Abstract
We present a simple and robust method to evaluate the transfection efficiency of commercially available transfection reagents intended to be established for use in nonmammalian cell lines. To illustrate the method, we compare the ability of four different reagents to transfect the embryonic zebrafish cell line Z3. Z3 cells were seeded in a 96-well plate and simultaneously transfected in several variations by using minimum volumes of transfection reagent and a vector DNA encoding an amplified version of green fluorescent protein (GFP). After 24 and 48 h, transfection efficiency was determined by a dual fluorescence plate reader measurement of GFP and Hoechst 33342 fluorescence, an indicator of cell density. Of the four different reagents tested, certain variations of JetPrime™ reagent and X-tremeGene™ HP reagent produced the highest fluorescence signal per cell after 24- and 48-h incubation, respectively. The simultaneous multivariate setup enables comparing different reagent/DNA combinations at different time points well, independent of cell growth variability or seeding density.
Key words: Hoechst 33342, maxGFP, transfection, zebrafish cell culture, Z3
Introduction
For the last three decades, the use of zebrafish (Danio rerio) as an important vertebrate model system has advanced our knowledge in many different fields of biology, such as developmental biology, genetics, and neurobiology. First and foremost, zebrafish researchers benefit from the fully sequenced genome, the fast development of the transparent embryos, and the comparison of mutants to well-characterized wild-type individuals. With zebrafish rising in popularity within the scientific community, the development of zebrafish cell lines, like the fibroblast-like ZF41 and PAC22 cell lines, pluripotent embryonic stem cell lines,3 or organ-specific cell lines, like the spleen cell line ZSSJ,4 commenced. Accordingly, in recent years an increasing number of studies have included zebrafish cell lines as tool to complete the explanation of organismic events; for example, by the detailed resolution of gene regulation and signaling processes in vitro5,6 or by implantation of cells back into the embryo.7 Still, the overall numbers of available zebrafish cell lines and tissue-specific zebrafish cell lines are low compared with cell lines from longer established vertebrate model organisms (e.g., rodents). So far, most of the zebrafish cell lines used by the scientific community originate from embryonic tissue and feature fibroblast-like morphology.8
With the use of zebrafish cell cultures, the need to stably or transiently introduce genetic constructs became evident. Besides emerging protocols for adenoviral transfection methods in zebrafish cell cultures,9 protocols for the use of nonviral transfection methods, now an essential cell biology tool, became available and proved the principal usability of zebrafish cell lines as transfection hosts.1,8,10 However, it is believed that fish cell cultures in general are very sensitive to mammalian transfection reagents,7 and to our knowledge, all of the major commercially available nonviral transfection reagents are optimized for mammalian cell cultures, whereas only few address specific difficulties or provide protocols when transfecting zebrafish cells cultured at near room temperature. Also, the scientific community is only beginning to evaluate certain transfection methods for their suitability in primary zebrafish cells and zebrafish cell lines.10–13 As a result, wanting to work with transfected zebrafish cells often means accepting weak transfection efficiencies or includes a strenuous and time-consuming optimization process. Here we provide a relatively simple method to measure transfection efficiency within the optimization ranges of vector DNA and reagent amount proposed by individual manufacturers. With our proposed multivariate 96-well plate setup, it is possible to compare several different transfection products simultaneously and thereby reduce result variability.
For this study, we optimized transfection conditions for the Z3 zebrafish cell line. The Z3 cell line was first established in 2001 by Pando and colleagues14 from embryonic zebrafish tissue. These adherent fibroblast-like cells grow at 26°C with atmospheric CO2 levels, are cultured in conventional media and serum, have a population doubling time of less than 24 h, and are, like PAC2 cells, light entrainable.
We used a commercially available vector holding the gene for an advanced version of enhanced green fluorescent protein (GFP) called maxGFP under the control of the cytomegalovirus (CMV) promoter (pmaxGFP vector, Amaxa) as an indicator for successful transfection. According to the manufacturer, maxGFP has an increased fluorescence quantum yield compared to conventional enhanced GFP and therefore may be used as sensitive positive control vector, while providing similar transfection efficiencies as well as cell mortalities. To be able to correlate maxGFP fluorescence signal to cell density, we established a calibration curve for Z3 cells stained with the nuclear dye Hoechst 33342 (Invitrogen). Hoechst 33342 is a cell-permeable live nuclear cell stain that binds to the minor groove of double-stranded DNA; it does not fluoresce until it is bound to DNA.
Out of the numerous transfection reagents available today from many of the major biochemical suppliers, we chose four nonviral transfection reagents which can be used in serum-containing media. The first one, Lipofectamine™ LTX (Invitrogen) transfection reagent, is based on the lipofection method, in which the negatively charged vector DNA is trapped in a cationic lipid vacuole, the liposome, formed by the reagent. The liposome fuses with the cell membrane and thereby releases vector DNA into the cell.15,16 According to its manufacturer, Lipofectamine reagents have become the most referenced transfection reagents since their launch in 1993. They are therefore considered the “gold-standard” of transfection reagents and are used as a basis of comparison for efficiencies of other transfection methods.11
The second reagent we tested was X-tremeGene™ HP (Roche) which, according to the manufacturer, is a sterile and nonliposomal blend of lipids and other components supplied in 80% ethanol. X-tremeGene HP is designed for usage on “hard-to-transfect” cells.
