Abstract
Neural information processed through the striatum of the basal ganglia is crucial for sensorimotor and psychomotor functions. Genes that are highly expressed in the striatum during development may be involved in neural development and plasticity in the striatum. We report in the present study the identification of a previously uncharacterized mammalian member of the nocA/elB/tlp-1 family, Nolz-1, that is preferentially expressed at high levels in the developing striatum. Nolz-1 mRNA was expressed as soon as striatal anlage began to form at embryonic day 13 in the rat. Nolz-1 mRNA was predominantly expressed in the lateral ganglionic eminence (striatal primordium) and was nearly absent in the adjacent structures of the medial ganglionic eminence and the cerebral cortex. Moreover, Nolz-1 was highly expressed in the subventricular zone of the lateral ganglionic eminence and was colocalized with the early neuronal differentiation markers of TuJ1 and Isl1 and the projection neuron marker of DARPP-32, suggesting that Nolz-1 was expressed in differentiating progenitors of striatal projection neurons. A time course study showed that Nolz-1 mRNA was developmentally regulated, as its expression was down-regulated postnatally with low levels remaining in the ventral striatum at adulthood. As the tagged Nolz-1 protein was localized in the nucleus, Nolz-1 may function as transcriptional regulator. In a model system for neural differentiation, Nolz-1 mRNA was dramatically induced on neural induction of P19 embryonal carcinoma cells by retinoic acid, suggesting that Nolz-1 activation may be involved in neural differentiation. Our study suggests that Nolz-1 is preferentially expressed in differentiating striatal progenitors and may be engaged in the genetic program for controlling striatal development.
The striatum is the major input information processing unit in the basal ganglia circuits of the mammalian forebrain. What makes the striatum an attractive and important system for neurobiological study is its involvement in multiple neurological functions ranging from motor control, cognition, emotion, and reinforcement to plasticity underlying learning and memory (1). The importance of the striatum is also reflected in a number of neurological disorders, including Parkinson's disease, Huntington's disease, and obsessive–compulsive disorders in which malfunction of neurotransmission or neurodegeneration are associated with the striatal system (2, 3). The study of development and function of the striatum is thus fundamentally important not only to the understanding of integrative brain function, but also to the development of therapeutic approaches to neurological diseases.
An experimental approach to study the development and function of the striatum is to identify genes that are preferentially expressed by striatal neurons. Based on the study of cDNA subtractive hybridization cloning, it has been estimated that there are about 100 genes that are preferentially expressed at high levels in the striatum (4). The striatum-enriched genes comprise at least two classes of genes with distinct function. One class of genes are transcriptional regulators that control neuronal development, including the Dlx family, Mash1, Gsh2, Ebf1/Olf1, Six3, Isl1, Foxp1, and Foxp2 (5–11). The other class of genes are signaling transduction molecules related to neurotransmission, including dopamine D1 and D2 receptors; the G protein, Golf; the adenylyl cyclase type V; the serine/threonine protein phosphatase-1 inhibitor, DARPP-32; the striatal-enriched tyrosine phosphatase, STEP; and the Rap guanine nucleotide exchange factor, CalDAG-GEF I (12–16). The enrichment of these signaling molecules in the striatum constitutes parts of the specificity of neurotransmission in the striatal system.
These two classes of striatum-enriched genes are important for setting up the cellular and molecular infrastructure of the striatal signaling system. We report in the present study the identification of a striatum-enriched gene, Nolz-1, that is a previously uncharacterized mammalian member of the nocA/elB/tlp-1 family. The spatiotemporal expression pattern of Nolz-1 mRNA suggests that it may regulate development and function of striatal neurons.
Materials and Methods
Subtractive Hybridization of cDNA Cloning. The lateral ganglionic eminence (LGE) and the overlying cerebral cortex were dissected from the embryonic day (E) 15 rat forebrain (Sprague–Dawley, National Yang-Ming University). Total RNA was extracted by using the guanidinium thiocyanate extraction method and was then reverse transcribed into cDNAs. The PCR-select cDNA subtraction was performed by following the manufacturer's instruction (Clontech). The subtracted cDNAs were subcloned into the pCRII vector (Invitrogen) and were sequenced. The protocol of animal use conformed to National Institutes of Health guidelines.
Northern Blotting. Total RNA was isolated from rat tissues and P19 embryonal carcinoma cells. The random hexamer method was used to synthesize [32P]dCTP-labeled probes from pCRII-LGEC-18 plasmid that contained 3′ UTR of rat Nolz-1 cDNA (506 bp). The procedures of hybridization and autoradiography were as described (17).
