Abstract
Humans perceive an immense variety of chemicals as having distinct odors. Odor perception initiates in the nose, where odorants are detected by a large family of olfactory receptors (ORs). ORs have diverse protein sequences but can be assigned to subfamilies on the basis of sequence relationships. Members of the same subfamily have related sequences and are likely to recognize structurally related odorants. To gain insight into the mechanisms underlying odor perception, we analyzed the human OR gene family. By searching the human genome database, we identified 339 intact OR genes and 297 OR pseudogenes. Determination of their genomic locations showed that OR genes are unevenly distributed among 51 different loci on 21 human chromosomes. Sequence comparisons showed that the human OR family is composed of 172 subfamilies. Types of odorant structures that may be recognized by some subfamilies were predicted by identifying subfamilies that contain ORs with known odor ligands or human homologs of such ORs. Analysis of the chromosomal locations of members of each OR subfamily revealed that most subfamilies are encoded by a single chromosomal locus. Moreover, many loci encode only one or a few subfamilies, suggesting that different parts of the genome may, to some extent, be involved in the detection of different types of odorant structural motifs.
The initial event in odor perception is the detection of odorants by olfactory (odorant) receptors (ORs), which are located on olfactory sensory neurons in the olfactory epithelium of the nose (1–4). ORs are seven-transmembrane domain G protein-coupled receptors, which are encoded by a large multigene family (1, 5). This family has been conserved during vertebrate evolution, but its estimated size varies from ≈100 genes in fish (6) to over ≈1,000 in mice (7, 8, 36).
Odor detection and coding by the OR family are combinatorial: each OR recognizes multiple odorants (9–18), but different odorants are detected, and thereby encoded, by different combinations of ORs (11). The OR family can be divided into subfamilies whose members have related protein sequences (1). Odorants detected by the same OR have related structures (9–18). In addition, ORs that belong to the same subfamily can detect odorants with similar structures (11, 14). This suggests that each subfamily may be dedicated to the detection of a particular class of odorant structures. It does not preclude the recognition of a particular class of odorants by different subfamilies, however (11). OR genes can be found at many chromosomal loci, but highly related ORs often reside at the same locus (7, 8, 19–23, 36), raising the possibility that different parts of the genome are, to some degree, involved in the recognition of different types of odorants.
Previous studies provided information on the number of human OR genes and some information on the chromosomal locations of OR genes (21, 24). However, the subfamily structure of the human OR family is unknown. Moreover, the reported composition of the human OR family was based on early unfinished versions of the human genome sequence and, in addition, precise chromosomal locations were reported for only some OR genes (21).
To obtain a fuller understanding of the human OR repertoire, we conducted a comprehensive analysis of the composition of the human OR gene family, its chromosomal organization, its subfamily structure, and the relationship between its chromosomal organization and subfamily structure. These studies make predictions regarding the functional complexity of the human OR family and the potential roles of different chromosomal loci in the perception of diverse odors.
Methods
Database Searches. Initial tblastn searches for human OR genes were performed using human genome sequences contained in the National Center for Biotechnology Information (NCBI) finished (nr) and draft (htgs) databases (build 32 data) (www.ncbi.nlm.nih.gov/BLAST). Conserved OR sequence motifs used as queries included MAYDRYVAIC transmembrane domain 3 (TM3) and its variants, MALDRYVAIC and MAFDRYVAIC, and KAFSTCASH (TM6). Seven diverse mouse ORs were also used as queries in separate tblastn searches. The short peptide sequences were used in tblastn searches until no new OR sequences were obtained. A nucleotide sequence (≈2 kb) containing each match was retrieved from the database and then translated using ORF Finder (www.ncbi.nlm.nih.gov/gorf/gorf.html) to obtain the encoded protein sequence.
A protein was considered an OR if it was encoded by a coding region of ≈1 kb and contained four OR sequence motifs (GN; MAYDRYVAIC, KAFSTCASH, and PMLNPFIY) or their variants at appropriate positions. When sequences satisfying these criteria were used as queries in blastp searches of the NCBI nr database, they invariably showed best matches to known ORs. Sequences with one or more, but not all four motifs, were used as queries in such searches and were considered to be ORs if their best matches were to known ORs. Coding sequences that contained stop codons or frameshifts were counted as pseudogenes, but extremely pseudogenized sequences and isolated gene fragments were excluded.
