Abstract
Caspases mediate apoptosis and have also been implicated in stem-cell biology. How caspases are linked to stem-cell biology is not known. Here, we show that the apoptotic blebs of cancer cells fuse together to form novel structures called ‘blebbishields'. Blebbishields form spheres by fusion. Both blebbishield formation and sphere formation involve active caspases and N-linked glycosylation. Sphere formation is enhanced by acidic pH and is counteracted by inhibitors of proton pump, caspases, and cholesterol. The blebbishields from VEGFR2High cells are capable of enhanced sphere formation. Blebbishields express transiently downregulated stem-cell markers and the sphere-forming blebbishield-derived cells are tumorigenic. Our study demonstrates that the cancer stem cells can survive after apoptosis by blebbishield formation and subsequent sphere formation.
Keywords: apoptosis, blebbishields, cancer stem cells, VEGFR2, p70S6K, caspase
Generation of induced pluripotent stem cells (iPSCs) using defined set of transcription factors can rewrite the stemness of cells.1 Caspases have been implicated in the generation of iPSCs after transduction of iPSC inducing transcription factors.2 It is not known whether iPSC generation is supported by the apoptotic or non-apoptotic functions of caspases. Hematopoietic stem cells (HSCs), iPSCs, and cancer stem cells (CSCs) share many characteristics in common.3 Therefore, caspases may also play a role in cancer stem-cell biology.
CSCs are found in various cancer types with distinct CSC marker expression.4 Although the CSCs between cancer types differ in marker expression, most CSCs share common features such as the ability to form spheres, dye exportation, drug resistance, heterogeneity, and tumorigenicity.4, 5, 6, 7, 8 However, not all CSCs exhibit all of these properties. Hence, tumorigenicity is considered as the important criteria for CSC characterization.9
Apoptosis is considered a terminal event in the biology of cells. During execution phase of apoptosis, the cell components are packed into small apoptotic bodies.10 Cancer cells employ a multitude of mechanisms to overcome apoptosis.8, 11 CSCs have been implicated in the resistance to therapeutic interventions in various cancers.4, 8 However, it is not completely understood whether the resistance and relapse of cancers are due to insufficient therapy or due to the intrinsic survival mechanisms of CSCs. Here, we show that cancer cells with apoptotic characteristics survive by evoking a novel emergency program involving fusion of apoptotic blebs to form spherical structures, which we term blebbishields. Blebbishields generate stem-cell spheres by fusion and the blebbishield-derived cells display heterogeneity, dye export properties, chemoresistance, and express many CSC markers. The sphere-forming cells are tumorigenic and we identify the factors that positively and negatively influence sphere formation from blebbishields.
Results and Discussion
Molecular alterations in isogenic cancer cell lines, which differ in tumorigenic aggressiveness
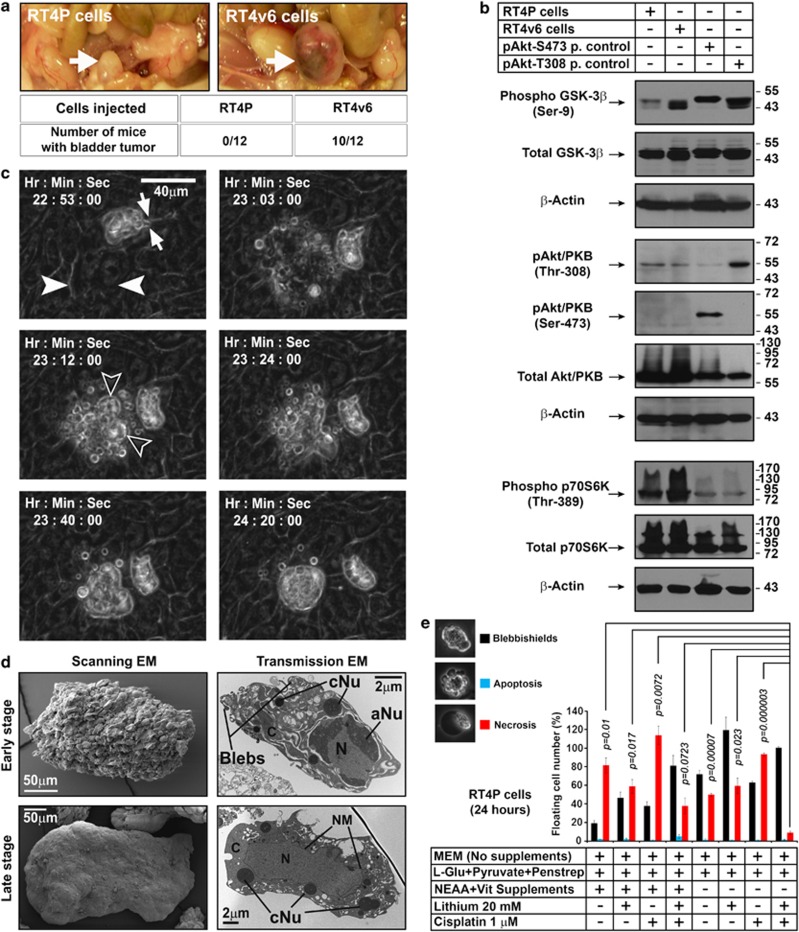
Serial in vivo passaging is known to increase the stemness of cancer cell lines.12, 13 To understand the key signaling pathways altered in this process, we used the human bladder cancer cell lines RT4P and its derivative RT4v6 (six serial passages in vivo). When orthotopically injected, RT4v6 cells formed tumors more rapidly, well before RT4P cells established visually detectable tumors, confirming the increased aggressiveness of RT4v6 cells (Figure 1a). Microarray analysis revealed that these cells had major differences in insulin-like growth factor binding protein-5 (IGFBP-5) mRNA expression (Supplementary Figure S1a). Since IGFBP-5 is involved in glucose metabolism, we examined these cells for differences in phosphorylation of Akt and GSK-3β, two main regulators of glucose metabolism.14 RT4P and RT4v6 cells differed in their GSK-3β Ser-9 phosphorylation but not in Akt activation (Figure 1b). We also examined the activation status of p70S6K, since p70S6K is known to phosphorylate GSK-3β at Ser-9 in TSC-2 null cells without Akt activation.15 Interestingly, p70S6K activation correlated with increased GSK-3β Ser-9 phosphorylation (Figure 1b).
Figure 1.
Inhibition of key signaling pathways that differ in isogenic bladder cancer cell lines RT4P and RT4v6 lead to blebbishield formation. (a) In vivo tumorigenicity of RT4P and RT4v6 cells determined by injecting 200 000 cells orthotopically into the bladder walls of nude mice (n=12 for each cell line). The tumor formation was assessed on day 34 post-injection. Arrow=bladder. (b) Western blotting analysis of regulators of glucose metabolism GSK-3β and Akt, and p70S6K. P. control=positive control; Akt-S473 positive control=UM-UC-3 cells; Akt-T308 positive control=UM-UC-14 cells. (c) RT4P cells at high density (1 × 107 cells/T–75 flask) were exposed to blebbishield ejection (BE) medium and imaged using time-lapse microscopy in DIC bright field settings. Solid arrowheads: the cell(s) starting the blebbishield emergency program. Note a preformed blebbishield in frame 22 : 53 : 00. Arrows in frame 22 : 53 : 00 indicate the thin fragile stalk that anchors the blebbishield with substratum (please also see Supplementary Movie S1). Hollow arrowheads: main body of blebbishield (also see Supplementary Figure S2). Note: The time is counted form the treatment with BE medium. (d) Scanning and transmission electron microscopy of early- and late-stage blebbishields. Note the transmission electron microscopy of early-stage blebbishields with discontinuous cytoplasm. N, nucleus; C, cytoplasm; aNu, active nucleolus; cNu, cytoplasmic nucleolus; NM, nuclear membrane (also see Supplementary Figure S1) (e) Requirement of cisplatin, lithium chloride, and NEAA+Vitamin deprivation to increase the number of blebbishields but to reduce necrosis
Microarray data indicated that the DNA-damage related genes were downregulated in RT4v6 cells (Supplementary Figure S1b). Hence, we examined the effect of combined inhibition of GSK-3β and p70S6K, and induction of DNA-damage. We used lithium chloride to inhibit GSK-3β, non-essential amino-acid (NEAA) restriction to inhibit p70S6K16 and cisplatin, a key chemotherapeutic drug for bladder cancer and a known inducer of DNA-damage and p70S6K cleavage, to induce DNA-damage.17
Blebbishield-mediated sphere formation after apoptosis and its dependence on amino acid-p70S6K signaling
In RT4P cells, upon combined inhibition of GSK-3β, p70S6K and induction of DNA-damage using Blebbishield Ejection medium (hereafter referred to as BE medium), a subset of cells underwent apoptosis. Instead of forming apoptotic bodies, the apoptotic blebs fused together to form a shield-like structure surrounding the DNA containing main body (hence we termed these structures blebbishields) (Figure 1c; Supplementary Figure S2a; Supplementary Movie S1). The blebbishields were ejected over the monolayer of cells, but remained anchored to the substratum through thin fragile stalks (Figure 1c; Supplementary Figure S1c). Scanning electron microscopic observations confirmed the bleb nature of early-stage blebbishield surface and relatively smoother cell surface in late-stage blebbishields (Figure 1d). Transmission electron microscopic observations revealed the bleb-natured cytoplasm and nearly intact nucleus in the early-stage blebbishields and relatively complete cytoplasm in the late-stage blebbishields. Notably, the nucleus had fenestrated nucleoli (active), and the cytoplasm had electron dense cytoplasmic nucleoli, intact mitochondria, rough-endoplasmic reticulum, and apoptotic Golgi-clusters (Figure 1d; Supplementary Figure S1c).