In Situ Hybridization. The isotope and nonisotope methods of in situ hybridization were performed as described (11, 18). The [35S]UTP-labeled and digoxigenin-UTP-labeled antisense and sense Nolz-1 riboprobes were synthesized by in vitro transcription with HindIII or XhoI linearized pCRII-LGEC-18 plasmids as templates. The rat Raldh-3 probe (461 bp) was derived from PCR cloning. After in situ hybridization, some of the sections were further processed for immunostaining for Mash1 (1:100, kindly provided by D. J. Anderson, California Institute of Technology, Pasadena), Isl1 (1:1,000, 3A4, Developmental Studies Hybridoma Bank), DARPP-32 (1:20,000, kindly provided by H. C. Hemmings, Weill Medical College, Cornell University, Ithaca, NY), and Tuj1 (1:5,000, Promega) with the tyramide amplification method, except Tuj1 (19, 20).
Plasmids. The full-length mNolz-1-FLAG cDNA was subcloned into pcDNA3.1 and pEF1/Myc-His (Invitrogen) vectors to generate the expression plasmids of pCMV-Nolz-1-FLAG and pEF1-Nolz-1-FLAG, respectively. The pGL3-mNolz-1-P1.4 and pGL3-mNolz-1-dRARE [RARE, retinoic acid (RA) response element] vectors were constructed by subcloning ≈1.4-kb (nucleotides -235 to -1648) and ≈1.27-kb (nucleotides -235 to -1509) 5′ flanking regions, respectively, of mNolz-1 into pGL3 vectors (Promega). The plasmids were verified by sequencing.
Cell Culture and Transfection. HEK293T cells were cultured in DMEM with 10% FBS. P19 cells were cultured with α-MEM containing 7.5% newborn calf serum and 2.5% FBS. The pCMV-Nolz-1-FLAG and pEF1-Nolz-1-FLAG expression plasmids were cotransfected with the reporter plasmid pGFP-C1 (5:1; Clontech) into HEK293T cells and P19 cells, respectively, by calcium phosphate-mediated transfection. Neural induction of P19 cells was performed by treating aggregated P19 cells with all-trans RA (1 μM) in α-MEM with 5% FBS. Four days after the RA induction, the cell aggregates were trypsinized and cultured on poly(D)-lysine-coated dishes with SF21 medium (19) to promote neural differentiation.
Electroporation. The pCMV-Nolz-1-FLAG plasmid was transfected into E13.5 mouse forebrains by using the method of in vivo electroporation (21). The brains were harvested for immunostaining 1 day after electroporation. The pGL3-mNolz-1-P1.4 or pGL3-mNolz-1-dRARE plasmids were coelectroporated with pCMV-β-gal plasmid into the LGE of E16 rats, and the transfected tissues were cultured as explants for 1 day in vitro. The luciferase activity was normalized with the β-gal activity.
Immunocytochemistry. The transfected HEK293T, P19 cells and the brain tissue were processed for immunocytochemistry with FLAG antibodies (monoclonal antibody, 1:8,000, Sigma; polyclonal antiserum, 1:10,000, kindly provided by F.-J. Lee, National Taiwan University) and Nkx2.1 antibody (1:100, NeoMarkers, Fremont, CA) as described (19). The immunostained cells and tissue were counterstained with the DNA dye Hoechst 33258.
Results
Identification of Nolz-1 Gene. To search for novel striatum-enriched genes, we used the method of cDNA subtractive hybridization cloning. We subtracted the cDNAs of the LGE (striatal primordium) from the cDNAs of the cerebral cortex of E15 rats. Of 86 subtracted clones, there were 60 independent cDNAs in which 24 were known genes, 16 were uncharacterized genes, 17 were unknown EST clones, and 3 were novel clones. The RT-PCR analysis of 53 subtracted clones showed that three clones including Dlx2 (5), Six3 (8), and a previously uncharacterized gene (clone LGEC-18) were preferentially expressed at high levels in the LGE. We identified and sequenced two clones containing the full-length cDNA of LGEC-18 from the RIKEN mouse full-length cDNA library (RIKEN 41-22401-M18, B830032-M18). The ORFs of these two clones were identical, but the clones differed at the 5′ and the 3′ UTR, where several nucleotide variations occurred (data not shown).
The mouse gene encoded a putative protein of 652 aa that contained a serine-rich and two glycine-rich domains in its amino terminus, and five stretches of alanine-rich domains and a C2H2 zinc-finger motif (amino acids 519–550) in its carboxyl terminus (Fig. 1 A and C). The zinc-finger motif of this protein shared 48%, 61%, 55%, 87%, and 94% identity with the proteins encoded by Caenorhabditis elegans tlp-1 (22), Drosophila l (2)35Ba/no ocelli (nocA) and elB (23, 24), Zebrafish Noz1/Nlz (25, 26), and Xenopus xNolz (GenBank accession no. BC047263), respectively (Fig. 1C). The alignments showed that the amino terminus (amino acids 66–144) and the carboxyl terminus (519–652) of this protein had high homology with nocA, elB, Noz1/Nlz, and xNolz proteins (Fig. 1 B and C). Thus, the murine homologue of nocA-like zinc-finger protein (Nolz-1) represented a previously uncharacterized murine member of the nocA/elB/tlp-1 family. The search of GenBank further identified its human homologue, hNolz-1 (BC011625) and its rat homologue, rNolz-1 (a composite cDNA-derived from EST clones AW915098, AI600276, and AI599428 and the genomic clone AC128846) (Fig. 1 B and C). The entire amino acid sequence of mouse Nolz-1 (mNolz-1) was 99% and 96% identical to rNolz-1 and hNolz-1 proteins, respectively.