Sequence Alignments. Nucleotide and amino acid sequences were aligned by using clustalw 1.83 (European Molecular Biology Laboratory–European Bioinformatics Institute, Cambridge, U.K.). The alignments were visually inspected and edited as necessary. The final alignment was used to generate unrooted phylogenetic trees (clustalw 1.83). Nucleotide and protein sequence identities were determined using the distances function of the Genetics Computer Group (GCG Wisconsin Package, Accelrys, San Diego). The uncorrected distance matrix was then used to assign ORs to subfamilies in which all members of a subfamily were at least 60% identical to all other members in protein sequence. Members of the same subfamily displayed strong phylogenetic grouping. Bootstrap values were generally ≥50%.
To identify the closest human homologs of rodent ORs with known odor ligands (9–14, 16–18), the sequence of each rodent OR was used as a query to search (tblastn) the NCBI human genome sequence database (build 32).
The unrooted phylogenetic tree in Fig. 2 was prepared using the protein sequences of the 339 human ORs, the 23 rodent ORs with known odor ligands, and 28 ORs from several species of fish (NCBI).
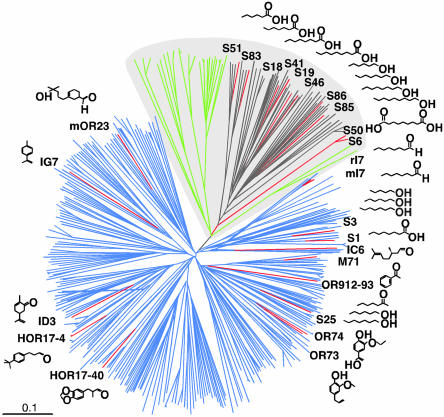
Fig. 2.
Phylogenetic tree of sequence relationships among ORs. This tree compares the 339 members of the human OR family, 23 rodent ORs of known function, and 28 fish ORs. Green branches represent fish ORs, and red branches represent human and rodent ORs with known odorant specificities. Odorants detected are indicated near the tip of each red branch. The majority of human homologs of rodent ORs for aliphatic odorants are located in one distinct branch of the tree. This branch (shaded in gray) also contains all of the fish ORs, suggesting a distant evolutionary relationship between receptors for aliphatic odorants and fish ORs.
Chromosome Localization. To determine the physical locations of all 636 human OR genes, the coding region sequence of each gene was used to search (blastn) the assembled NCBI human genome database. Using the NCBI map viewer program (www.ncbi.nlm.nih.gov/cgi-bin/Entrez/map_search), it was possible to determine the chromosomal locations of 630 of the 636 human OR genes.
Results
Composition of the Human OR Gene Family. We first sought to identify the full repertoire of human ORs. Using diverse ORs and OR sequence motifs as query sequences, we exhaustively searched for sequences encoding ORs in the 93% of the human genome available in finished and draft NCBI databases (25) (see Methods). We then retrieved DNA sequence in the area of each match and examined the protein it encoded. OR genes have intronless coding regions of ≈1 kb, facilitating their analysis (1). Classification as an OR gene was based on the presence of conserved OR sequence motifs in the encoded protein and, in equivocal cases, on results of blastp searches of the NCBI protein database. Small fragments of OR genes, including those that might be pieces of highly pseudogenized OR genes, were not included in our analysis. To exclude allelic variants, the chromosomal locations of pairs of genes with ≥99% sequence identity were compared using the NCBI assembled human genome sequence database.
These studies identified 636 human OR genes, 339 of which have open reading frames of ≈1 kb that encode full length ORs. Sequence identity among the 339 intact OR genes is 34–99%. To assess the efficacy of our search methods, we asked whether human OR genes that were in the OR database (http://senselab.med.yale.edu/senselab/ORDB) and had been identified by other methods (e.g., PCR) were included among those we had identified. Ninety-seven percent (41/42) of the genes tested were included in our set, indicating that our search strategy was highly efficient. Given that the human genome assembly was ≈93% complete at the time of our searches, the 339 intact OR genes we identified are likely to represent nearly the full human OR repertoire.
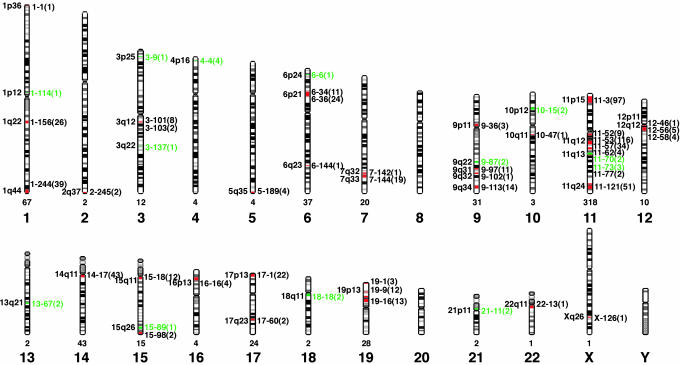
Chromosomal Organization of Human OR Genes. We next determined the chromosomal location of each intact OR gene and OR pseudogene by examining the chromosomal assignment of the clone in which it was found. In these studies, we identified 51 different OR gene loci, which are distributed among 21 different chromosomes (Fig. 1, Table 1). The only chromosomes that appear to lack OR genes are chromosomes 8 and 20 and the Y chromosome.