Both Lipofectamine and a previous version of X-tremeGene HP, Fugene™ were already successfully used for transfecting Z3 cells,17,18 although the former one had a reportedly weak performance.
The third transfection reagent evaluated in this study was JetPrime™ (Polyplus transfection, France), which is a nonliposomal, polymer-based transfection reagent for which we were not granted detailed chemical information from the company. In preliminary tests, we also used another polymer product from the same company called JetPEI™. JetPEI is based on polyethyleneimine (PEI) which is a cationic polymer, a so-called “proton sponge,” that complexes with DNA and is subsequently introduced into the cell via endocytosis.19 PEI polymers are widely used as cheap and well-performing alternatives to lipofection methods, and it was shown in a previous publication that a higher transfection efficiency could be achieved with PEI (27%) compared to Lipofectamine (25%) in ZF4 zebrafish cells.11 However, in preliminary tests on Z3 cells and another group using epithelioma papulosum cyprini (EPC) and rainbow trout gonad (RTG2) cell lines,20 we found that the commercially available JetPEI led to low transfection efficiencies with high cytotoxic effects that, according to this latter study, may be overcome by using fractioned PEI polymers of intermediate molecular weight. Since we were not able to apply such fractioning, we used JetPrime reagent, which showed very low cytotoxic effects in preliminary tests.
Both lipofection and polymer transfections, like JetPEI and JetPrime, involve extensive membrane interaction in the form of liposome fusion or charge binding and subsequent endocytosis, respectively. Since zebrafish cells are cultured at lower temperatures compared to cells of warm-blooded mammalian hosts, we hypothesized that lower temperatures may affect the necessary membrane interactions, thus altering liposome fusion or polymer endocytosis and reducing overall transfection performance. This hypothesis is supported by observations that show fish, due to their poikilotherm nature, experience temperature-dependent changes in membrane turnover and membrane lipid composition,21 which underlie a fundamental difference from cell membranes of homeotherm organisms. Considering these temperature-related differences that may affect transfection efficiency, we chose Matra-A™ reagent (IBAfect, Germany) as the fourth transfection method since this reagent is based on DNA linked to magnetic beads, which when placed onto a magnetic plate are pulled into the cells by magnetic attraction.
While these four different transfection reagents can only represent a small fraction of available products on the market, our description of the simultaneous multivariate setup in 96-well plates may generally prove to be a valuable tool for future evaluation of transfection efficiencies when establishing newly introduced transfection reagents in rapidly expanding new types of cell lines.
Material and Methods
Cell culture
Z3 cells were a kind gift from Dr. Jun Hirayama (University of California, Irvine, CA). Cells were free of mycoplasma and were cultured in Leibovitz L15 medium supplemented with 15% fetal bovine serum, 2 mM glutamine, penicillin, streptomycin, and gentamycin (all Invitrogen) in sealed 75-cm2 flasks (Sarstedt) and were split once a week. Cells were cultured in a Peltier-cooled incubator (Memmert) at 26°C under atmospheric CO2 concentrations. For transfection experiments, cells between passages 10 and 20 were taken from the flask stock and seeded into 10-cm petri dishes for 1 week before the final seeding of 18,000 cells per well in 96-well plates (Sarstedt). At this density, cells were attached to the well bottom after 24 h at a confluency of 70%–80%.
Transfection
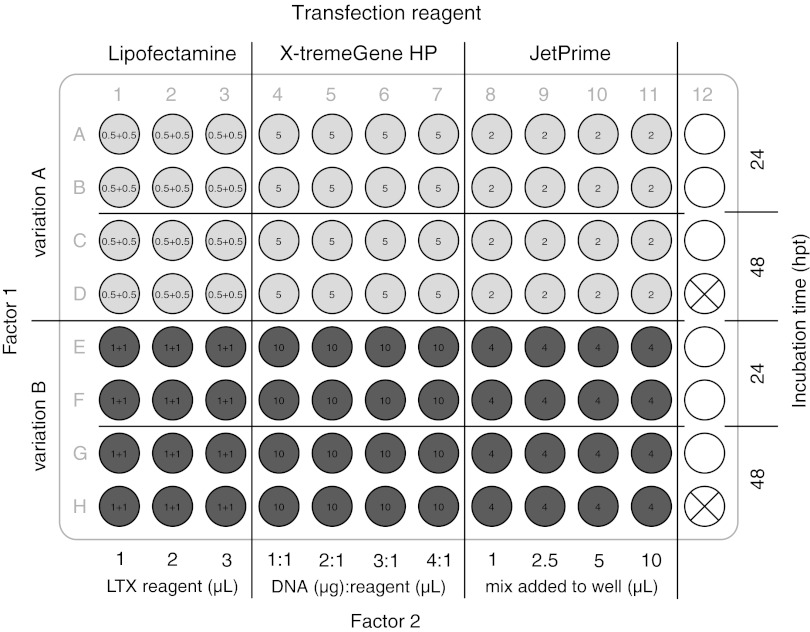
Twenty-four hours after seeding, growth medium in all wells was exchanged for 100 μL of fresh growth medium following transfection with the pmaxGFP vector (1 μg/μL, Amaxa) according to the multivariate layout (Fig. 1). Variations were chosen according to the manufacturers' manuals (Table 1). For each 24- and 48-h post transfection (hpt) incubation time point and for each individual variation, two wells were treated identically. Transfection volumes were scaled down to fit 96-well plate dimensions in accordance with the protocols provided in the manuals. The transfection procedure was performed according to the individual manuals and was planned so that time waiting on steps in one reagent's protocol were spent working on steps in another reagent's protocol.