Fig. 1.
(A) The deduced amino acid sequences of mNolz-1. The following domains are marked: glycine (G)-rich domains (17–56 and 146–328; underlined); serine (S)-rich domain (320–340; italic and bold); alanine (A)-rich domains (398–496 and 570–576; italic); Groucho binding sequence (bold); C2H2 zinc-finger motif (bold and underlined); Zn2+ binding motif [two cysteine (C) and two histidine (H); bold, italic, and underlined]; amino acids that differ from hNolz-1 (shading). (B and C) Amino acid sequence alignments of the nocA/elB family in amino-terminal (B) and carboxyl-terminal (C) domains showing high homology among the family members. The identical residues are indicated by black and shaded boxes. The zinc-finger motifs are in italic, and the consensus residues are shown in bold below (C). The sequences of mNolz-1 shown in B and C are identical for rNolz-1 and hNolz-1.
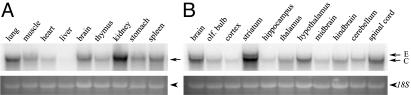
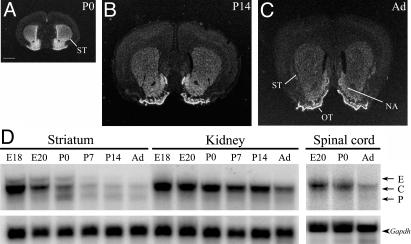
Expression of Nolz-1 Transcripts in Developing Rat Embryos. The Nolz-1 transcript appeared as a major band of ≈3.5 kb in different tissues of E20 rat embryos, including the brain, lung, muscle, heart, thymus, kidney, stomach, and spleen (Fig. 2A). Within the E20 rat brain, Nolz-1 was highly expressed in the striatum, confirming Nolz-1 as a striatum-enriched gene (Fig. 2B). Notably, the striatal Nolz-1 transcripts comprised a doublet of a major ≈3.5-kb band and a minor ≈4-kb band. More interestingly, the doublet transcripts were present only in the striatum. A single ≈3.5-kb transcript was detected in other examined brain regions (Fig. 2B).
Fig. 2.
Northern blots of Nolz-1 transcripts in E20 rat embryo. Shown is expression of Nolz-1 in different organ tissues of E20 rat embryo (A) and in different regions of E20 rat brain (B). Note that a doublet of transcripts consisting of the E and C forms are detected in the striatum.
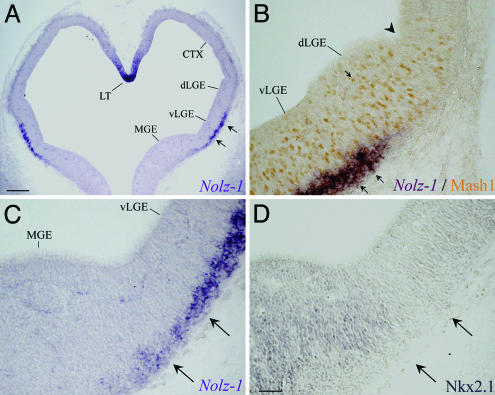
Preferential Expression of Nolz-1 in the Developing Striatum. The detailed expression pattern of Nolz-1 mRNA in the developing rat brains was examined by in situ hybridization with the same 3′ UTR probe of rat Nolz-1 for Northern blotting. Nolz-1 mRNA was detected in the lateral parts of the ventral forebrain at E13 (Fig. 3A). Nolz-1 mRNA was expressed in the Mash1-positive and Nkx2.1-negative LGE, except that the dorsal part of LGE lacked Nolz-1 (Fig. 3 B–D). Nolz-1 was not expressed in the adjacent Nkx2.1-positive medial ganglionic eminence (MGE), except for a strip of Nolz-1-positive cells across the borders between the MGE and the LGE (Fig. 3 C and D).
Fig. 3.
Expression of Nolz-1 mRNA in E13 rat telencephalon. (A) Nolz-l mRNA (double arrows) is expressed in the lateral part of ventral LGE (vLGE). (B) Nolz-1 is not expressed in the dorsal LGE (dLGE) containing Mash1-positive nuclei (arrow), nor is it expressed in the Nkx2.1-positive MGE (D) or the cortical primordium (CTX). The arrowhead in B indicates the border between the dLGE and CTX. The sections in C and D are adjacent sections, and the two arrows point to the Nolz-1-positive streak slightly extending into the Nkx2.1-poor region in the MGE. LT, lamina terminalis. (Scale bars: A, 200 μm; D for B–D, 50 μm.)