Fig. 1.
Chromosome locations of human OR genes. Six hundred thirty OR genes were localized to 51 different chromosomal loci distributed over 21 human chromosomes. OR loci containing one or more intact OR genes are indicated in red; loci containing only pseudogenes are indicated in green. The cytogenetic position of each locus is shown on the left, and its distance in megabases from the tip of the small arm of the chromosome is shown on the right (chromosome-Mb). The number of OR genes at each locus is indicated in parentheses, and the number of OR genes on each chromosome is indicated below. Most human homologs of rodent ORs for n-aliphatic odorants are found at a single locus, chromosome 11p15.
Table 1. Composition of individual OR gene loci.
| Cytogenetic position | Chromosomal locus* | No. of intact genes | No. of pseudogenes | Percent pseudogenes | No. of subfamilies |
|---|---|---|---|---|---|
| 1p36.33 | 1-1 | 1 | 0 | 0 | 1 |
| 1p12 | 1-114 | 0 | 1 | 100 | 0 |
| 1q22 | 1-156 | 15 | 11 | 42 | 9 |
| 1q44 | 1-244 | 29 | 10 | 26 | 17 |
| 2q37.3 | 2-245 | 2 | 0 | 0 | 1 |
| 3p25.3 | 3-9 | 0 | 1 | 100 | 0 |
| 3q12.2 | 3-101 | 2 | 6 | 75 | 1 |
| 3q12.3 | 3-103 | 2 | 0 | 0 | 1 |
| 3q22.1 | 3-137 | 0 | 1 | 100 | 0 |
| 4p16.2 | 4-4 | 0 | 4 | 100 | 0 |
| 5q35.3 | 5-189 | 2 | 2 | 50 | 2 |
| 6p24.3 | 6-6 | 0 | 1 | 100 | 0 |
| 6p21.31 | 6-34 | 4 | 7 | 64 | 3 |
| 6p21.31 | 6-36 | 13 | 11 | 46 | 10 |
| 6q23.1 | 6-144 | 1 | 0 | 0 | 1 |
| 7q32.3 | 7-142 | 1 | 0 | 0 | 1 |
| 7q33 | 7-144 | 12 | 7 | 37 | 5 |
| 9p11.2 | 9-36 | 2 | 1 | 33 | 2 |
| 9q22.32 | 9-87 | 0 | 2 | 100 | 0 |
| 9q31.2 | 9-97 | 7 | 4 | 36 | 3 |
| 9q32 | 9-102 | 1 | 0 | 0 | 1 |
| 9q34.11 | 9-113 | 13 | 1 | 7 | 7 |
| 10p12.33 | 10-15 | 0 | 2 | 100 | 0 |
| 10q11.23 | 10-47 | 1 | 0 | 0 | 1 |
| 11p15.4 | 11-3 | 56 | 41 | 42 | 35 |
| 11q12.1 | 11-52 | 6 | 3 | 33 | 4 |
| 11q12.1/.3 | 11-53 | 49 | 67 | 58 | 23 |
| 11q12.2/.3 | 11-57 | 17 | 17 | 50 | 10 |
| 11q13.1 | 11-62 | 2 | 2 | 50 | 1 |
| 11q13.3 | 11-70 | 0 | 2 | 100 | 0 |
| 11q13.3 | 11-73 | 0 | 3 | 100 | 0 |
| 11q13.5 | 11-77 | 1 | 1 | 50 | 1 |
| 11q24.1/2 | 11-121 | 24 | 27 | 53 | 10 |
| 12p11.1 | 12-46 | 1 | 0 | 0 | 1 |
| 12q12 | 12-56 | 2 | 3 | 60 | 2 |
| 12q12 | 12-58 | 3 | 1 | 25 | 2 |
| 13q21.31 | 13-67 | 0 | 2 | 100 | 0 |
| 14q11.1 | 14-17 | 23 | 20 | 47 | 15 |
| 15q11.1 | 15-18 | 4 | 8 | 67 | 2 |
| 15q26.1 | 15-89 | 0 | 1 | 100 | 0 |
| 15q26.3 | 15-98 | 2 | 0 | 0 | 1 |
| 16p13.11 | 16-16 | 3 | 1 | 25 | 2 |
| 17p13.3 | 17-1 | 13 | 9 | 41 | 5 |
| 17q23.3 | 17-60 | 1 | 1 | 50 | 1 |
| 18q11.1 | 18-18 | 0 | 2 | 100 | 0 |
| 19p13.3 | 19-1 | 1 | 2 | 67 | 1 |
| 19p13.2 | 19-9 | 8 | 4 | 33 | 4 |
| 19p13.13 | 19-16 | 11 | 2 | 15 | 2 |
| 21p11.1 | 21-11 | 0 | 2 | 100 | 0 |
| 22q11.1 | 22-13 | 1 | 0 | 0 | 1 |
| Xq26.1 | X-126 | 1 | 0 | 0 | 1 |
Locus designation indicates chromosome-Mb from tip of small arm of chromosome