To examine the requirement of BE medium for blebbishield generation, we treated RT4P cells with various combinations of LiCl, cisplatin, and NEAA restriction and found that, although RT4P cells spontaneously generate low numbers of blebbishields at baseline (∼20% of the floating cells outnumbered by 80% of the necrotic cells), the combination of LiCl, cisplatin, and NEAA restriction is required to increase the yield and purity of blebbishields (Figure 1e).
To characterize the blebbishields, we fractionated the RT4P cells treated with BE medium into a blebbishield fraction and a blebbishield-depleted fraction (the adherent cells without blebbishields). We found that the p70S6K–45 kDa cleavage band and GSK-3β Ser-9 inhibitory phosphorylation were detected in the blebbishields, but not in the blebbishield-depleted cells (Figure 2a). Consequently, the p70S6K downstream target 4EBP1 phosphorylation at Thr-70 was lost (18-kDa band) and the total 4EBP1 was cleaved only in blebbishields (Figure 2a). These results suggest that the GSK-3β inhibition and p70S6K cleavage are specific for blebbishields. Of note, both p70S6K and 4EBP1 are caspase substrates known to undergo cleavage during apoptosis.17, 18
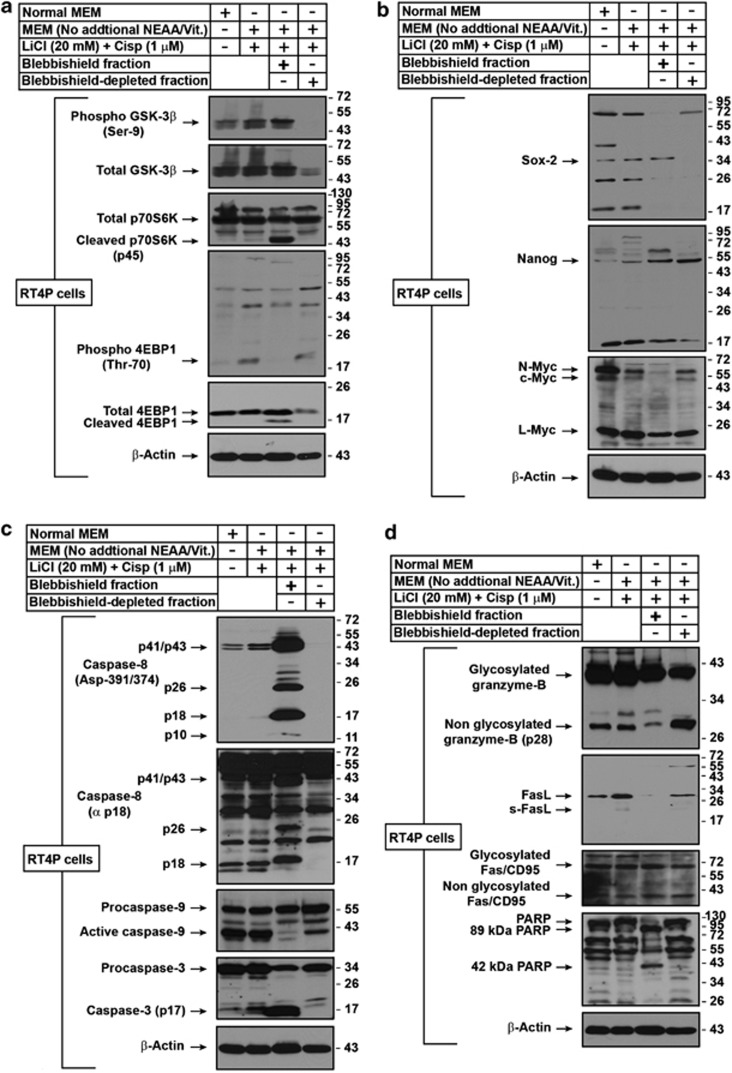
Figure 2.
Analysis of blebbishields for GSK3-β, p70S6K, 4EBP1, stem-cell transcription factors, and apoptotic signatures. (a) Effects of combined GSK-3β and p70S6K inhibition and DNA-damage (p70S6K cleavage) were studied by western blotting. The blebbishield and blebbishield-depleted fractions were isolated as described in Materials and methods. Lanes 3 and 4 are fractions of lane 2. Note the cleavages of 4EBP1 and p70S6K and phosphorylation of GSK-3β in blebbishields. (b) Characterization of blebbishields for the expression of core stem-cell transcription factors by western blotting. Note that N-myc and c-myc are downregulated in blebbishields. (c) Evaluation of caspase activation in blebbishields by western blotting. Caspase-8 Asp-391/372 was detected by an antibody specific to cleavage site, and the constitutively activated p18 fragment was detected by anti-p18 antibody (not specific to cleavage site). Note the inhibition of caspase-9 processing and induction of caspase-3 p17 in blebbishields. (d) Examining the role of caspases and granzyme-B in blebbishields. Note that the proportion of non-glycosylated granzyme-B versus glycosylated granzyme-B differed between blebbishields and blebbishield-depleted fraction (top panel). Fas but not FasL was expressed in blebbishields (Note that FasL was expressed in blebbishield-depleted fraction). PARP cleavage fragments in blebbishields indicating the involvement of both caspases and granzyme-B in blebbishield generation. MEM, minimal essential medium; NEAA, non-essential amino acid (except ℒ-glutamine); Vit., vitamins; Cisp, cisplatin
Since GSK-3β inhibition is known to maintain the pluripotency of stem cells,19 and caspases are implicated in iPSC generation,2 we examined the blebbishields for expression of core stem-cell transcription factors. Blebbishields expressed Sox-2, Nanog, L-myc (but not c-Myc or N-myc) and hard-to-detect low levels of Oct4, suggesting that the blebbishields possess stemness (Figure 2b; Supplementary Figures S2b and c).
Since the blebbishields showed visual signs of apoptosis, we examined the activation of caspases in blebbishields. Blebbishields had caspase-8 Asp-391/374 cleavage mediated p26 and p18 fragments (which are distinct from the constitutive caspase-8 activation) and active caspase-3 p17 fragment, but reduced constitutive processing of caspase-9 (Figure 2c).
Caspase-8 activation can occur due to death-receptor activation and also due to granzyme-B, a pro-apoptotic protease, enriched in self-renewing HSCs.20 Blebbishields expressed lower levels of native granzyme-B (non-glycosylated p28 form), reduced levels of FasL and normal levels of Fas receptor compared with untreated RT4P cells, suggesting the involvement of both granzyme-B, and death-receptor-mediated caspase activation in blebbishields (Figure 2d). Since no available small-molecule inhibitors can discriminate caspases and granzyme-B, and since the granzyme B inhibitor z-AAD-fmk also inhibits caspase-3,21 we chose PARP to investigate the involvement of granzyme-B and caspases. The 89-kDa cleavage band of PARP is specific for caspases whereas 72 and 42-kDa PARP cleavage bands are specific for granzyme-B.22 Blebbishields had both 89 and 42-kDa bands, suggesting the involvement of both caspases and granzyme-B, in blebbishield formation (Figure 2d). These results demonstrate the presence of caspase and granzyme-B-mediated apoptotic signatures, and stem-cell transcription factors in blebbishields.
Next we asked, whether the blebbishields can survive? If survive, whether they can resist cisplatin similar to CSCs, which are known to resist chemotherapeutics?12 We examined both issues by plating blebbishields in parallel with blebbishield-depleted adherent RT4P cells in high-dose (10 μM) cisplatin containing BE medium. Within 24 h, the blebbishields but not blebbishield-depleted cells formed large spheres similar to CSC spheres (Figure 3a), demonstrating that blebbishields can survive, form spheres, and resist high-dose cisplatin.