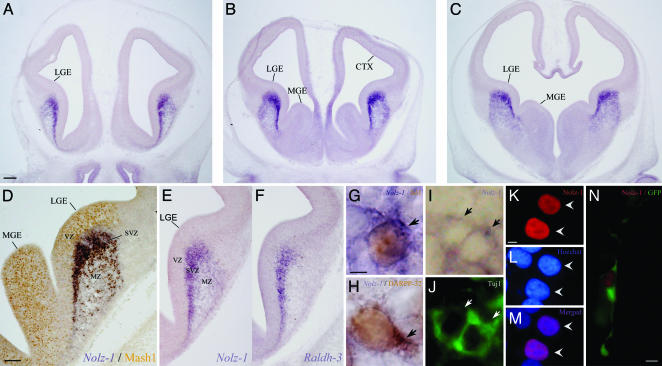
At E15, high levels of Nolz-1 mRNA were present in the LGE (striatal anlage) (Fig. 4 A–E). There were two interesting features of Nolz-1 expression in the ventral telencephalon at this developmental stage. First, Nolz-1 mRNA was not detectable in the Mash1-positive ventricular zone (VZ) (Fig. 4D), whereas it was dramatically up-regulated in the adjacent subventricular zone (SVZ), in which cells coexpressing Nolz-1 and Isl1 could be found (Fig. 4G). The expression of Nolz-1 mRNA was decreased in the differentiated mantle zone. Double labeling showed that Nolz-1-positive cells coexpressed TuJ1, a marker of early differentiating neurons (27), in the SVZ (Fig. 4 I and J), suggesting that Nolz-1 was expressed in early differentiating neurons. Second, in contrast to the LGE, Nolz-1 was nearly absent in the MGE (Fig. 4 B and C). Notably, Nolz-1 was highly expressed in the SVZ in which retinaldehyde dehydrogenase-3 (Raldh-3), a synthetic enzyme of RA, was also expressed (Fig. 4 E and F) (28). Nolz-1 mRNA was not detectable in the dorsal telencephalon.
Fig. 4.
Expression of Nolz-1 mRNA in E15 rat telencephalon. (A–C) Nolz-1 mRNA is primarily expressed in the LGE at rostral (A), middle (B), and caudal (C) levels. (D) Double labeling of Nolz-1 mRNA and MASH1 protein. Nolz-1 is highly expressed in the SVZ but is not expressed in the MASH1-positive ventricular zone (VZ, brown nuclei). Nolz-1 expression is decreased in the mantle zone (MZ). (E and F) Adjacent sections showing spatial correspondence of Nolz-1 and Raldh-3 mRNA expression in the SVZ of LGE. (G) Coexpression of Nolz-1 mRNA (purple cytosolic ring) and Isl1 protein (brown nucleus) in early differentiating striatal cells (arrow) in the SVZ of LGE. (H) Double labeling of Nolz-1 mRNA (purple cytosolic ring) and DARPP-32 protein (brown nucleus and cytosol) in E20 striatal neurons (arrow, dark brown cytosol). (I and J) Coexpression of Nolz-1 mRNA (I) and TuJ1 protein (J) in differentiating neurons (arrows) in SVZ of LGE. (K–N) Nuclear localization of FLAG-tagged Nolz-1 protein in HEK 293T cells (K–M, arrowheads) and mouse E13.5 forebrain (N). (K) FLAG immunostaining. (L) Hoechst 33258 nuclear staining. (M) Merged images of K and L.(N) A cotransfected GFP-positive cell with neuronal morphology in the brain shows nuclear localization of tagged Nolz-1 protein (red nucleus). (Scale bars: A for A–C, 200 μm; D for D–F, 200 μm; G for G–J, 2 μm; K for K–M, 5 μm; N, 5 μm.)
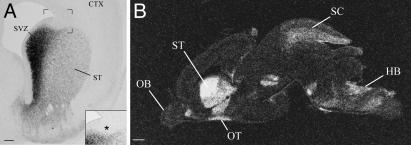
At E18–E20, the LGE and the MGE were fused to form a single bulge of ganglionic eminence that became the developing striatum. High levels of Nolz-1 mRNA were maintained in the SVZ of the ganglionic eminence except the dorsalmost region, which lacked Nolz-1 expression (Fig. 5A). Nolz-1 was still not detectable in the VZ. Double labeling of Nolz-1 mRNA and DARPP-32 protein, a marker for striatal projection neurons (29), showed that many DARPP-32-immunoreactive neurons coexpressed Nolz-1 mRNA in the mantle zone of E18 and E20 striatum (Fig. 4H). In addition to the developing striatum, Nolz-1 mRNA was expressed in a number of developing brain regions, including the medial septum, superior colliculus, and hindbrain (Fig. 5B).
Fig. 5.