There is wide variation in the number of OR genes at individual OR gene loci (1–116 OR genes) and on different chromosomes (0–318 OR genes) (Fig. 1, Table 1). The percentage of OR genes that are pseudogenes also varies among loci (0–100%) (Table 1). Of the 51 OR gene loci, 13 have only pseudogene(s), 38 have at least one intact OR gene, and 27 have more than one intact OR gene. Thus, 38 loci are potentially functional.
Consistent with previous reports of OR genes at many subtelomeric and pericentromeric locations (20), subtelomeric and pericentromeric loci account, respectively, for 12/51 and 14/51 of the OR gene loci and 8/38 and 11/38 of loci with at least one intact OR gene. The ratio of intact OR genes to OR pseudogenes at these loci does not differ significantly from other OR loci, arguing against the idea that genes at subtelomeric and pericentromeric loci may be more susceptible to mutation and pseudogenization (26).
Subfamily Structure of the Human OR Family. We next analyzed the subfamily structure of the human OR family. We used the criterion that members of the same subfamily are ≥60% identical in amino acid sequence (27), because ORs with 60% or more sequence identity have been found to recognize structurally related odorants (refs. 11 and 14; K. Nara, P.A.G., and L.B.B., unpublished work).
These studies showed that the human OR family can be divided into 172 subfamilies (see Table 5, which is published as supporting information on the PNAS web site). The large number of subfamilies emphasizes the extensive variability of ORs and is consistent with the ability of the OR family to interact with odorous chemicals with diverse structures. The sizes of human OR subfamilies ranges from one to nine ORs (Table 2). Of the 172 OR subfamilies, 94 (55%) contain only one OR. Of the remaining subfamilies, 73 have two to six members, and five have eight to nine members. Thus OR subfamilies can vary up to 9-fold in size.
Table 2. Size distribution of human OR subfamilies.
| Number ORs | Number subfamilies* |
|---|---|
| 1 | 94 |
| 2 | 40 |
| 3 | 18 |
| 4 | 8 |
| 5 | 4 |
| 6 | 3 |
| 7 | 0 |
| 8 | 4 |
| 9 | 1 |
Number of subfamilies that contain number of ORs on left
OR Gene Loci and OR Subfamilies. To investigate possible associations between the subfamily structure of the human OR family and the chromosomal organization of OR genes, we determined the chromosomal locations of genes encoding members of each of the 172 human OR subfamilies (Table 5).
These studies showed that the majority of subfamilies are encoded by genes at a single chromosomal locus (Table 3). Of the 78 subfamilies with more than one member, 79% (62/78) are encoded by genes at one locus. An additional 8% (six subfamilies) are encoded by genes at adjacent loci. Only 10/78 subfamilies (13%) are encoded by genes located on different chromosomes or at widely separated loci on the same chromosome. This organization highlights the important role of local gene duplication and divergence in the evolution of the OR gene family. As a result of these processes, different chromosomal loci encode different subfamilies of ORs and might therefore be involved in the perception of different odors.