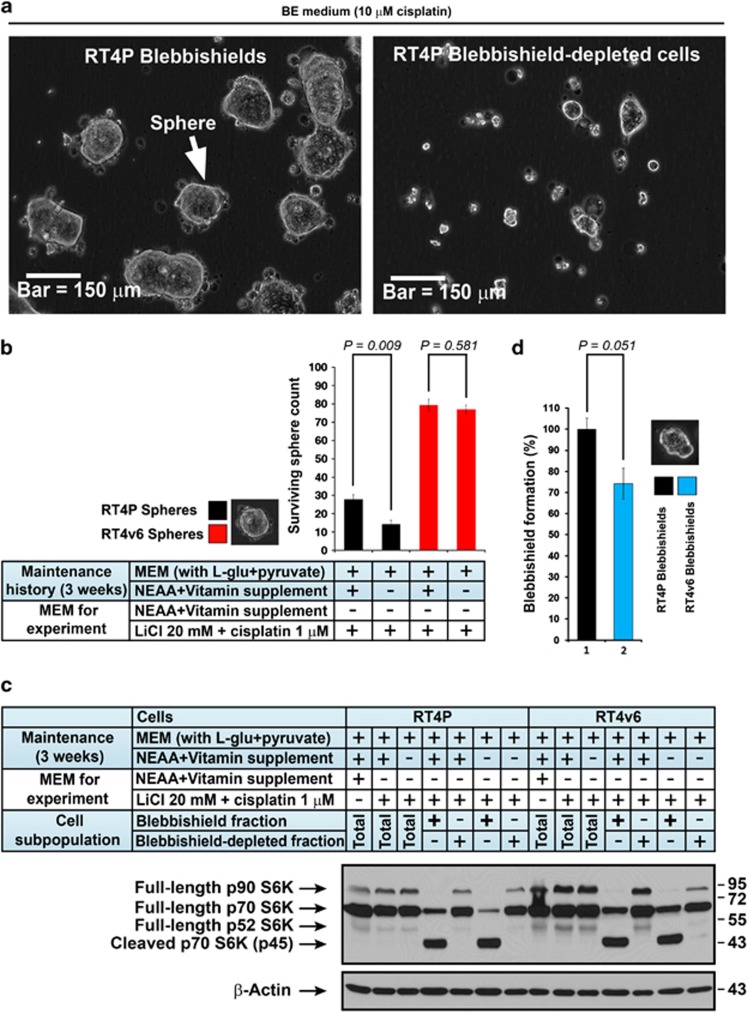
Figure 3.
Blebbishields are capable of forming spheres; role of amino acids and vitamins in blebbishield survival. (a) Blebbishields are capable of survival and form spheres. Freshly isolated blebbishields (using BE medium with 1 μM cisplatin) and cells from blebbishield-depleted fraction were plated separately in BE medium with higher dose of cisplatin (10 μM cisplatin) for 24 h. Note that the blebbishield fractions formed larger spheres by fusion (see also Supplementary Movie S2 for fusion events). (b) RT4P and RT4v6 cells were maintained with or without the indicated supplements for a minimum of 3 weeks and then subjected to blebbishield isolation and the blebbishields were plated in BE medium with 1 μM cisplatin. After 24 h, the surviving spheres were counted and plotted (n=4). Inset figure: morphological criteria for sphere count. (c) Correlation of p70S6K with blebbishield survival. RT4P and RT4v6 cells were maintained with or without the indicated supplements for a minimum of 3 weeks and then treated as indicated. Fresh blebbishields, blebbishield-depleted fractions, and control total cell populations were subjected to western blotting. Note that the full-length p70S6K from panel c correlated with the survival of blebbishields in panel b. (d) RT4P and RT4v6 cells were subjected to BE medium (with 1 mM cisplatin) treatment and the blebbishields were isolated at 24 h and counted (no amino acid and vitamin restriction were done for cell maintenance before the experiment). Vit., vitamins; ℒ-Glu, ℒ-Glutamine; MEM, minimal essential medium; NEAA, non-essential amino acid (except ℒ-glutamine)
We utilized this sphere-forming property to examine the role of amino acid-p70S6K signaling in blebbishield survival. We maintained the RT4P and RT4v6 cells with or without non-essential amino acids for 2–3 weeks and then examined sphere formation. The sphere formation of blebbishields from RT4P cells but not RT4v6 was significantly affected by NEAA deprivation (Figure 3b). Interestingly, full-length p70S6K levels correlated with the number of surviving spheres (Figure 3c correlated to Figure 3b). Since RT4v6 cells were more tumorigenic than RT4P cells, we examined whether these isogenic cell lines differ in the number of blebbishield formation. RT4v6 cells formed substantially less blebbishields than RT4P cells, indicating that the RT4v6 cells are more resistant to BE medium and p70S6K cleavage but the RT4v6 blebbishields have more survival capacity (Figure 3d compared with Figures 3b and c). These results suggest the requirement of intact 70S6K for blebbishield survival.
Differential expression profiling of floating (dead) and reattaching (sphere-forming) blebbishields and sphere characterization
Since blebbishield-derived spheres are several-fold larger than the original blebbishields (Figure 3a), we examined sphere formation by live time-lapse microscopy. Freshly isolated blebbishields fuse between themselves to form spheres (Supplementary Movie S2) and underwent morphological differentiation in normal medium (Supplementary Movie S3). In response to 10 μM cisplatin, the spheres pinched-off vesicles from the periphery (Supplementary Movie S4).
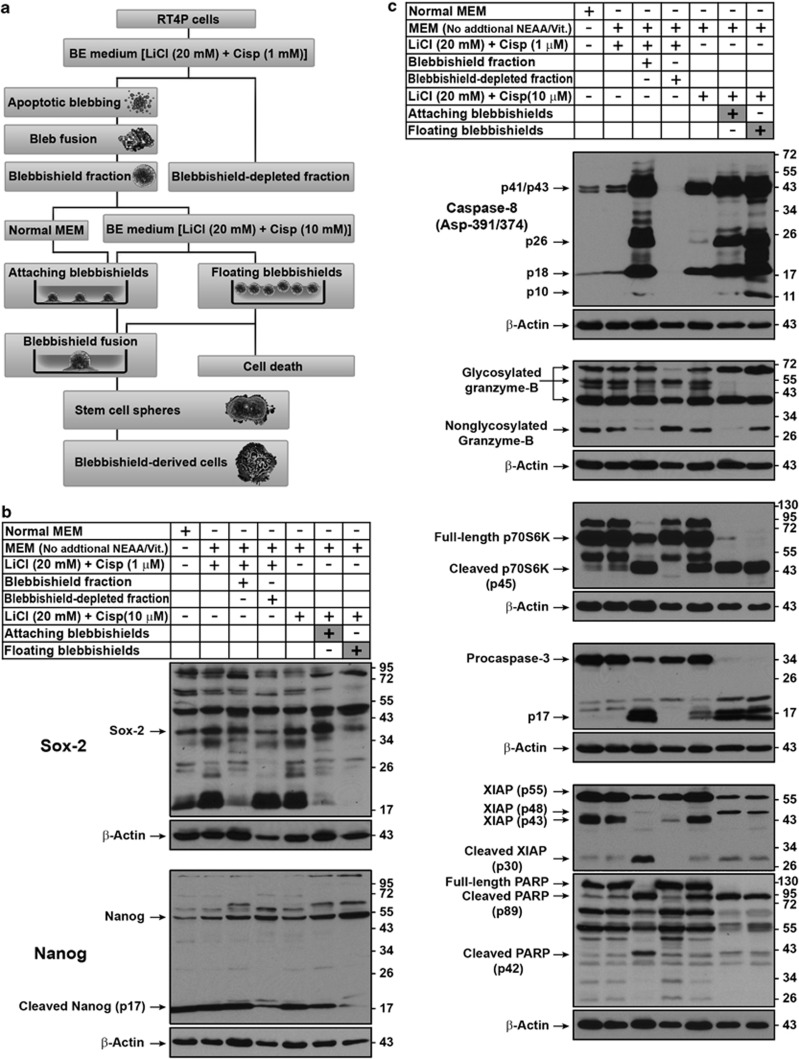
Since only a subset of blebbishields formed spheres by fusion (Supplementary Movie S2), we compared the floating and sphere-forming blebbishields to refine the key molecular signatures that are involved in sphere formation by western blotting (the scheme of the various experiments is shown in Figure 4a). Sox-2 and cleaved Nanog (p17) were expressed more in sphere-forming blebbishields (Figure 4b) whereas caspase-8 p10 and non-glycosylated granzyme-B (p28 form) were expressed more in floating blebbishields (Figure 4c), suggesting a role of caspases and protein glycosylation in blebbishield biology (examined below). Consistent with the data in Figure 3c, full-length p70S6K was feebly detected in sphere-forming blebbishields but not in floating blebbishields (Figure 4c). We found no difference in caspase-3 fragments between the floating and sphere-forming blebbishields; however, procaspase-3 was downgraded to p17 in both subpopulations (Figure 4c). The caspase-3 inhibitor XIAP had a distinct 48-kDa band in both floating and sphere-forming blebbishields (Figure 4c). Of note, caspase-3 is known to cleave XIAP into XIAP-p30 during apoptosis.23 Both sphere-forming and floating blebbishields had 89-kDa PARP fragment (Figure 4c).