Expression of Nolz-1 mRNA in E20 rat forebrain. (A) Nolz-1 mRNA is highly expressed in the SVZ of the developing striatum (ST) except the dorsal cap, which lacks Nolz-1 (asterisk). The bracketed region is shown at high magnification in Inset.(B) Film autoradiogram of a parasagittal section showing enriched expression of Nolz-1 mRNA in the striatum of E20 rat brain with 35S-labeled probes. CTX, cortex; HB, hindbrain; OB, olfactory bulb; OT, olfactory tubercle; SC, superior colliculus. (Scale bars: A, 200 μm; B, 500 μm.)
Developmental Regulation of Nolz-1 in the Striatum. After birth, the expression of Nolz-1 mRNA in the striatum was gradually down-regulated from postnatal day 0 and 14 to adulthood (Fig. 6 A–C). In the adult striatum, the levels of Nolz-1 mRNA were very low in the dorsal striatum, but low levels of Nolz-1 expression remained in the nucleus accumbens and high levels remained in the olfactory tubercle (Fig. 6C). Interestingly, the Northern blotting analysis indicated that there were dynamic changes of Nolz-1 transcripts during striatal development (Fig. 6D). At postnatal day 0 striatum, instead of the doublet transcripts found at the E20 striatum, a triplet of transcripts was detected, i.e., in addition to the major ≈3.5-kb transcript, two transcripts (≈4 kb and ≈3 kb) with lower expression levels were found. Similar results were obtained by using another probe (nucleotides 1240–1627 of RIKEN clone 41-22401-M18) that recognized partial coding sequences in the second exon of Nolz-1 gene (data not shown). It was notable that the expression of these two transcripts was under different temporal control. The ≈4-kb transcript was transiently present in the embryonic striatum from E15 to postnatal day 0 (Figs. 6D and 7A), possibly representing an embryonic isoform of Nolz-1 transcript [embryonic (E) form]. By contrast, the ≈3-kb transcript was not significantly detected in the striatum until after birth, which might represent a postnatal isoform (P form; Fig. 6D). The major ≈3.5-kb transcript [constitutive (C) form] appeared constitutively through striatal development, albeit with gradually decreasing expression level during postnatal maturation. No such complex regulation of different Nolz-1 transcripts was observed in the development of the spinal cord and the kidney (Fig. 6D).
Fig. 6.
Developmental regulation of Nolz-1 mRNA in the striatum during development. Expression of Nolz-1 mRNA is gradually decreased from postnatal day 0 (A) and postnatal day 14 (B) to adult (Ad) striatum (ST; C) as assayed by in situ hybridization with 35S-labeled probes. The Nolz-1 expression remains in the nucleus accumbens (NA) and olfactory tubercle (OT) at adulthood. (D) Dynamic regulation of different Nolz-1 transcripts in the striatum during development. A triplet of transcripts consisting of the E, C, and P forms are present only in the postnatal day 0 striatum. No such complex regulation of Nolz-1 transcripts is detected in the development of the kidney and the spinal cord where Nolz-1 expression is also abundant. (Scale bar: A for A–C, 1 mm.)
Fig. 7.
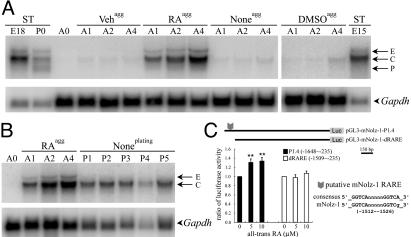
Regulation of Nolz-1 in P19 cells and the LGE culture. (A) A doublet of Nolz-1 transcripts similar to the E and C forms found in E15, E18, and postnatal day 0 striatum (ST) are induced in aggregated (agg) P19 cells with RA treatment (RAagg). Little or no Nolz-1 induction is found with vehicle (Vehagg) or DMSO (DMSOagg) treatments, or without any treatment (Noneagg). (B) After the cell aggregates are dissociated for plating and cultured without RA (Noneplating), only the C form remains in the dissociated cells. (C) The pGL3-mNolz-1-P1.4 luciferase reporter gene vector was transfected into E16 rat LGE explant culture. All-trans RA increases the luciferase activity by 1.3-fold. This inductive effect is abolished when the RARE-containing region is deleted from the construct (pGL3-mNolz-1-dRARE). **, P < 0.01, Student's t test. A0–A4, 0–4 days after cell aggregation; P1–P5, 1–5 days after dissociating cell aggregates for dissociated cells culture.
Subcellular Distribution of Nolz-1 Protein. To study the subcellular distribution of Nolz-1 protein, we expressed a FLAG epitope-tagged Nolz-1 recombinant protein in HEK293T cells. The FLAG-tagged recombinant protein was immunocytochemically detected in the nuclei of transfected HEK293T cells (Fig. 4 K–M). A similar result of nuclear localization was obtained in the transfected P19 embryonal carcinoma cells (data not shown). To further confirm these results in a physiologically relevant context, we electroporated the expression plasmids into E13.5 mouse forebrains in vivo (21). Consistently, the tagged mNolz-1 protein was detected in the nucleus of transfected cells in the forebrain (Fig. 4N).