Table 3. Chromosomal distribution of genes encoding individual OR subfamilies.
| Intact OR genes per locus*
|
Loci per subfamily†
|
Subfamilies per locus‡
|
|||
|---|---|---|---|---|---|
| No. of OR genes | No. of loci | No. of subfamilies | No. of loci | No. of subfamilies | No. of loci |
| 0 | 13 | 156 | 1 | 1 | 16 |
| 1 | 11 | 14 | 2 | 2 | 7 |
| 2 | 8 | 2 | 3 | 3 | 2 |
| 3-8 | 7 | 4 | 2 | ||
| 11-17 | 7 | 5 | 2 | ||
| 23 | 1 | 6 | 0 | ||
| 24 | 1 | 7 | 1 | ||
| 29 | 1 | 9-17 | 6 | ||
| 49 | 1 | 23 | 1 | ||
| 56 | 1 | 35 | 1 | ||
Number of OR gene loci with 0-56 intact OR genes
Number of subfamilies whose members are encoded at 1-3 loci
Number of loci that encode members of 1-35 subfamilies
We next determined the subfamily composition of individual OR gene loci (Table 3). These studies showed that 42% (16/38) of the OR loci with one or more intact OR genes encode only one OR subfamily, including 11 loci encoding only one OR. An additional 29% of these loci (11/38) encode members of two to four subfamilies; the remaining 29% code for ORs that belong to 5–35 subfamilies. Thus, the majority of OR loci encode only one or a few subfamilies. Moreover, each locus is unique in its subfamily composition.
Potential Functions for OR Subfamilies and Gene Loci. To explore the potential roles of individual OR subfamilies and OR gene loci in odor perception, we turned to ORs with known odorant specificities. Ligands have been reported for a total of two human ORs (13, 18) and 23 rodent ORs (9–12, 14, 16, 17). This set includes ORs that recognize odorants with diverse structures as well as a variety of perceived odors, including, sweet, floral, herbal, woody, rancid, and sweaty. Fifteen of the rodent ORs detect n-aliphatic odorants, but the aliphatic odorants they recognize vary in carbon chain length, functional group, and perceived odor.
In initial studies, we searched for the closest human homolog of each rodent OR of known function by using the rodent OR as a search query, first against the set of 339 human ORs we had identified and then against the translated human genome sequence database. For two rodent ORs, there was no significant match and for another the closest match was the product of a pseudogene, but for the other 19 rodent ORs, the closest homolog was a human OR that was 62–87% identical to the rodent OR (Table 4). It is possible that the human and mouse homologs are true orthologs, but the complexity of the OR family does not permit this determination. However, these interspecies homologs are as highly related as members of the same subfamily and are therefore likely to have a similar functional relationship (28). In other words, human and mouse ORs with 70% identity are as likely to recognize the same type of odorants as are two mouse ORs with 70% identity.
Table 4. Potential associations between OR gene loci and odorant recognition.
| Locus | OR (MOR#)* | Amino acid identity, % | Odorant(s) recognized | Perceived odor |
|---|---|---|---|---|
| 1q22 | OR23(267-13) | 87 | Lyral | Lemony, green |
| 5q35.3 | IG7(276-1) | 83 | Limonene | Lemon |
| 9q34.11 | ID3(136-6) | 71 | I-carvone | Spearmint, caraway |
| 11q12.1 | OR73(174-9)† | 82 | Eugenol | Spicy |
| 11q12.1 | OR74(174-4)† | 76 | Ethyl vanillin | Vanilla |
| 11q12.2 | OR912-93(175-1) | 66 | 2-Heptanone | Fruity |
| 11p15.4 | S25(204-32) | 74 | n-aliphatic alcohols | Herbal, woody, orange, rose |
| 11p15.4 | S46(32-4) | 69 | n-aliphatic acids | Rancid, sour, sweaty, fatty |
| 11p15.4 | S85(13-6) | 60 | n-aliphatic acids | As above |
| 11p15.4 | S86(8-2) | 67 | n-aliphatic acids | As above |
| 11p15.4 | S18(31-2) | 70 | n-aliphatic acids/alcohols | As above |
| 11p15.4 | S19(33-1) | 62 | n-aliphatic acids/alcohols | As above |
| 11p15.4 | S41(22-2) | 81 | n-aliphatic acids/alcohols | As above |
| 11p15.4 | S51(40-1) | 83 | n-aliphatic acids/alcohols | As above |
| 11p15.4 | S83(40-4) | 81 | n-aliphatic acids/alcohols | As above |
| 11p15.4 | 17(103-15) | 87 | n-aliphatic aldehydes | Fatty |
| 11q24.2 | M71(171-2) | 62 | Heptanol | Violet/woody |
| 14q11.1 | S3(106-13P)† | 81 | n-aliphatic alcohols | Herbal, woody, orange, rose |
| 14q11.1 | S1(106-1)† | 87 | n-aliphatic acids | Rancid, sour, sweaty, fatty |
| 17p13.3 | hOR17-4 | 87 | Bourgeonal | Lily of the valley |
| 17p13.3 | hOR17-40 | 81 | Helional | Sweet, hay-like |
Mouse OR designation according to ref 7
OR73 and OR74 are in the same subfamily as are S1 and S3
Fig. 2 shows a phylogenetic tree of sequence relationships among the 339 human ORs, the 23 rodent ORs with known ligands (20 with close human homologs and three without), and 28 ORs identified in fish. Strikingly, most (10/15) rodent ORs for aliphatic odorants are located in one distinct branch of the tree. Consistent with previous reports (27, 29, 30), this branch also contains all of the fish ORs. The mammalian ORs in this branch, including the receptors for aliphatic odorants, may therefore have a distant evolutionary relationship to fish ORs. Remarkably, the 46 human ORs in this branch are all encoded at the same locus, chromosome 11p15.4.