Figure 4.
Refining the molecular profile of floating (dead) and sphere-forming (attaching) blebbishields. (a) Experimental plan to refine the molecular profile of floating versus sphere-forming blebbishields. See text for more details (the blebbishield-derived cells are characterized in Figures 5, 6, 7, 8). Cisp., cisplatin. (b) Western blotting analysis to compare the differences in stem-cell transcription factors Sox-2 and Nanog between floating and attaching blebbishields. (c) Western blotting analysis to compare the differences in crucial apoptotic regulators and p70S6K between freshly isolated blebbishields, reattaching blebbishields (sphere-forming), and floating (dead) blebbishields. Note the differences in expression of caspase-8 p10, non-glycosylated granzyme-B (∼28 kDa band) full-length p70S6K, XIAP (p48 fragment), and PARP (p42) bands. MEM, minimal essential medium; NEAA, non-essential amino acid (except ℒ-glutamine); Vit., vitamins; Cisp, cisplatin
The presence of active caspase-3, active caspase-8, 89-kDa cleaved PARP, increased Sox-2, and cleaved Nanog p17 in the reattaching blebbishields demonstrated that the apoptotic cells (blebbishields) are the initiators of spheres. Together, these data demonstrate that blebbishields survive by fusion to form spheres, resist cell death in response to higher-dose cisplatin, and the blebbishields with apoptotic signature are the initiators of spheres.
Involvement of caspases and N-linked glycosylation in blebbishield biology
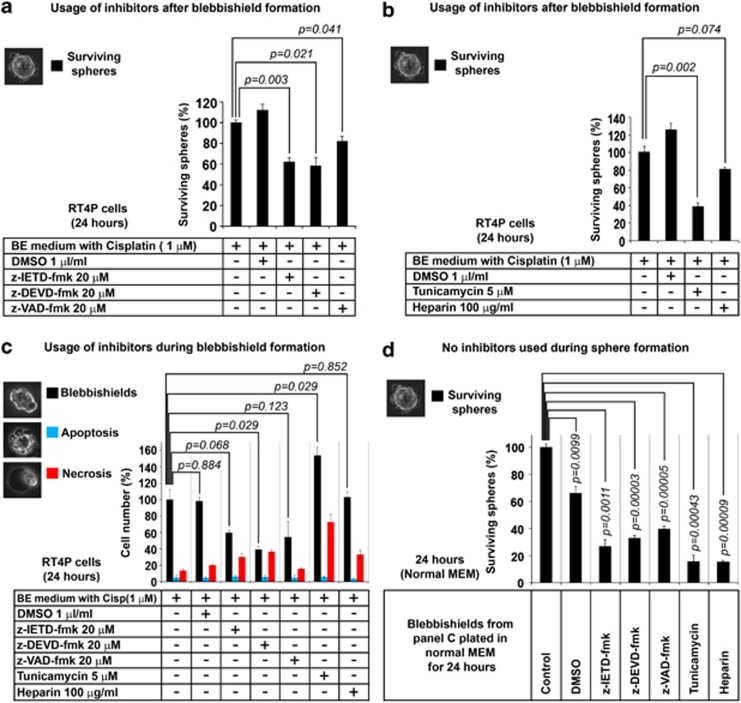
Since the sphere-forming blebbishields express active caspases and reduced levels of non-glycosylated granzyme-B, we examined the role of caspases and glycosylation inhibitors in sphere formation. Caspase inhibitors z-IETD-fmk, z-DEVD-fmk, and z-VAD-fmk reduced the sphere formation from blebbishields by 38.1, 41.4, and 18%, respectively (Figure 5a). To study the effect of glycosylation we used tunicamycin and heparin, both of which are known to act on granzyme-B glycosylation.24, 25 Both tunicamycin and heparin reduced the sphere formation by 61.4 and 19.3%, respectively (Figure 5b). These data demonstrate that caspases and N-linked glycosylation are involved in blebbishield to sphere formation stage.
Figure 5.
Role of caspases and N-linked glycosylation during sphere formation and blebbishield formation in RT4P cells. (a) Role of caspases in sphere formation from blebbishields. Blebbishields were incubated with caspase inhibitors for 24 h, and the surviving spheres (inset photomicrograph) were counted. (b) Role of tunicamycin (N-linked glycosylation inhibitor) and heparin (non-specific granzyme-B inhibitor) in sphere formation from blebbishields. Blebbishields were incubated with tunicamycin or heparin for 24 h and the surviving spheres (inset photomicrograph) were counted. (c, d) Effects of caspase inhibitors, tunicamycin and heparin on blebbishield generation (c, treatments in BE medium as indicated) and subsequent sphere formation in normal MEM without inhibitors (d). Blebbishield generation in the presence of inhibitors was measured 24 h after treatment and subsequent sphere formation was measured 24 h after plating in normal MEM. During counting, blebbishields, apoptotic cells and necrotic cells were determined based on morphology shown in the inset photomicrographs. P-values in c are for blebbishields only
Next, we examined the role of caspases and N-linked glycosylation in blebbishield formation by using these inhibitors during blebbishield generation and then allowing sphere formation in normal MEM. The caspase inhibitors z-IETD-fmk, z-DEVD-fmk, and z-VAD-fmk reduced blebbishield generation by 40.4, 61, and 45.9%, respectively (Figure 5c) and reduced subsequent sphere formation by 73.5% (z-IETD-fmk), 67.4% (z-DEVD-fmk), and 60.4% (z-VAD-fmk), respectively (Figure 5d). These data demonstrate that caspases are involved in blebbishield generation and that their inhibition during blebbishield generation reduces sphere formation from blebbishields.
In contrast, usage of tunicamycin but not heparin, during blebbishield generation increased blebbishield formation by 53.4% (Figure 5c). Tunicamycin also resulted in an increased proportion of necrotic cells (one-third of floating cells) (Figure 5c). However, the sphere-forming efficiencies of these blebbishields were significantly reduced (84.3% reduction by tunicamycin and 84.8% reduction by heparin) (Figure 5d), suggesting a predominant role of N-linked glycosylation in blebbishield generation than sphere formation (Figure 5b versus d). Together these results demonstrate that, caspases and protein N-linked glycosylation are involved in blebbishield generation and sphere formation.
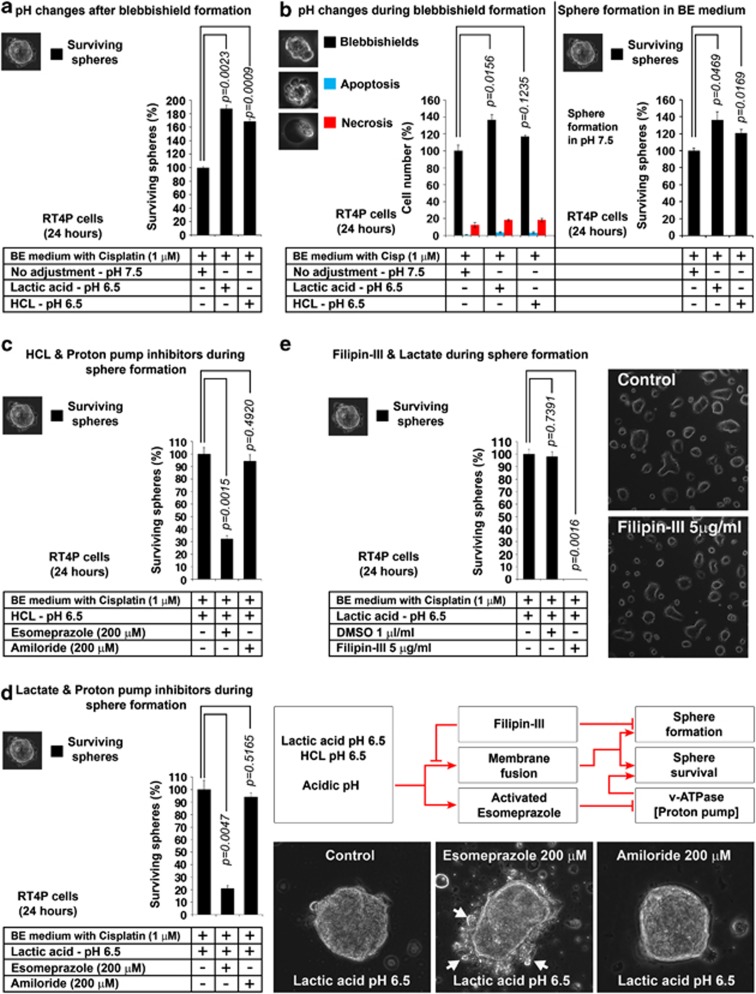
Acidic pH enhances sphere formation from blebbishields and is interfered by v-ATPase inhibitor and blocked by cholesterol inhibitor
Although fusion is facilitated by proteins and carbohydrates in the form of glycosylated proteins/lipids, membrane fusion primarily involves lipids. Acidic pH is known to facilitate lipid-mediated membrane fusion.26, 27 In vivo, the tumor microenvironment pH is known to be as low as 6.5 and the tumor cells are capable of avoiding intracellular acidification by proton pumps.28, 29 It has also been reported that the pH 6.5 promotes CSC sphere formation in vitro.29 Thus, we isolated blebbishields at pH 7.5 and allowed sphere formation in pH 6.5 (pH adjusted using HCl and lactic acid). Both acids significantly promoted sphere formation from blebbishields at pH 6.5 (HCl, 68.4% increase; lactic acid, 87.1% increase) (Figure 6a). We examined the effect of pH 6.5 during blebbishield formation and then subsequent sphere formation in pH 7.5 and found that, both acids promoted blebbishield formation at pH 6.5 (HCl, 17.1% increase; lactic acid 37% increase) and subsequent sphere formation (HCl, 20.9% increase; lactic acid, 36.2% increase) (Figure 6b).