FIG. 1.
Example for a multivariate optimization layout. Schematic layout of different transfection variations (see Table 1) used for optimizing three transfection methods in one 96-well plate simultaneously. Variations of factor 1 are represented as wells filled with light (variation A) and dark (variation B) gray color. In the case of Lipofectamine LTX, the amount of DNA (μg) and PLUS reagent (μL) varied with X-tremeGene HP, the volume of transfection mix added to the wells varied; and with JetPrime the volume of reagent (μL) used varied. Variations of factor 2 are organized in well columns. At 24 and 48 h post transfection (hpt) incubation time points are organized in double rows. White wells held nontransfected cells used as controls for the cell viability assay. The crossed-out wells represent empty wells used for holding the Hoechst staining blank. The layout for the Matra-A transfection was similarly organized on an additional 96-well plate (layout not shown).
Table 1.
Simultaneous Transfection Variations
| Reagent, manufacturer | Factors recommended to vary | Variation rangea | Variations |
|---|---|---|---|
| Lipofectamine LTX, Invitrogen | Factor 1: DNA (μg) and PLUS reagent (μL) | A: 0.5 and 0.5 | 6 |
| B: 1 and 1 | |||
| Factor 2: LTX reagent (μL) | 1, 2, 3 | ||
| X-tremeGene HP, Roche | Factor 1: transfection mix added (μL) | A: 5 | 8 |
| B: 10 | |||
| Factor 2: ratio DNA (μg):reagent (μL) | 1:1, 2:1, 3:1, 4:1 | ||
| JetPrime, Polyplus transfection | Factor 1: JetPrime reagent (μL) | A: 2 | 8 |
| B: 4 | |||
| Factor 2: transfection mix added (μL) | 1, 2.5, 5, 10 | ||
| Matra-A, IBAfect | Factor 1: media change after magnet incubation | A: yes | 6 |
| B: no | |||
| Factor 2: ratio DNA (μg):reagent (μL) | 0.5:1, 1:1, 2:1 |
In the case of Matra-A transfection, cells from the same cell solution were seeded in a separate 96-well plate. After 24 h, the cells were transfected and were then placed on a magnetic plate for 15 min according to the manufacturer's manual. One variation of this transfection method received a second washing step after the magnetic incubation with fresh growth medium to test for detrimental effects of the transfection solution.
Fluorescence measurement
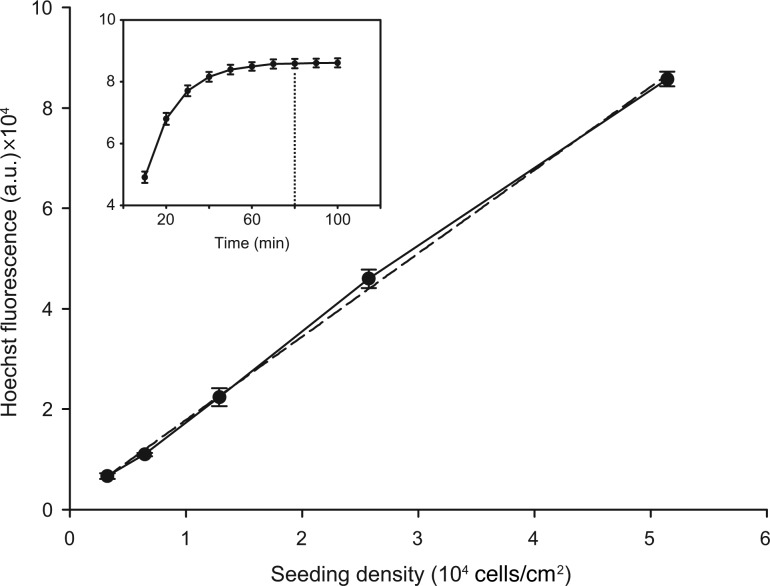
Twenty-four hours after transfection, one replicate sample was measured, whereas the other two corresponding wells of each transfection variation were replenished with 100 μL of fresh growth medium and otherwise were left untouched. To measure maxGFP fluorescence in relation to cell density, transfected cells were stained with the fluorescent nuclear dye Hoechst 33342 (Invitrogen, Germany). For this, wells holding the 24-h-old transfected cells were washed two times with Hank's buffered salt solution (HBSS, Invitrogen, Germany) containing Ca2+ and Mg2+ at pH 7.600 and were incubated for 80 min in the incubator with 100 μL of Hoechst 33342 in HBSS at a final concentration of 1 μg/mL. The incubation time for saturated Hoechst-stained Z3 cells was optimized in a preliminary test measuring Hoechst fluorescence in 10-min steps in the plate reader (Fig. 2, inset). After incubation with the nuclear stain, wells holding the 24-h cells were measured from the bottom in a fluorescence plate reader (Victor X4, Perkin Elmer) using a 355-nm excitation filter and a 460-nm emission filter for measurement of the Hoechst 33342 signal and a 485-nm excitation filter and 535-nm emission filter for measurement of the maxGFP signal. We included blanks containing only Hoechst staining solution as well as blanks of nontransfected cells stained with the Hoechst dye (untreated control group). After the measurement, medium in the wells holding the 24-hpt cells was aspirated before the 96-well plate was incubated for another 24 h. After 48 h from the time of transfection, the remaining duplicate wells holding the 48-hpt cells of each transfection variation were stained and measured identically as the cells 1 day before.