We next examined the chromosomal locations of genes encoding the two human ORs with known odor specificities and the closest human homologs of rodent ORs with known odor ligands (Table 4). The human OR that recognizes helional, an aromatic methylketone methylether with a sweet hay-like odor, is located at chromosome 17p13.3. Genes encoding the closest human homologs of rodent receptors for lyral, an alkyl-substituted cyclohexane aldehyde; limonene, an aromatic hydrocarbon; and carvone, a substituted cyclohexone, are each found at a different chromosomal locus (chromosomes 1q22, 5q35.3, and 9q34.11, respectively). In contrast, genes encoding 10 of the 12 homologs of rodent ORs for n-aliphatic odorants are found at the same locus (chromosome 11p15.4). These results add further support to the idea that different loci are, at least to some extent, involved in the detection of different classes of odorants.
Of the 10 OR genes at chromosome 11p15.4 that encode homologs of rodent ORs for n-aliphatic odorants, eight are clustered in a distal 2.0-Mb segment of the locus, whereas two are located more proximally. Altogether, the distal locus contains 56 intact OR genes, which belong to 35 different subfamilies. Genes encoding homologs of rodent ORs for aliphatic odorants belong to eight of these subfamilies, which together have 30 members. Surprisingly, although the 43 genes in the distal segment of this locus code for ORs belonging to 29 different subfamilies, those ORs are all located in the same branch of the phylogenetic tree described above (Fig. 2). In addition, the 43 ORs all have charged residues at specific positions in transmembrane domains 4 and 5 (data not shown), an unusual feature shared by many rodent ORs for aliphatic odorants (11) as well as many fish ORs. These findings suggest that many, perhaps even all, of the ORs encoded at chromosome 11p15.4 may recognize n-aliphatic odorants.
Discussion
In these studies, we defined the full repertoire of OR genes in the NCBI human genome database and determined the chromosomal location of each OR gene. We then analyzed the subfamily structure of the human OR family, the chromosome locations of genes encoding members of each subfamily, and the subfamily composition of each chromosomal locus that contains intact OR genes. Finally, we used information on mammalian ORs with known odorant specificities to explore potential relationships between odor detection and OR subfamilies and gene loci.
These studies indicate that humans have 636 OR genes, 339 of which are intact and therefore likely to encode functional odorant receptors in the nose. Similarly, studies of earlier less complete versions of the human genome sequence identified 322 and 347 intact OR genes, respectively (21, 24). Because the human genome sequence was 93% complete at the time of our analyses, our results argue against the proposal that humans could have up to 1,000 different OR genes (21).
Our studies localized human OR genes to 51 different loci on 21 human chromosomes. They also defined the number of intact OR genes and pseudogenes at each locus. The results indicate that 38 chromosomal loci have one or more intact OR genes and are therefore likely to function in odor perception. In defining the megabase coordinates and compositions of each OR locus, these studies significantly extend the findings of previous studies, which gave an overview of the distribution of human OR genes, but provided precise locations for only some OR loci (8, 20, 21, 26, 27, 29–35).
Analysis of sequence relationships among human ORs showed that the human OR family is composed of 172 subfamilies whose members are 60% or more identical in protein sequence. The definition of subfamilies used here was based on observations that ORs that are ≥60% identical can recognize odorants with related structures (refs. 11 and 14; K. K. Nara, P.A.G., and L.B.B., unpublished work). The subfamilies defined by this criterion are therefore likely to be more relevant to a functional understanding of the OR family than are “families” of ORs defined by a 40% identity cutoff and/or by phylogenetic clustering (24, 27).
The identification of 172 human OR subfamilies emphasizes the extreme diversity of the human OR family. Current information on odorants detected by individual ORs suggests a model in which each subfamily recognizes a particular class of odorant structures or structural features (11, 14). In this model, members of the same subfamily would recognize partially overlapping sets of odorants, thereby allowing for the fine discrimination of odorants with highly related structures. Although some odorants could conceivably be detected by ORs in only a single subfamily, previous studies indicate that at least some odorants are recognized by ORs that are <60% identical and therefore belong to different subfamilies (11).