Figure 6.
Effects of low pH, PPIs, and cholesterol inhibitors in blebbishield-mediated sphere formation (a) Effect of pH on sphere formation from blebbishields. Blebbishields isolated at pH 7.5 were plated in indicated pH for sphere formation. (b) Effect of pH on blebbishield formation (histogram on left) and determining the survival capacity of blebbishields from these conditions in normal pH 7.5 (histogram on right). (c) Proton pump v-ATPase inhibitor reduces sphere formation in the presence of HCl. (d) Proton pump v-ATPase inhibitor reduces sphere formation in the presence of lactic acid. Right panel shows visual indication of stress induced by esomeprazole and schematic diagram. (e) Filipin-III (freshly prepared) abolishes the lactate induced high frequency sphere formation. Photomicrographs shows that Filipin-III by itself is not toxic to RT4P cells
Cells are protected from acidic pH by proton pumps.28 Hence, we examined whether proton pump inhibitors (PPIs) such as esomeprazole (v-ATPase inhibitor) and amiloride (Na+/H+ exchanger inhibitor) could inhibit sphere formation. Esomeprazole reduced the sphere formation by 70% in the presence of HCl and 80% in the presence of lactate whereas, amiloride could not do so (Figures 6c and d). Importantly, the surviving spheres under esomeprazole but not amiloride treated conditions showed signs of stress by pinching off vesicles, indicating that the esomeprazole induces cell death in spheres (Figure 6d). Of note, PPIs are known to induce cell death.30 Next, we examined whether the sphere formation from blebbishields is simple aggregation of cells or involves membrane fusion. The cholesterol inhibitor Filipin-III could completely abolish the sphere formation from blebbishields (Figure 6e). Similar concentration of Filipin-III in RT4P cells had no apparent cell death (Figure 6e, photomicrograph), indicating that Filipin-III indeed prevented fusion. Together these results demonstrate that the pH 6.5 increases sphere formation from blebbishields and the same can be inhibited using esomeprazole by cell death induction and Filipin-III by preventing fusion.
Stemness, heterogeneity, and tumorigenicity of blebbishields
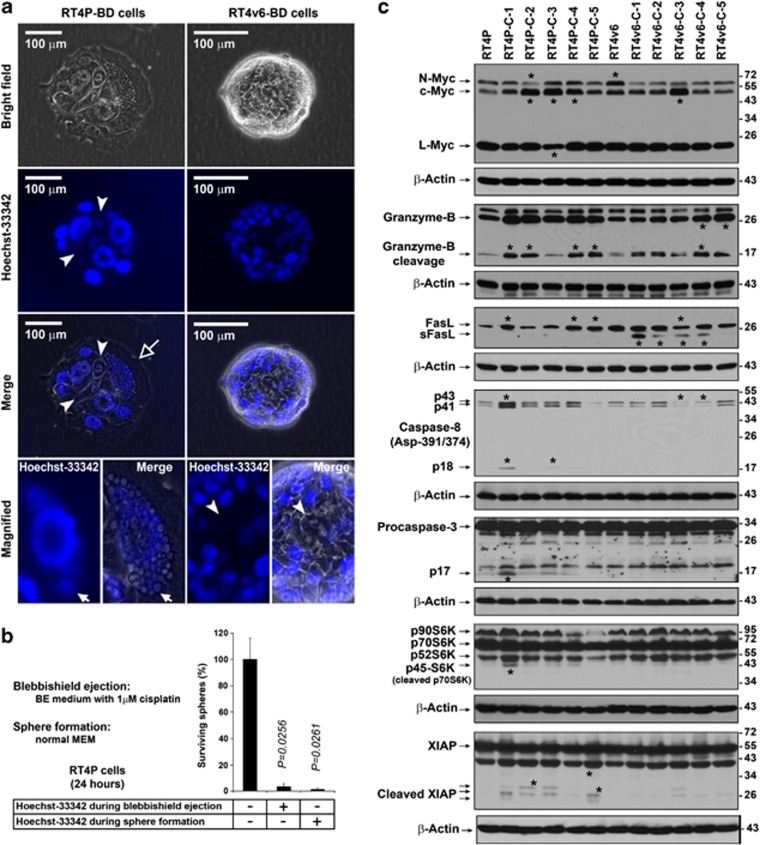
To examine the stemness of blebbishields, we cultured the blebbishield-derived spheres in normal MEM to allow morphological differentiation (Supplementary Movie S3). The colonies derived from RT4P blebbishields (but not from RT4v6 blebbishields) had at least one highly vacuolated cell per colony, with DNA-containing vacuoles (Figure 7a, magnified panel). Both RT4P and RT4v6 colonies derived from blebbishields had a few cells per colony with low Hoechst-33342 uptake, similar to side-population cells observed in cancer and normal stem cells5 (Figure 7a). The fresh blebbishield-derived spheres exhibited vesicles budding out from plasma membrane in response to Hoechst-33342 in normal MEM (Supplementary Movie S5).
Figure 7.
Blebbishield-derived cells (BD Cells) are susceptible to Hoechst-33342 and are heterogeneous in molecular profile. (a) Low Hoechst-33342 positivity and vacuolation in colonies derived from blebbishields. Blebbishields were allowed to differentiate in normal MEM and the colonies were stained with live stain Hoechst-33342 and imaged. Note the nuclei stained with low intensity similar to that of side-population cells (arrowheads). RT4P blebbishield-derived cells had at least one highly vacuolated cell per colony (hollow arrow). Note the DNA content in vacuoles of RT4P blebbishield-derived cells (solid arrows). The interior of the colony displayed no intense Hoechst-33342 positivity even after 90 min of incubation in Hoechst-33342 containing medium. (b) Effect of Hoechst-33342 in sphere survival. Hoechst-33342 (10 nM) was treated during or after blebbishield generation (24 h) and the surviving spheres were plotted. (c) Heterogeneity analysis of colonies (C1–C5) derived from single blebbishields of RT4P and RT4v6 cells by western blotting. Note the difference in levels of expression of myc isoforms, cleaved granzyme-B, s-FasL, active caspase-8, active caspase-3, cleaved p70S6K and cleaved XIAP. Asterisks, bands with major differences between colonies
Treatment with Hoechst-33342 either during blebbishield generation or during sphere formation drastically reduced the survival of spheres (reduced from 100 to 3.3% and from 100 to 1.5%, respectively), indicating that spheres derived from blebbishields are susceptible to Hoechst-33342 (Figure 7b). Interestingly, the RT4P colonies derived from blebbishields were able to undergo second round of blebbishield formation (second generation blebbishields; Blebbishield Serial Ejection: BSE-2) with increased frequency (minimum of one blebbishield per every colony) (Supplementary Figure S3a). The RT4v6 blebbishield-derived colonies had Hoechst-33342High cells at the periphery and Hoechst-33342Low cells in the middle of the colony. In response to BSE-2 in the presence of Hoechst-33342, the Hoechst-33342High cells underwent pyknosis first (due to Hoechst-33342), and the Hoechst-33342Low cells underwent blebbishield formation later (Supplementary Figure S3b). These results suggest the presence of blebbishield forming cells (stem cells) in all colonies derived from blebbishields.
Aggressive cancers have intratumoral heterogeneity due to CSC heterogeneity.7, 31 Analysis of BSE-2 colonies derived from single blebbishields of RT4P and RT4v6 cells showed that the blebbishield-derived colonies are heterogeneous in expression of myc isoforms, granzyme-B cleavage, FasL, caspase-8 Asp-391/374 cleavage, caspase-3, S6K isoforms, and XIAP, as the colonies had different levels of these proteins or their cleaved forms (Figure 7c). The increased expression of c-Myc in colonies derived from blebbishields (Figure 7c) indicates that, the c-Myc downregulation observed in freshly isolated blebbishields (Figure 2b) was a transient phenomenon.