Regulation of Nolz-1 in Neural Induction of P19 Cells and in the LGE. We used the P19 cell line as a paradigm for neural differentiation to study whether Nolz-1 was associated with neural induction and differentiation. Little Nolz-1 mRNA was detected in the naive P19 cells. On the neural induction of aggregated P19 cells by RA, a doublet of Nolz-1 transcripts similar to striatal E and C forms were induced (Fig. 7 A and B). After dissociating the inductive cell aggregates to facilitate neural differentiation, only a single transcript similar to the striatal C form was detected (Fig. 7B). Little Nolz-1 was induced with the DMSO treatment, which promoted P19 cell differentiation into nonneural cells (Fig. 7A) (30). These findings suggest that the induction of striatal E form transcript may require aggregation of cells in the presence of activated RA signaling, a situation that may occur in the SVZ of LGE where cells are tightly packed and Raldh-3 is highly expressed (Fig. 4 E and F).
The analysis of the genomic sequence of mNolz-1 indicated that a putative direct repeat 5 (DR5) RARE was present at nucleotides -1512 to -1526 upstream of the first ATG translation start site (Fig. 7C). We then transfected into E16 rat LGE explant culture a luciferase reporter gene driven by an ≈1.4-kb Nolz-1 promoter region including the putative RARE (nucleotides -235 to -1648). The results showed that all-trans RA induced 1.3-fold increases of luciferase activity (5 μM, 1.31 ± 0.07, P < 0.01, n = 12; 10 μM, 1.34 ± 0.08, P < 0.01, n = 12; Fig. 7C). Notably, deletion of the putative RARE-containing region (nucleotides -1648 to -1508) from the construct abolished the RA inductive effects, demonstrating the specificity of RA effect (5 μM, 0.98 ± 0.06, n = 8; 10 μM, 1.07 ± 0.05, n = 9). These results suggest that retinoid signaling may regulate Nolz-1 expression in the developing striatum. Although the activation of the Nolz-1 reporter gene in the LGE by the exogenous RA was small, it is possible that high levels of endogenous RA in the LGE might have prevented further increases of Nolz-1 reporter gene activity by the exogenous RA (31). Alternatively, RA may regulate only the E form of Nolz-1 transcripts in the LGE, and the weak inductive effect may reflect the minor representation of the E form in the entire Nolz-1 transcripts of the LGE (Fig. 7A).
Discussion
Despite the importance of the striatal system in neurological function (1), the developmental process by which striatal neurons acquire their special neurochemical properties and establish specific neural connectivity with other brain regions remains elusive. In an attempt to explore this issue, we searched for genes that are preferentially expressed in the developing striatum. We reasoned that developmentally regulated striatum-enriched genes may be involved in neural development and plasticity of the striatal system. The zinc-finger gene Nolz-1 that we identified represents an interesting molecule for understanding the molecular mechanisms underlying striatal development and function.
Nolz-1 as a Previously Uncharacterized Mammalian Member of the nocA/elB/tlp-1 Family. A salient feature of the Nolz-1 protein is that it contains an atypical C2H2 zinc-finger motif. In contrast to the typical C2H2 zinc-finger motif, X (2)-Cys-X (2, 4)-Cys-X (12)-His-X (3–5)-His, in which the two cysteine residues are spaced by conserved two or four amino acid residues (32), the two cysteine residues are spaced by eight residues in Nolz-1. This atypical arrangement of zinc-finger motif is conserved in the nocA/elB/tlp-1 family (22–26). The mNolz-1 protein contained five stretches of alanine residues and a serine-rich domain, all of which were also found in nocA protein (23). The alignments of nocA, elB, Noz1/Nlz, xNolz, and mNolz-1 indicated that, in addition to the zinc-finger motif-containing carboxyl terminus, a domain of ≈79 aa in the amino terminus was also highly conserved, suggesting that these two domains may be of functional significance.
Nolz-1 as a Developmental Marker for Differentiating Striatal Progenitors. Our study showed that Nolz-1 mRNA was highly expressed in the SVZ, a transitional zone between the ventricular and mantle zones. This expression pattern of Nolz-1 classifies it into a group of striatum-enriched genes that are primarily expressed in the SVZ of the ganglionic eminences, including Dlx5, Brn4, RAR-alpha, SCIP, EphA4, and Raldh-3 (7, 28). The finding that Nolz-1 mRNA was coexpressed with the early postmitotic neuronal markers TuJ1 and Isl1 suggests that Nolz-1 is likely to be expressed by striatal neurons at early phases of differentiation (9, 27, 33). Nolz-1 is selectively expressed in the LGE, together with a small group of Nolz-1-positive cells extending ventromedially into the MGE at caudal levels. This nearly selective expression of Nolz-1 in the LGE distinguishes Nolz-1 from other striatum-enriched genes such as Dlx1, Dlx2, Mash1, and Gsh2 that are generally expressed in both the LGE and MGE (5–7). The LGE contains progenitors that give rise to striatal projection neurons and GABAergic interneurons destined for the olfactory bulb and the cerebral cortex (34). The expression levels of Nolz-1 mRNA were, however, low in the developing olfactory bulb and cerebral cortex. Moreover, Nolz-1 was not expressed in the most dorsal part of the SVZ in which progenitors of olfactory bulb interneurons resided (35). The MGE, which is the main source of GABAergic interneurons tangentially migrating to the striatum and the cerebral cortex (34), also lacked Nolz-1 expression. Therefore, Nolz-1 may be primarily expressed by differentiating progenitors of striatal projection neurons. Consistent with this hypothesis, Nolz-1 was colocalized with DARPP-32, a marker for striatal projection neurons (29). Nolz-1 may prove to be a useful marker for identifying differentiating striatal progenitors.