The delineation of OR subfamilies provides a template to investigate the functional organization of the OR repertoire. The present studies make an initial effort in this direction by identifying subfamilies that contain human ORs with known odor ligands or the closest human homologs of rodent ORs with defined odorant specificities. This analysis predicts types of odorant structures that might be detected by 20 different human OR subfamilies and their 54 members, 15% of the total repertoire. It should be noted, however, that further studies are needed to verify the reliability of this prediction method and to determine the extent to which individual ORs might interact with odorants with different structures.
These studies showed that the vast majority of OR subfamilies are encoded by genes at a single locus. Moreover, 42% of functional OR loci encode only one OR subfamily, and an additional 29% encode only two to four subfamilies. These findings suggest that different chromosomal loci may, to some extent, be dedicated to the recognition of different types of odorant structural features. An odorant detected by a single subfamily might involve a single locus, whereas an odorant detected by a combination of subfamilies might involve a combination of loci.
Although an OR gene locus might be specialized for the recognition of odorants with particular types of structures, the locus need not be correlated with a specific class of perceived odors. As a group, the rodent homologs of ORs encoded at one locus, chromosome 11p15.4 detect n-aliphatic odorants with similar carbon chains but varied functional groups. Humans perceive these odorants as having odors as different as rancid vs. orange. In the case that all members of an odorant structural class have related odors, however, the hypothetical OR gene locus involved might be associated not only with the recognition of a given class of odorant structure but also with a particular type of odor, such as minty or sweet.
Note Added in Proof. As of January 2004, the following genes are no longer present in the human genome sequence database: hOR1-51, hOR11-10, hOR11-163, hOR12-NP1, and hOR15-4.
Supplementary Material
Acknowledgments
We thank Xiaolan Ye and Manjula Subramanian for help with sequence analyses and other members of the Buck laboratory for helpful suggestions. This project was supported by the Howard Hughes Medical Institute and by grants from the National Institutes of Health (National Institute on Deafness and Other Communication Disorders), the Department of Defense (Army Research Office), and Fundação de Amparo à Pesquisa do Estado de São Paulo (B.M.).
Abbreviations: OR, olfactory receptor; NCBI, National Center for Biotechnology Information.
Data deposition: The sequence, reported in this paper have been deposited in the GenBank database (accession no. BK004190–BK004820).
References
- 1.Buck, L. & Axel, R. (1991) Cell 65, 175-187. [DOI] [PubMed] [Google Scholar]
- 2.Mombaerts, P. (1999) Annu. Rev. Neurosci. 22, 487-509. [DOI] [PubMed] [Google Scholar]
- 3.Buck, L. B. (2000) Cell 100, 611-618. [DOI] [PubMed] [Google Scholar]
- 4.Firestein, S. (2001) Nature 413, 211-218. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts, P. (1999) Science 286, 707-711. [DOI] [PubMed] [Google Scholar]
- 6.Ngai, J., Dowling, M. M., Buck, L., Axel, R. & Chess, A. (1993) Cell 72, 657-666. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, X. & Firestein, S. (2002) Nat. Neurosci. 5, 124-133. [DOI] [PubMed] [Google Scholar]
- 8.Young, J. M., Friedman, C., Williams, E. M., Ross, J. A., Tonnes-Priddy, L. & Trask, B. J. (2002) Hum. Mol. Genet. 11, 535-546. [DOI] [PubMed] [Google Scholar]
- 9.Zhao, H., Ivic, L., Otaki, J. M., Hashimoto, M., Mikoshiba, K. & Firestein, S. (1998) Science 279, 237-242. [DOI] [PubMed] [Google Scholar]
- 10.Krautwurst, D., Yau, K. W. & Reed, R. R. (1998) Cell 95, 917-926. [DOI] [PubMed] [Google Scholar]