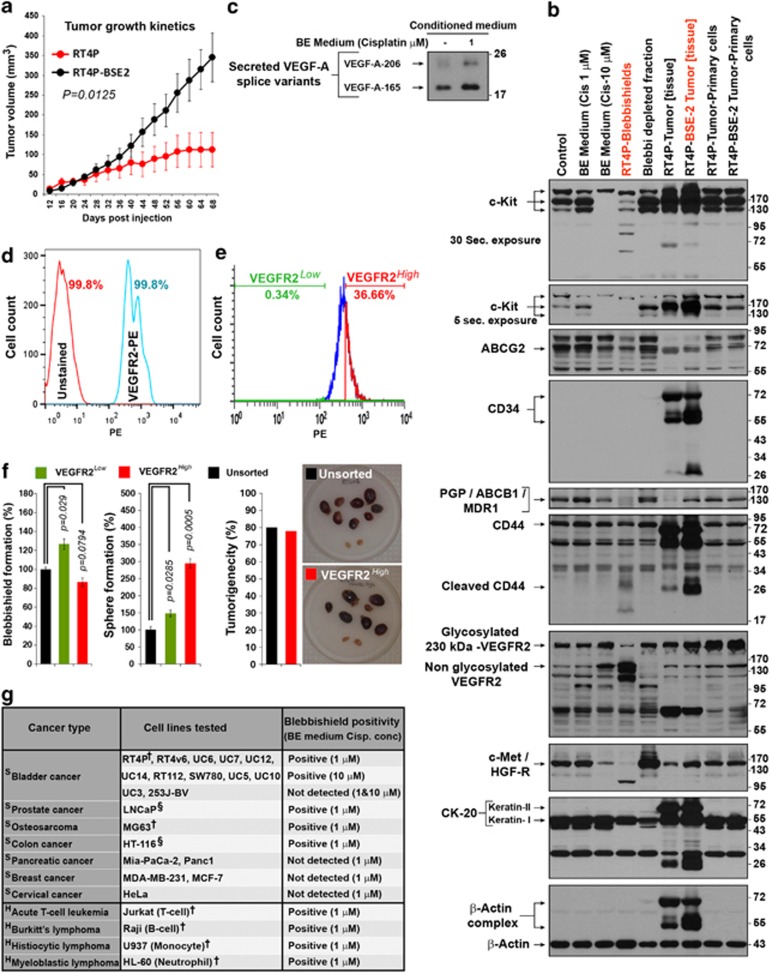
Tumorigenicity is the gold-standard to confirm stemness. Subcutaneous injection of RT4P and RT4P-BSE-2 cells (blebbishield-derived cells) in nude mice show that, the blebbishield-derived cells are tumorigenic (Supplementary Figure S4a) and that, the RT4P-BSE-2 tumors grew faster than the RT4P-tumors (Figure 8a).
Figure 8.
Determination of tumorigenic potential and stem-cell marker profiling of blebbishield-derived cells. (a) Comparative tumor growth pattern of RT4P and RT4P-BSE-2 cells. Note that, the RT4P-BSE-2 tumors grow faster than RT4P cells (representative of two independent experiments; error bars: S.E.; n=6; P=0.0125). (b) Stem-cell marker profiling of RT4P cells, RT4P-blebbishields, RT4P-BSE-2 tumor and RT4P-BSE-2 primary cells from tumor by western blotting. Though blebbishields had downregulation of PGP/ABCB1, glycosylated VEGFR2, and c-Met, they retained feeble bands and hence were not negative for expression. Also note, these markers were restored in RT4P-BSE-2 tumor (Frozen tissue). Please see text for more details. (Note: CK-20 is superficial bladder wall marker, also note the SDS-resistant actin complexes). (c) VEGF-A secretion is increased in BE medium treated conditioned medium detected by western blotting. (d) Flowjo merged FACS histogram of RT4v6 viable single cells showing complete positivity of VEGFR2 (PE-conjugated VEGFR2 stained RT4v6 cells compared with unstained cells) (e) FACS sorting of VEGFR2High and VEGFR2Low populations from PE stained RT4v6 viable single cells of panel d. (f) Sphere formation, blebbishield formation, and tumorigenicity of sorted and unsorted RT4v6 cells mice per group (n=10). Note the hemorrhagic nature of tumors and one mice from VEGFR2High cell injected group was dead by cannibalism with unknown tumor status hence n=9 for this group. (g) List of cell lines that are capable of forming blebbishields. Note: The efficiency of blebbishield varies greatly between cell lines and the purity is relatively high in RT4 (RT4P) cells. The cells with more mitotic cells (exposed from the monolayer) are not suitable for isolation of blebbishields. †Blebbishields were confirmed by time-lapse microscopy; §Less prominent blebbing; H Hematologic cancers; S Solid tumor. The Lithium concentration of BE medium was 20 mM
The data on expression of stem-cell transcription factors, low Hoechst-33342 positivity, chemoresistance, capability for BSE and in vivo tumorigenicity together confirm that the sphere-forming blebbishields are true stem cells.
Stem-cell marker profiling of blebbishields
Western blotting had revealed that the RT4P blebbishields expressed lower levels of c-Kit, ABCB1/P-glycoprotein, CD44, 230-kDa VEGFR2 and c-Met than RT4P-BSE-2 tumor tissue, indicating the transient downregulation of these markers (Figure 8b). The RT4P-BSE-2 tumor tissue expressed higher levels of c-Kit, CD34, ABCB1/P-glycoprotein, 230-kDa VEGFR2, cleaved CD44, and c-Met than the RT4P tumor (Figure 8b). Notably, the high expression of CD34, c-Kit, and CD44 in vivo are substantially reduced or lost upon adaptation back to in vitro by primary culture (Figure 8b). Of note, these markers are reported in HSCs and various CSCs.4, 5, 32 The superficial bladder wall marker CK-20 was also detected in RT4P and blebbishields (Figure 8b).
Newly synthesized VEGFR2 (Flk-1/KDR) is 150 kDa, partially glycosylated VEGFR2 is from 180 to 200 kDa and heavily glycosylated mature VEGFR2 is 230 kDa in SDS–PAGE analysis.33 Although the 230-kDa glycosylated VEGFR2 was transiently downregulated in blebbishields, the 150-kDa VEGFR2 was significantly increased in blebbishields (Figure 8b), suggesting a role of VEGFR2-glycosylation in blebbishield biology. VEGFR2 is known to get downregulated during apoptosis.34 Alternatively, VEGF stimulation can downregulate VEGFR2.35 230-kDa VEGFR2 downregulation in blebbishields is already in the context of apoptosis and hence we examined the conditioned BE medium and detected the presence of VEGF-A autocrine loop during blebbishield generation (Figure 8c).
To examine whether VEGFR2 is a specific marker for blebbishields that are tumorigenic, we examined the VEGFR2 positivity of RT4v6 cells and found that, RT4v6 cells are completely positive for VEGFR2 (Figure 8d). We sorted the RT4v6 cells into VEGFR2High and VEGFR2Low populations and found that, both populations are capable of forming blebbishields, but VEGFR2Low cells formed more blebbishields than VEGFR2High cells (P=0.006) (Figures 8e and f). However, the sphere-forming potential of VEGFR2High cells were 146% more than VEGFR2Low cells (P=0.00155) (Figure 8f). These data are analogous to the sphere-forming and tumorigenic potential differences between RT4P and RT4v6 cells (Figures 1a and 3b–d versus Figure 8f), indicating that the stem cells with more tumorigenic capacity are capable of more sphere formation. VEGFR2High cells were equally tumorigenic as unsorted RT4v6 cells when injected orthotopically (Figure 8f; Supplementary Figures S4b and c). The hemorrhagic nature of tumors further supports the role of VEGFR2 (Figure 8f). Interestingly, VEGFR2 is already implicated in tumorigenicity and stemness.32, 36 These data together demonstrate that, VEGFR2 is clearly linked to blebbishield-mediated sphere formation and tumorigenesis.
We had identified additional cell lines that formed, and that not formed blebbishields (Figure 8g), indicating that the blebbishield emergency program is not restricted to RT4P and RT4v6 cells.
Concluding remarks
Although our study identified multiple aspects of blebbishield biology, the fusion of apoptotic blebs, retention of full-length p70S6K, pH, and lipid-dependent fusion between blebbishields to form spheres and VEGFR2-mediated sphere survival are the key aspects. Multiple evidences from our study link VEGFR2 with blebbishield biology such as, tunicamycin inhibit sphere formation (Figure 5d) and also reported to downregulate VEGFR2.37 Heparin binds VEGFR2 at the same motif where VEGF-A binds,38 hence heparin might block sphere formation by interfering VEGF-A engagement with VEGFR2. Furthermore, acidic pH is known to influence VEGF production and bioavailability.39, 40
In summary, our study has identified a novel aspect of CSC biology in which cancer cells evoke an emergency cell survival program after activation of morphological (membrane blebbing) and biochemical apoptosis (caspase activation and multiple caspase/granzyme-B substrate cleavages).