FIG. 2.
Calibration curve of Hoechst 33342-saturated zebrafish Z3 cells seeded at various cell densities. Hoechst fluorescence was recorded after 80-min incubation with the Hoechst 33342 nuclear stain (1 μg/mL) in Z3 cells which were seeded at various densities in triplicates 24 h earlier. The curve fitted dashed line has a correlation factor R2=0.994. Inset: The incubation time until Hoechst fluorescence signal becomes saturated. Fluorescence was recorded every 10 min after incubation started in triplicate wells holding 5×104 zebrafish Z3 cells/cm2. The dotted line marks the 80-min incubation time point. All values represent blank-corrected means±SEM of three independent cell culture preparations.
Transfection efficiency
Transfection efficiency was calculated as percentage of transfected cells from all cells by counting transfected cells holding a maxGFP signal as well as Hoechst-stained nuclei in exemplary images recorded 24 or 48 hpt. Images were taken on a Zeiss Axiovert 100 microscope equipped with a 20× lens, a CCD camera, and fluorescence filter sets for fluorescein isothiocyanate or 4′,6-diamidino-2-phenylindole excitation/emission, respectively.
Data analysis
Results of the Hoechst 33342 fluorescence measurement were blank-corrected with background signal from cell-free wells holding only staining solution. The blank-corrected values were transformed into cells per square centimeter using a calibration curve that was established with Hoechst-stained cells seeded at various densities (Fig. 2).
Results of the maxGFP fluorescence measurements were blank-corrected with signal from nontransfected but Hoechst-stained cells and normalized to 10,000 cells with the corresponding cell density value from the same well. Values are presented as maxGFP relative fluorescence units (rfu) per 10 cells.
Cell viability at 24 and 48 hpt was calculated from cell density numbers from each well, normalized to the mean cell density calculated from the three nontransfected control wells.
Statistics
Values reported in the figures are means±SEM. To exclude pseudoreplicates, values of the two corresponding wells from each biological replicate were combined to form one mean value. Experiments were performed in five independent cell culture preparations (e.g., biological replicates) unless stated otherwise in the figures. Differences of maxGFP rfu or cell density between different transfection variations were analyzed by two-way analysis of variance (ANOVA) with a post hoc Holm–Sidak multiple comparison, or when appropriate by paired Student's t-test. Transformation of data was applied to meet assumptions for homogeneity and normality. Statistical analyses were performed using Sigmaplot 12.0 (Systat Systems Inc.). Significant differences were accepted when p≤0.05.
Results
Hoechst cell quantification
Hoechst staining of live cells showed a clear saturation curve after 60 min (Fig. 2, inset). Therefore, we measured Hoechst fluorescence after 80 min in all following experiments and also for establishing the calibration curve. The calibration curve (Fig. 2) delivered a linear relationship between seeding density and Hoechst rfu with a correlation of R2=0.994.