Potential Involvement of Nolz-1 Gene in Neural Development of the Mammalian Telencephalon. The possibility that Nolz-1 is involved in regulating mammalian neural development is consistent with the functional study of Drosophila nocA. Mutation of l(2)35Ba/nocA results in hypertrophy of the embryonic supraesophageal ganglion (Drosophila brain). This hypertrophy is due to incorporation of nonneural cells in the procephalic lobe into the supraesophageal ganglion, and many of the misplaced nonneural cells become neurons (23). Mutation of tlp-1 affects asymmetric cell fate determination and morphogenesis of tail (22), whereas elB is involved in cell fate specification of tracheal branches (24). The zebrafish Noz1/Nlz is expressed mainly in the posterior parts of the brain, and it is inducible by the RA signaling (25, 26). Consistent with the reports that Tlp-1 and ElB proteins are nuclear transcription regulators (22, 24), the transfected Nolz-1 protein was localized in the nucleus. Notably, ElB protein can associate with Grouch protein to function as transcriptional repressor (24), and its Grouch interacting motif FKPY is conserved in Nolz-1 protein. It is possible that Nolz-1 may also function as transcriptional repressor.
The developing telencephalon is compartmentally organized into three progenitor domains including the dorsal cortex and the ventral LGE and MGE. The boundaries between these three domains have been shown to be specified and maintained by the balanced activity of domain-specific genes (36–38). The specific expression of Nolz-1 in the LGE domain and the possibility that Nolz-1 may function as transcriptional repressor suggest that Nolz-1 may regulate the formation and maintenance of the LGE domain by interacting with other domain-specific genes in the developing telencephalon. Future study of Nolz-1 function may provide insight into the genetic programs underlying development of the telencephalon.
Acknowledgments
We thank Drs. D. J. Anderson, H. C. Hemmings, and F.-J. Lee for providing reagents, and C.-K. Chou for help with DNA sequencing. We are grateful to Dr. A. M. Graybiel for her support. This work was funded in part by the National Health Research Institutes (NHRI-EX92-9010NL), the National Science Council (NSC92-2311-B-010-007), the Ministry of Education (89-B-FA22-1-4-03), and the Brain Research Center at the University System of Taiwan.
Abbreviations: C form, constitutive form; En, embryonic day n; E form, embryonic form; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; RA, retinoic acid; RARE, RA response element; SVZ, subventricular zone.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY281290 and AY281291).
References
- 1.Graybiel, A. M. (1995) Curr. Opin. Neurobiol. 5, 733-741. [DOI] [PubMed] [Google Scholar]
- 2.Wichmann, T. & DeLong, M. R. (1996) Curr. Opin. Neurobiol. 6, 751-758. [DOI] [PubMed] [Google Scholar]
- 3.Graybiel, A. M. & Rauch, S. L. (2000) Neuron 28, 343-347. [DOI] [PubMed] [Google Scholar]
- 4.Usui, H., Falk, J. D., Dopaza, A., de Lecea, L., Erlander, M. G. & Sutcliffe, J. G. (1994) J. Neurosci. 14, 4915-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porteus, M. H., Bulfone, A., Ciaranello, R. D. & Rubenstein, J. L. R. (1991) Neuron 7, 221-229. [DOI] [PubMed] [Google Scholar]
- 6.Toresson, H., Potter, S. S. & Campbell, K. (2000) Development (Cambridge, U.K.) 127, 4361-4371. [DOI] [PubMed] [Google Scholar]
- 7.Garel, S., Marin, F., Grosschedl, R. & Charnay, P. (1999) Development (Cambridge, U.K.) 126, 5285-5294. [DOI] [PubMed] [Google Scholar]
- 8.Oliver, G., Mailhos, A., Wehr, R., Copeland, N. G., Jenkins, N. A. & Gruss, P. (1995) Development (Cambridge, U.K.) 121, 4045-4055. [DOI] [PubMed] [Google Scholar]
- 9.Wang, H.-F. & Liu, F.-C. (2001) Neuroscience 103, 999-1016. [DOI] [PubMed] [Google Scholar]