Materials and Methods
Cells and maintenance
Human bladder cancer cells RT4 (ATCC, Manassas, VA, USA, Cat. number HTB-2; referred to in this study as RT4P to denote parental cells) and RT4v6 (generated by AMK by six serial passages of RT4 cells in nude mice and identity confirmed by DNA fingerprinting) were maintained in MEM with 10% fetal bovine serum, ℒ-glutamine, pyruvate, non-essential amino acids, vitamins, penicillin, and streptomycin, unless and otherwise specified (for other cell lines see Figure 7g and Supplementary Figure S2c). The RT4v6 cells were maintained in vitro for more than a year allowing adaptation back to tissue culture and their identity was periodically verified by DNA fingerprinting.
Reagents and antibodies
Lithium chloride (L-4408), Tunicamycin (T7765), Heparin (H3393), Filipin-III (F4767), Amiloride (A7410), Esomeprazole (E7906), and Lactic acid (L1875) were from Sigma (St Louis, MO, USA). Cisplatin (NDC 0015–3220–22) was purchased from Bristol Laboratories (Princeton, NJ, USA). Phospho-antibodies to Akt-Thr-308 (9275), Akt-Ser-473 (587F11), total Akt (9272), c-Myc (D84C12), phospho Thr-389 p70S6K (9206), p70S6K (2708), phospho GSK-3β (Ser-9) (9336), GSK-3β (9332), Nanog (3580), Sox-2 (2748), Oct4 (4286), phospho 4EBP1 Thr-70 (9455), 4EBP1 (9644), Granzyme-B (4275), FasL (4273), Caspase-8 Asp-391/374 (9496), c-Kit (3074P), ABCG2 (4477S), and CD44 [8E2] (5640) were from Cell Signaling Technology, Beverly, MA, USA. Antibodies to Caspase-8 anti-p18 (Sc-7890), Caspase-9 (Sc-7885), Caspase-3 (Sc-7148), PARP-1 (Sc-7150), CD34 (Sc-65261), P-glycoprotein/MDR1 (Sc-8313), VEGFR2 (Sc-504), VEGF-A (Sc-7269), and c-Met (C28) (Sc-161) were from Santa Cruz Biotechnology, Santa Cruz, CA, USA. CD95/Fas (610197) antibody was purchased from Transduction Laboratories, Lexington, KY, USA. PE-conjugated VEGFR2 (FAB357P), antibody was purchased from R&D Systems (Minneapolis, MN, USA).
Microarray analysis
Transcriptome data were generated from RT4P and RT4v6 cells as described below. Total RNA was isolated from RT4P and RT4v6 cells (MirVana kit, Ambion, Austin, TX, USA) and was subjected to biotinylated cRNA synthesis (Illumina RNA amplification kit, Ambion). The cRNAs were then hybridized to Illumina Human-HT12v3 chips (Illumina, San Diego, CA, USA). After washing, the chips were scanned with BeadStation (Illumina) and the signal intensities were quantified with Genome Studio (Illumina). Quantile normalization was used to normalize the data. To generate heat map, each gene's values adjusted to be a mean of zero, were used with Cluster and Tree View.41
Blebbishield isolation, blebbishield serial ejection (BSE), and blebbishield heterogeneity analysis
RT4P and RT4v6 cells were plated at super-high densities (1 × 107 to 8 × 107 cells/T-75 flask/15 ml medium) so that the cells became complete monolayer in 24 h while minimizing their tendency to cause clumped overgrowth (RT4v6 cells do not form complete monolayer unless plated at super-high densities). Twenty-four hours after plating, the cells were exposed to Blebbishield Ejection Medium (BE medium: 10% FBS containing MEM with 20 mM freshly prepared LiCl and 1 μM cisplatin, with essential amino acids (Invitrogen 11700–077) without additional non-essential amino acids (NEAA) except ℒ-glutamine and without additional vitamin supplements). The cisplatin concentration was shifted to 10 μM for certain experiments as indicated in the text. At 24 h post-treatment, blebbishields (apoptotic cells with bleb fusion) were collected by gently streaming the existing medium over the monolayer using a 10-ml wide-mouth pipette three times (touching the monolayer lawn with pipette was avoided to prevent dislodging the adherent cells) and the medium was collected in 50 ml tubes (the remaining adherent cells were washed with ice-cold PBS and scrapped in PBS, pelleted, and lysed to get blebbishield-depleted fractions). The blebbishields were pelleted down at 1200 r.p.m. for 3 min, resuspended gently with wide-mouthed pipette tips, counted in 96- or 24-well plates and plated as indicated.
To ensure purity, two sequential blebbishield ejections (BSE-1 and then BSE-2 from BSE-1 cells) were performed. Briefly, isolated blebbishields (BSE-1) were cultured in normal MEM to expand cells for 30 days. On day 30, the BSE-1 cells were subjected to a second round of blebbishield ejection to get BSE-2 cells. This fresh blebbishield preparation (BSE-2) was subjected to limiting dilution in 96-well plates and single blebbishields were allowed to undergo colony expansion. Five individual colonies per cell line (RT4P and RT4v6) were expanded and analyzed for heterogeneity by western blotting.
Evaluation of VEGFR2 in blebbishield, sphere, and tumor formation
Phycoerythrin-conjugated VEGFR2 antibody stained RT4v6 cells were filtered for single cells, gated for viable single cells, and sorted into VEGFR2High and VEGFR2Low cells using BD Influx sorter. The sorted cells were cultured for 1–2 weeks for blebbishield and sphere-formation assays. The sorted cells were cultured for 5 days before orthotopic mice injections (described below).
Scanning electron microscopy of blebbishields
Blebbishields were isolated as described above, fixed (3% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer pH 7.3) and post-fixed in 1% cacodylate buffered osmium tetroxide followed by 1% tannic acid and 1% uranyl acetate. The blebbishields were dehydrated using graded series of ethanol followed by hexamethyldisilazane and air dried before mounting and platinum and flash carbon coatings. The blebbishields were imaged at an accelerating voltage of 5 kV using JSM-5910 scanning electron microscope (JEOL, USA Inc., Peabody, MA, USA).
Transmission electron microscopy of blebbishields
Blebbishields were isolated as described above fixed (3% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer pH 7.3) and post-fixed in 1% cacodylate buffered osmium tetroxide followed by 1% tannic acid and 1% uranyl acetate. The blebbishields were dehydrated using graded series of ethanol followed by infiltration and embedding in LX-112 medium and polymerization. Ultra-thin sections were cut using Leica Ultracut microtome (Leica, Deerfield, IL, USA), stained with uranyl acetate and lead citrate and examined in JEM 1010 transmission electron microscope (JEOL, USA Inc.) at an accelerating voltage of 80 kV.
Determination of amino-acid dependency of blebbishields for survival
RT4P and RT4v6 cells were maintained in MEM with or without non-essential amino acids (NEAA: except ℒ-glutamine) supplement for a minimum of 2–4 weeks and then were induced to undergo blebbishield ejection using BE medium as described above. The blebbishields were counted in 24- or 96-well plates, normalized and plated on six-well plates in BE medium for 24 h and the surviving spheres were counted and plotted (n=4).
Evaluation of the role of caspases, N-linked glycosylation, Hoechst-33342, pH, PPIs, and cholesterol inhibitor in blebbishield generation and/or sphere formation
To evaluate the effects of caspase inhibitors, tunicamycin and heparin in blebbishield generation, 3 × T–75 confluent flasks of RT4P cells per condition were subjected to blebbishield isolation in the presence or absence of various inhibitors as indicated in the figures for 24 h (15 ml medium/T–75 flask). The freshly isolated blebbishields from each flask were resuspended in 2 ml of normal MEM and plated in 96-well plates (100 μl/well) in triplicate. The cells were allowed to settle down for 5 min, and then blebbishields, apoptotic cells and necrotic cells, were counted based on morphology depicted in the inset photomicrographs in Figure 5. The remaining 1.7 ml of resuspended blebbishields/from-each-flask were plated in six-well plates with an additional 1.3 ml of normal MEM (the sphere formation phase was allowed in the absence of inhibitors) to evaluate the ability of these blebbishields to form spheres (in triplicate). Throughout the study, the caspase inhibitors were kept constant (20 μM) since high doses of caspase inhibitors can induce cell death.
To evaluate the role of caspases and N-linked glycosylation in sphere formation from blebbishields, 16 confluent T–75 flasks of RT4P cells (for each agent) were subjected to blebbishield isolation as described above. The freshly isolated blebbishields were pooled and then equally distributed to various treatment conditions (caspase inhibitors, Hoechst-33342, N-glycosylation/non-specific granzyme-B inhibitors) in BE medium with 1 μM cisplatin and plated in six-well plates in triplicate. After 24 h, the floating cells were washed off, and the adherent spheres were counted.