Transfection and cell viability
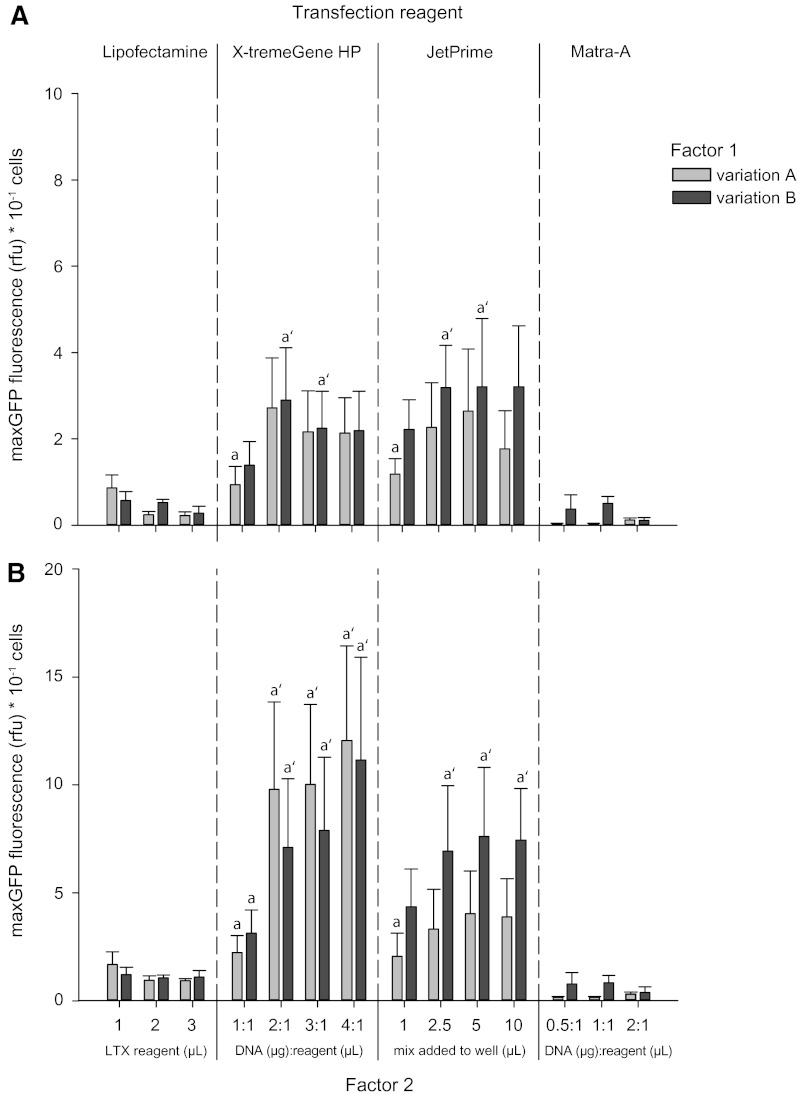
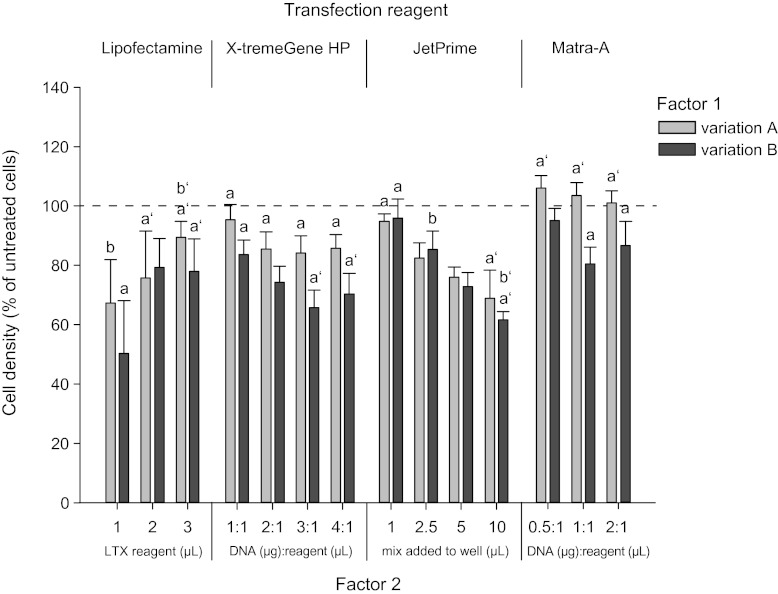
Lipofectamine LTX transfection resulted in overall weak maxGFP expression with less than 1 fluorescence unit per 10 cells after 24 hpt (Fig. 3A) and less than 2 units 48 hpt (Fig. 3B). Within the variations of this transfection we could not find a significantly stronger subgroup that could be discriminated from weaker signals. Interestingly, increasing the amount of Lipofectamine LTX reagent significantly increased cell viability (Fig. 4), whereas the lowest concentration of the compound seemed to have the most detrimental effects on these cells. In preliminary tests, we found that the application of DNA together with PLUS™ reagent, an enhancing compound supplied with the reagent kit, increased transfection success. However, increasing the amount of DNA and PLUS reagent together as suggested in the manual did not lead to better results.
FIG. 3.
Quantified transfection. Bar charts show maxGFP fluorescence normalized to cell density at 24 h (A) and 48 h (B) post transfection. For each of the four transfection reagents factors 1 and 2 were varied according to Table 1. Significant differences (p≤0.05) between variations within each transfection reagent are discerned by letters with or without a prime symbol. Values represent means±SEM of five independent cell culture preparations.
FIG. 4.
Cell viability 48 h post transfection. Bar charts represent cell densities of different transfection variations translated from Hoechst measurements 48 h post transfection and displayed as a percentage of nontransfected control cell density (dashed line). Cell viability was not significantly altered after 24 h post transfection (data not shown). Significant differences (p≤0.05) between variations within each transfection reagent are discerned by letters with or without a prime symbol. Values represent means±SEM of five independent cell culture preparations.
