguski, M. S. (1993) Nat. Genet. 3, 193-194. [DOI] [PubMed] [Google Scholar]
- 19.Kane, D. A. & Kimmel, C. B. (1993) Development (Cambridge, U.K.) 119, 447-456. [DOI] [PubMed] [Google Scholar]
- 20.Riley, B. B., Zhu, C., Janetopoulos, C. & Aufderheide, K. J. (1997) Dev. Biol. 191, 191-201. [DOI] [PubMed] [Google Scholar]
- 21.Gompel, N., Cubedo, N., Thisse, C., Thisse, B., Dambly-Chaudiere, C. & Ghysen, A. (2001) Mech. Dev. 105, 69-77. [DOI] [PubMed] [Google Scholar]
- 22.Pereira-Leal, J. B. & Seabra, M. C. (2001) J. Mol. Biol. 313, 889-901. [DOI] [PubMed] [Google Scholar]
- 23.Seabra, M. C., Goldstein, J. L., Südhof, T. C. & Brown, M. S. (1992) J. Biol. Chem. 267, 14497-14503. [PubMed] [Google Scholar]
- 24.Alory, C. & Balch, W. E. (2000) J. Cell Biol. 150, 89-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cremers, F. P., van de Pol, D. J., van Kerkhoff, L. P., Wieringa, B. & Ropers, H. H. (1990) Nature 347, 674-677. [DOI] [PubMed] [Google Scholar]
- 26.Seabra, M. C., Brown, M. S., Slaughter, C. A., Südhof, T. C. & Goldstein, J. L. (1992) Cell 70, 1049-1057. [DOI] [PubMed] [Google Scholar]
- 27.Ayazi, S. (1981) Am. J. Ophthalmol. 92, 63-69. [DOI] [PubMed] [Google Scholar]
- 28.Merry, D. E., Lesko, J. G., Sosnoski, D. M., Lewis, R. A., Lubinsky, M., Trask, B., van den Engh, G., Collins, F. S. & Nussbaum, R. L. (1989) Am. J. Hum. Genet. 45, 530-540. [PMC free article] [PubMed] [Google Scholar]
- 29.van den Hurk, J. A., Hendriks, W., van de Pol, D. J., Oerlemans, F., Jaissle, G., Ruther, K., Kohler, K., Hartmann, J., Zrenner, E., van Bokhoven, H., et al. (1997) Hum. Mol. Genet. 6, 851-858. [DOI] [PubMed] [Google Scholar]
- 30.Cremers, F. P., Armstrong, S. A., Seabra, M. C., Brown, M. S. & Goldstein, J. L. (1994) J. Biol. Chem. 269, 2111-2117. [PubMed] [Google Scholar]