- 11.Malnic, B., Hirono, J., Sato, T. & Buck, L. B. (1999) Cell 96, 713-723. [DOI] [PubMed] [Google Scholar]
- 12.Touhara, K., Sengoku, S., Inaki, K., Tsuboi, A., Hirono, J., Sato, T., Sakano, H. & Haga, T. (1999) Proc. Natl. Acad. Sci. USA 96, 4040-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wetzel, C. H., Oles, M., Wellerdieck, C., Kuczkowiak, M., Gisselmann, G. & Hatt, H. (1999) J. Neurosci. 19, 7426-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kajiya, K., Inaki, K., Tanaka, M., Haga, T., Kataoka, H. & Touhara, K. (2001) J. Neurosci. 21, 6018-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araneda, R. C., Kini, A. D. & Firestein, S. (2000) Nat. Neurosci. 3, 1248-1255. [DOI] [PubMed] [Google Scholar]
- 16.Gaillard, I., Rouquier, S., Pin, J. P., Mollard, P., Richard, S., Barnabe, C., Demaille, J. & Giorgi, D. (2002) Eur. J. Neurosci. 15, 409-418. [DOI] [PubMed] [Google Scholar]
- 17.Bozza, T., Feinstein, P., Zheng, C. & Mombaerts, P. (2002) J. Neurosci. 22, 3033-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spehr, M., Gisselmann, G., Poplawski, A., Riffell, J. A., Wetzel, C. H., Zimmer, R. K. & Hatt, H. (2003) Science 299, 2054-2058. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan, S. L., Adamson, M. C., Ressler, K. J., Kozak, C. A. & Buck, L. B. (1996) Proc. Natl. Acad. Sci. USA 93, 884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouquier, S., Taviaux, S., Trask, B. J., Brand-Arpon, V., van den Engh, G., Demaille, J. & Giorgi, D. (1998) Nat. Genet. 18, 243-250. [DOI] [PubMed] [Google Scholar]
- 21.Glusman, G., Yanai, I., Rubin, I. & Lancet, D. (2001) Genome Res. 11, 685-702. [DOI] [PubMed] [Google Scholar]
- 22.Strotmann, J., Hoppe, R., Conzelmann, S., Feinstein, P., Mombaerts, P. & Breer, H. (1999) Gene 236, 281-291. [DOI] [PubMed] [Google Scholar]
- 23.Xie, S. Y., Feinstein, P. & Mombaerts, P. (2000) Mamm. Genome 11, 1070-1078. [DOI] [PubMed] [Google Scholar]
- 24.Zozulya, S., Echeverri, F. & Nguyen, T. (2001) Genome Biol. 2, RESEARCH0018. [DOI] [PMC free article] [PubMed]
- 25.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 26.Trask, B. J., Friedman, C., Martin-Gallardo, A., Rowen, L., Akinbami, C., Blankenship, J., Collins, C., Giorgi, D., Iadonato, S., Johnson, F., et al. (1998) Hum. Mol. Genet. 7, 13-26. [DOI] [PubMed] [Google Scholar]
- 27.Glusman, G., Bahar, A., Sharon, D., Pilpel, Y., White, J. & Lancet, D. (2000) Mamm. Genome 11, 1016-1023. [DOI] [PubMed] [Google Scholar]
- 28.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- 29.Bulger, M., van Doorninck, J. H., Saitoh, N., Telling, A., Farrell, C., Bender, M. A., Felsenfeld, G., Axel, R., Groudine, M. & von Doorninck, J. H. (1999) Proc. Natl. Acad. Sci. USA 96, 5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulger, M., Bender, M. A., van Doorninck, J. H., Wertman, B., Farrell, C. M., Felsenfeld, G., Groudine, M. & Hardison, R. (2000) Proc. Natl. Acad. Sci. USA 97, 14560-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brand-Arpon, V., Rouquier, S., Massa, H., de Jong, P. J., Ferraz, C., Ioannou, P. A., Demaille, J. G., Trask, B. J. & Giorgi, D. (1999) Genomics 56, 98-110. [DOI] [PubMed] [Google Scholar]
- 32.Trask, B. J., Massa, H., Brand-Arpon, V., Chan, K., Friedman, C., Nguyen, O. T., Eichler, E., van den Engh, G., Rouquier, S., Shizuya, H., et al. (1998) Hum. Mol. Genet. 7, 2007-2020. [DOI] [PubMed] [Google Scholar]
- 33.Buettner, J. A., Glusman, G., Ben-Arie, N., Ramos, P., Lancet, D. & Evans, G. A. (1998) Genomics 53, 56-68. [DOI] [PubMed] [Google Scholar]
- 34.Glusman, G., Clifton, S., Roe, B. & Lancet, D. (1996) Genomics 37, 147-160. [DOI] [PubMed] [Google Scholar]
- 35.Sosinsky, A., Glusman, G. & Lancet, D. (2000) Genomics 70, 49-61. [DOI] [PubMed] [Google Scholar]
- 36.Godfrey, P. A., Malnic, B. & Buck, L. B. (2004) Proc. Natl. Acad. Sci. USA 101, 2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.