X-tremeGene HP transfection yielded strong maxGFP fluorescence signals after 24 h and the strongest signal of all four tested compounds 48 h post transfection. Here, the best result was achieved with the 4 to 1 ratio of micrograms of DNA to microliters of transfection reagent, resulting in a maxGFP signal as strong as 24.6 fluorescence units per 10 cells. Increasing factor 1, the volume of the transfection mix added to the cells, from 5 to 10 μL did not increase the maxGFP signal but led to significantly reduced viability when these two groups were compared in a paired t-test.
JetPrime reagent produced the strongest maxGFP fluorescence signals of all four tested compounds after 24 h with values up to 9.3 rfu per 10 cells in the variant using 4 μL of reagent (factor 1) and 5 μL transfection mix added to the cells (factor 2). However, increasing factor 2 also caused a significant decline in cell viability after 48 h. maxGFP signal after 48 h dropped below the signal observed in cells treated with X-tremeGene HP reagent.
Matra-A transfection led to the weakest maxGFP signal of all four transfection reagents tested. Data were also very heterogenous and could not be statistically analyzed because of a lack of homogeneity. Adding a media change after magnetic incubation (factor 1) to the transfection procedure did not lead to better transfection results but increased cell viability.
Transfection efficiency
Representative images of transfection variations (Fig. 5) that yielded strong maxGFP signals per cell number were used for cell counting. The strongest maxGFP plate reader signals could be translated into transfection efficiencies in the range of 25%–30%.
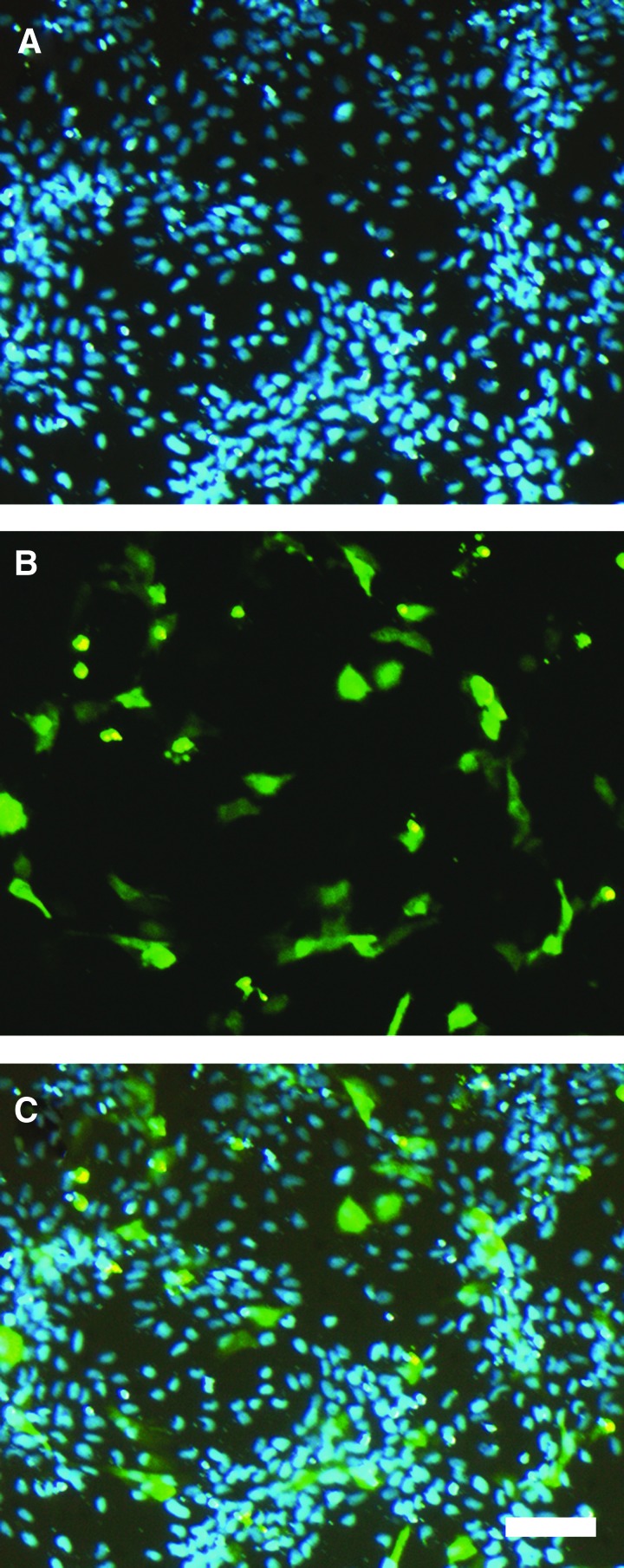
FIG. 5.
Hoechst 33342–stained nuclei and maxGFP expression recorded after 24 h in Z3 cells. Representative images of Z3 cells 24 h after transfection (18.4% efficiency) show fluorescence signals of Hoechst 33342 (A), maxGFP expression (B), and an overlay of both (C). Scale bar: 100 μm.