- 10.Ferland, R. J., Cherry, T. J., Preware, P. O., Morrisey, E. E. & Walsh, C. A. (2003) J. Comp. Neurol. 460, 266-279. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi, K., Liu, F.-C., Hirokawa, K. & Takahashi, H. (2003) J. Neurosci. Res. 73, 61-72. [DOI] [PubMed] [Google Scholar]
- 12.Drinnan, S. L., Hope, B. T., Snutch, T. P. & Vincent, S. R. (1991) Mol. Cell. Neurosci. 2, 66-70. [DOI] [PubMed] [Google Scholar]
- 13.Glatt, C. E. & Snyder, S. H. (1993) Nature 361, 536-538. [DOI] [PubMed] [Google Scholar]
- 14.Ouimet, C. C., Miller, P. E., Hemmings, H. C., Jr., Walaas, S. I. & Greengard, P. (1984) J. Neurosci. 4, 111-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombroso, P. J., Murdoch, G. & Lerner, M. (1991) Proc. Natl. Acad. Sci. USA 88, 7242-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawasaki, H., Springett, G. M., Toki, S., Canales, J. J., Harlan, P., Blumenstiel, J. P., Chen, E. J., Bany, I. A., Mochizuki, N., Ashbacher, A., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 13278-13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai, T. F., Chen, K. S., Weber, J. S., Justice, M. J. & Beaudet, A. L. (2002) Hum. Mol. Genet. 11, 1659-1668. [DOI] [PubMed] [Google Scholar]
- 18.Simmons, D., Arriza, J. & Swanson, L. (1989) J. Histotechnol. 12, 169-181 [Google Scholar]
- 19.Liu, F.-C., Takahashi, H., McKay, R. D. G. & Graybiel, A. M. (1995) J. Neurosci. 15, 2367-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams, J. C. (1991) J. Histochem. Cytochem. 40, 1457-1463 [DOI] [PubMed] [Google Scholar]
- 21.Saito, T. & Nakatsuji, N. (2001) Dev. Biol. 240, 237-246. [DOI] [PubMed] [Google Scholar]
- 22.Zhao, X., Yang, Y., Fitch, D. H. & Herman, M. A. (2002) Development (Cambridge, U.K.) 129, 1497-1508. [DOI] [PubMed] [Google Scholar]
- 23.Cheah, P. Y., Meng, Y. B., Yang, X., Kimbrell, D., Ashburner, M. & Chia, W. (1994) Mol. Cell. Biol. 14, 1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorfman, R., Glazer, L., Weihe, U., Wernet, M. F. & Shilo, B. Z. (2002) Development (Cambridge, U.K.) 129, 3585-3596. [DOI] [PubMed] [Google Scholar]
- 25.Andreazzoli, M., Broccoli, V. & Dawid, I. B. (2001) Mech. Dev. 104, 117-120. [DOI] [PubMed] [Google Scholar]
- 26.Sagerstrom, C. G., Kao, B. A., Lane, M. E. & Sive, H. (2001) Dev. Dyn. 220, 402-408. [DOI] [PubMed] [Google Scholar]
- 27.Menezes, J. R. & Luskin, M. B. (1994) J. Neurosci. 14, 5399-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, H., Wagner, E., McCaffery, P., Smith, D., Andreadis, A. & Drager, U. C. (2000) Mech. Dev. 95, 283-289. [DOI] [PubMed] [Google Scholar]
- 29.Ouimet, C. C., Langley-Gullion, K. C. & Greengard, P. (1998) Brain Res. 808, 8-12. [DOI] [PubMed] [Google Scholar]
- 30.Bain, G., Ray, W. J., Yao, M. & Gottlieb, D. I. (1994) BioEssays 16, 343-348. [DOI] [PubMed] [Google Scholar]
- 31.Toresson, H., Mata de Urquiza, A., Fagerstrom, C., Perlmann, T. & Campbell, K. (1999) Development (Cambridge, U.K.) 126, 1317-1326. [DOI] [PubMed] [Google Scholar]
- 32.Wolfe, S. A., Nekludova, L. & Pabo, C. O. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 183-212. [DOI] [PubMed] [Google Scholar]
- 33.Ericson, J., Thor, S., Edlund, T., Jessell, T. M. & Yamada, T. (1992) Science 256, 1555-1560. [DOI] [PubMed] [Google Scholar]
- 34.Marin, O. & Rubenstein, J. L. (2001) Nat. Rev. Neurosci. 2, 780-790. [DOI] [PubMed] [Google Scholar]
- 35.Stenman, J., Toresson, H. & Campbell, K. (2003) J. Neurosci. 23, 167-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuurmans, C. & Guillemot, F. (2002) Curr. Opin. Neurobiol. 12, 26-34. [DOI] [PubMed] [Google Scholar]
- 37.Rallu, M., Corbin, J. G. & Fishell, G. (2002) Nat. Rev. Neurosci. 3, 943-951. [DOI] [PubMed] [Google Scholar]
- 38.Campbell, K. (2003) Curr. Opin. Neurobiol. 13, 50-56. [DOI] [PubMed] [Google Scholar]