Discussion
The measure of transfection efficiency, the percentage of cells transfected from cells nontransfected, is a subjective measure prone to many variable factors, such as cell cycle progression, circadian rhythm of gene expression activity, promoter activity, and general activity of a given cell type. All of these factors can inhibit a cell from actually expressing the transfected protein. We found the intensity of maxGFP expression in our cells to vary highly from cell to cell in the same well at the same time point, some showing faint signal, others very strong signal. Our results also show that transfection results after 24 h can be very different from results obtained after 48 h, favoring different reagents at different time points; for example, JetPrime after 24 h and X-tremeGene HP after 48 h. Therefore, instead of presenting our results as a percentage of transfected cells, our approach was to measure overall abundance of the target protein, in our case by measuring maxGFP fluorescence per given cell number. This approach provides an objective measure of how well a reagent is able to deliver an introduced vector to be expressed in a target cell. Other publications using luciferase reporter constructs22 or β-galactosidase assays12 to compare different transfection reagents have to revert to arbitrary units per protein content instead of percentage of transfected cells as well.
However, we also saw that between biological replicates the signal level could be very different from one time point to another, while the overall signature of the response was similar, and significant responses could be extracted by multiple comparison procedures following ANOVA tests. Here, our multivariate setup in a 96-well plate may have helped to conserve similar response signatures between biological replicates by excluding many factors that can affect transfection levels in a given system (e.g., cell seeding density, cell passage number, or growth medium stability). By using the same starting conditions in each well, the comparison of effects induced by the reagent becomes more reliable and data become more robust. Variability within wells that received the same treatment over the same incubation time could be further reduced by increasing the number of replicate wells; however, this would also increase costs and preparation time as well as reduce the number of different variations present on one 96-well plate.
Hoechst 33342 staining of the live cells proved a stable and simple method to quantify adherent cells. That said, adherence of the cells is a prerequisite since the dye also stains dead cells. When working with adherent cells, dead cells can simply be removed by two washing steps prior to the staining. The incubation time until Hoechst 33342 staining reaches a stable plateau which should be separately assessed for each new cell type and incubation temperature.
While transfection protocols of most available commercial products are not optimized for them, cultured zebrafish cells do have significant advantages over mammalian cell culture, notably ambient air without the need to raise CO2 tension and culture at near–room temperature. However, if cultivation temperature of a cell line affects transfection efficiency, zebrafish cells with their higher cultivation temperature may also have advantages over other fish cell cultures. A preliminary comparison of the transfection ability of a commercially available trout gill cell line (RTgill W1) cultured at 18°C using our multivariate protocol and the same reagent variations showed that these cells had very low transfection signals after 24 and 48 h. Again, such limitations may be overcome by individual optimizations in a multivariate setup, since a study on trout liver cells (RTH-149) could show improving results with certain combinations of cationic lipid formulations.22
Of the four tested reagents, X-tremeGene HP delivered the best transfection results, which were comparable to those of other studies that used the reagent or its predecessor Fugene on Z3 embryonic fibroblasts18 or EPC carp cells.23 Compared with the other tested reagents, X-tremeGene HP also has the simplest and shortest protocol and uses vector DNA most efficiently. However, in recent publications the use of another nonviral transfection method, the Nucleofector method (Lonza), was shown to deliver extraordinary transfection efficiencies up to 43% in primary zebrafish fibroblasts13 and up to 90% in an Atlantic salmon head kidney cell line.24 However, given the comparison of these transfection efficiencies, it should be considered that the kind of cell type and whether cells are primary isolated or maintained as a cell line are major factors that influence general transfection ability itself.
Taken together, our results show that Z3 cells can be transfected well, and optimization of the protocol prior to the experiments is advisable and may increase transfection results significantly as well as identify detrimental conditions that affect cell viability in the long run. The relatively simple and simultaneous setup of various transfection conditions at low volumes in 96-well plates reduces variability, and the quick read-out of results in a fluorescence plate reader at various time points may help to identify factors which increase transfection efficiency.
Acknowledgments
We thank Martina Höckner (Institute of Zoology, University of Innsbruck) for helpful discussions, and the manufacturers for providing us with free samples of their transfection reagents as well as their technical support.
Disclosure Statement
No competing financial interests exist.
References
- 1.Driever W. Rangini Z. Characterization of a cell-line derived from zebrafish (Brachydanio rerio) embryos. In Vitro Cell Dev Biol Anim. 1993;29:749–754. doi: 10.1007/BF02631432. [DOI] [PubMed] [Google Scholar]
- 2.Culp P. Dissertation. Cambridge, MA: Massachusetts Institute of Technology; 1994. Random DNA integrations as an approach to insertional mutagenesis in the zebrafish (Brachydanio rerio) [Google Scholar]
- 3.Fan LC. Crodian J. Collodi P. Culture of embryonic stem cell lines from zebrafish. Methods Cell Biol. 2004;76:151–160. doi: 10.1016/s0091-679x(04)76009-4. [DOI] [PubMed] [Google Scholar]
- 4.Xing JG. El-Sweisi W. Lee LEJ, et al. Development of a zebrafish spleen cell line, ZSSJ, and its growth arrest by gamma irradiation and capacity to act as feeder cells. In Vitro Cell Dev Biol Anim. 2004;45:163–174. doi: 10.1007/s11626-008-9159-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirayama J. Cho S. Sassone-Corsi P. Circadian control by the reduction/oxidation pathway: Catalase represses light-dependent clock gene expression in the zebrafish. Proc Natl Acad Sci USA. 2007;104:15747–15752. doi: 10.1073/pnas.0705614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogstrand C. Zheng D. Feeney G, et al. Zinc-controlled gene expression by metal-regulatory transcription factor 1 (MTF1) in a model vertebrate, the zebrafish. Biochem Soc Trans. 2008;36:1252–1257. doi: 10.1042/BST0361252. [DOI] [PubMed] [Google Scholar]
- 7.Villalobos P. Rojas M. Conejeros P, et al. Lipopolyamine-mediated transfection of reporter plasmids into a fish cell line. Electron J Biotech. 1999;2:88–98. [Google Scholar]
- 8.He S. Salas-Vidal E. Rueb S, et al. Genetic and transcriptome characterization of model zebrafish cell lines. Zebrafish. 2006;3:441–453. doi: 10.1089/zeb.2006.3.441. [DOI] [PubMed] [Google Scholar]
- 9.Kawasaki T. Saito K. Mitsui K, et al. Introduction of a foreign gene into zebrafish and medaka cells using adenoviral vectors. Zebrafish. 2009;6:253–258. doi: 10.1089/zeb.2009.0596. [DOI] [PubMed] [Google Scholar]
- 10.Senghaas N. Koster RW. Culturing and transfecting zebrafish PAC2 fibroblast cells. Cold Spring Harb Protoc. 2009;6:prot5235. doi: 10.1101/pdb.prot5235. [DOI] [PubMed] [Google Scholar]
- 11.Ouyang SD. Pei YY. Weng SP, et al. Effective polyethyleneimine-mediated gene transfer into zebrafish cells. Zebrafish. 2009;6:245–251. doi: 10.1089/zeb.2009.0589. [DOI] [PubMed] [Google Scholar]
- 12.Vallone D. Santoriello C. Gondi SB, et al. Basic protocols for zebrafish cell lines: maintenance and transfection. Methods Mol Biol. 2007;362:429–441. doi: 10.1007/978-1-59745-257-1_35. [DOI] [PubMed] [Google Scholar]
- 13.Badakov R. Jazwinska A. Efficient transfection of primary zebrafish fibroblasts by nucleofection. Cytotechnology. 2006;51:105–110. doi: 10.1007/s10616-006-9018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pando MP. Pinchak AB. Cermakian N, et al. A cell-based system that recapitulates the dynamic light-dependent regulation of the vertebrate clock. Proc Natl Acad Sci USA. 2001;98:10178–10183. doi: 10.1073/pnas.181228598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felgner PL. Gadek TR. Holm M, et al. Lipofection—A highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felgner PL. Ringold GM. Cationic liposome-mediated transfection. Nature. 1089;337:387–388. doi: 10.1038/337387a0. [DOI] [PubMed] [Google Scholar]
- 17.Hirayama J. Fukuda I. Ishikawa T, et al. New role of zCRY and zPER2 as regulators of sub-cellular distributions of zCLOCK and zBMAL proteins. Nucleic Acids Res. 2003;31:935–943. doi: 10.1093/nar/gkg174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirayama J. Miyamura N. Uchida Y, et al. Common light signaling pathways controlling DNA repair and circadian clock entrainment in zebrafish. Cell Cycle. 2009;8:2794–2801. doi: 10.4161/cc.8.17.9447. [DOI] [PubMed] [Google Scholar]
- 19.Boussif O. Lezoualch F. Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo—polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falco A. Encinas P. Carbajosa S, et al. Transfection improvements of fish cell lines by using deacylated polyethylenimine of selected molecular weights. Fish Shellfish Immunol. 2009;26:559–566. doi: 10.1016/j.fsi.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Hazel JR. Effects of temperature on the structure and metabolism of cell membranes in fish. Am J Physiol Reg Int Comp Physiol. 1984;246:R460–R470. doi: 10.1152/ajpregu.1984.246.4.R460. [DOI] [PubMed] [Google Scholar]
- 22.Romoren K. Thu BJ. Bols NC, et al. Transfection efficiency and cytotoxicity of cationic liposomes in salmonid cell lines of hepatocyte and macrophage origin. Biochim Biophys Acta. 2004;1663:127–134. doi: 10.1016/j.bbamem.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Lopez A. Fernandez-Alonso M. Rocha A, et al. Transfection of epithelioma papulosum cyprini (EPC) carp cells. Biotechnol Lett. 2001;23:481–487. [Google Scholar]
- 24.Schiotz BL. Rosado EG. Baekkevold ES, et al. Enhanced transfection of cell lines from Atlantic salmon through nucleofection and antibiotic selection. BMC Res Notes. 2011;4:136. doi: 10.1186/1756-0500-4-136. [DOI] [PMC free article] [PubMed] [Google Scholar]