The effect of pH was evaluated in similar fashion where the media pH was adjusted using HCl or lactic acid. PPIs were evaluated for both pH adjustment by HCL and lactic acid. Filipin was evaluated at the pH of 6.5 (using lactic acid).
Animal experiments
Determination of in vivo tumorigenic potential of RT4P versus RT4v6 cells and RT4v6 versus RT4v6-VEGFR2High cells
Male nude mice (athymic NCr-nu/nu, NCI, Bethesda, MD, USA) were injected with 2 × 105 RT4P or RT4v6 cells (in 40 μl of HBSS) orthotopically into the bladder wall (n=12 for each cell line) as previously described.42 The mice were observed for tumor formation by palpation and morbidity signs such as hematuria, loss of weight, hunched posture, etc. Both groups of mice were euthanized at day 34 post-injection, which is the time at which the RT4v6 cell injected mice (but not RT4P cell injected mice) reached the euthanization point as per Institutional Animal Care and Usage Committee (IACUC) guidelines. Tumorigenicity experiments on RT4v6 versus RT4v6-VEGFR2High cells were performed similarly but with 3.5 × 105 cells/40 μl/mice (n=10 for each cell type) injections orthotopically. The mice were euthanized on day 23 post-injection due to mortality in VEGFR2High group, as per IACUC guidelines on tumor burden and the bladders were evaluated for tumor formation, and tumor weight.
Determination of in vivo tumorigenic potential of RT4P blebbishield-derived cells
RT4P cells were subjected to two BSEs as described above (typically obtained 200–400 spheres/isolation). RT4P-BSE-2 and RT4P cells (3 × 106 cells in HBSS representing the original 200–400 spheres) were injected subcutaneously in the thigh of nude mice. The tumors were measured to study the growth kinetics, and the tumor volume was calculated as described previously.43
All animal experiments performed in this study were approved and in accordance with theprotocols of MD Anderson Cancer Center Institutional Animal Care and Use Committee (IACUC).
Western immunoblotting
Cells or blebbishields were lysed in lysis buffer (50 mM Tris–HCl, pH 7.4; 150 mM NaCl; 5 mM EDTA; 25 mM NaF; 1% Triton-X 100; 1% NP-40; 0.1 mM Na3VO4; 12.5 mM β-glycerophosphate; 1 mM PMSF and complete protease inhibitor cocktail (Roche). The lysates were clarified at 13 000 r.p.m. for 10 min, and the supernatants were quantified and subjected to SDS-PAGE and western blotting on nitrocellulose membranes.
For differential expression profiling of floating and sphere-forming blebbishields by western blotting, freshly isolated blebbishields from 40–60 × T–75 flasks of RT4P cells were allowed to attach for a short period of 1 h in fresh medium and the floating blebbishields and attaching blebbishields were collected separately. To ensure purity of the attaching blebbishields, we rinsed the flask twice with 15 ml of PBS each before scrapping off the attached blebbishields.
Stem-cell marker profiling
Various stages of RT4P-derived blebbishield biology such as, fresh blebbishields, blebbishield-derived tumors (snap frozen tissue), and primary cells derived from tumors (maintained for 30 days) were analyzed for well-established stem-cell markers by western blotting.
Analysis of VEGF secretion
A total of 1 × 107 RT4P cells in 15 ml of MEM were plated and 24 h later challenged with or without BE medium for 24 h. The conditioned media were collected, clarified at 3500 r.p.m. for 5 min and stored at −20 oC until analysis by western blotting. In all, 30 μl of conditioned media were subjected to western blotting.
2D imaging and time-lapse microscopy
All imaging studies were performed with an Olympus IX81 microscope (Leeds Precision Instruments Inc., Irving, TX, USA (currently, Olympus America Inc., Miami, FL, USA)), equipped with a heating stage (37 oC) and CO2 chamber (5%). The images were exported to TIFF files using SlideBook software (version SB 4.2.0.12) (Intelligent Imaging Innovations, Inc., Denver, CO, USA) and were converted to QuickTime movie format using Lightwave 3D (NewTek, Inc., San Antonio, TX, USA) or Adobe AfterEffects CS5 (15 fps, 1000 kbps, 480 × 360, NTSC, mpeg) or composited using Adobe Photoshop CS5 softwares (Adobe Systems Inc., San Jose, CA, USA).
Statistical analyses
Statistical analyses were performed using Microsoft Excel 2010. The statistical significance was determined based on Student's t-test with two-tailed distribution and two-sample unequal variance and the P-values <0.05 were considered significant. The error bars represent standard errors.
Acknowledgments
We sincerely thank Dr. David J McConkey for generously providing microarray data, cell lines, and for intellectual and editorial help with the manuscript, and Drs Bharat B. Aggarwal, Gomez-Manzano Candelaria, Felipe Samaniego, Isaiah J Fidler, Sun-Jin Kim, Guillermo Garcia-Manero, Xiongbin Lu, Chun Li, Gordon B Mills, and Molina Jennifer for providing cell lines/reagents, Dr. Robert R Langley and Mr. Kenneth Dunner Jr. for helping with electron microscopy, Mr. Kunal Mandal and Ms. Karen Ramirez for helping in cell sorting, Dr. Aron Mobley, Dr. Neema Navai and Ms. I-Ling Lee for various helps, Ms. Deming Stephanie for editorial assistance with the manuscript. GJG was supported by MD Anderson Cancer Center and the Clayton Foundation for Research. This research was supported in part by the National Institutes of Health through MD Anderson Cancer Center's Support Grant, CA016672.
Author contributions
GJG designed, performed all experiments (except microarray), interpreted and documented results, and wrote the manuscript. WC contributed to the microarray experiment. JBS, EKL, DLW, and GJG performed the orthotopic mice experiments. AMK provided expert guidance in the design of experiments and interpretation of data, writing the manuscript, and generating the RT4v6 cell line.
Glossary
- p70S6K
70-kDa ribosomal protein S6 kinase
- VEGF-A
vascular endothelial growth factor-A
- VEGFR2
vascular endothelial growth factor receptor-2
- GSK-3β
glycogen synthase kinase-3β
- BSE-2
blebbishield serial ejection-2
- CSCs
cancer stem cells
- TSC-2
tuberous sclerosis complex-2
- NEAA
non-essential amino acids
- 4EBP1
4E binding protein-1
- PARP
poly-ADP ribose polymerase
- BE medium
blebbishield ejection medium
- XIAP
X-linked inhibitor of apoptosis
- PPI
proton pump inhibitor
- ABCB1
ATP-binding cassette subfamily B member-1
- CD44
cluster of differentiation 44
- CK-20
cytokeratin 20
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death & Differentiation website (http://www.nature.com/cdd)
Edited by R De Maria
Supplementary Material
References
- Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2:18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Li F, He Z, Shen J, Huang Q, Li W, Liu X, et al. Apoptotic caspases regulate induction of iPSCs from human fibroblasts. Cell Stem Cell. 2010;7:508–520. doi: 10.1016/j.stem.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K, et al. Cancer stem cell markers in common cancers—therapeutic implications. Trends Mol Med. 2008;14:450–460. doi: 10.1016/j.molmed.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Golebiewska A, Brons NH, Bjerkvig R, Niclou SP. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell. 2011;8:136–147. doi: 10.1016/j.stem.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Maugeri-Sacca M, Vigneri P, De Maria R. Cancer stem cells and chemosensitivity. Clin Cancer Res. 2011;17:4942–4947. doi: 10.1158/1078-0432.CCR-10-2538. [DOI] [PubMed] [Google Scholar]
- Cho RW, Clarke MF. Recent advances in cancer stem cells. Curr Opin Genet Dev. 2008;18:48–53. doi: 10.1016/j.gde.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Nunez R, Sancho-Martinez SM, Novoa JM, Lopez-Hernandez FJ. Apoptotic volume decrease as a geometric determinant for cell dismantling into apoptotic bodies. Cell Death Differ. 2010;17:1665–1671. doi: 10.1038/cdd.2010.96. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- Gleason CE, Ning Y, Cominski TP, Gupta R, Kaestner KH, Pintar JE, et al. Role of insulin-like growth factor-binding protein 5 (IGFBP5) in organismal and pancreatic beta-cell growth. Mol Endocrinol. 2010;24:178–192. doi: 10.1210/me.2009-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell. 2006;24:185–197. doi: 10.1016/j.molcel.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JL, Bonavaud SM, Toole BJ, Yeaman SJ. Regulation of glycogen synthesis by amino acids in cultured human muscle cells. J Biol Chem. 2001;276:952–956. doi: 10.1074/jbc.M004812200. [DOI] [PubMed] [Google Scholar]
- Dhar R, Persaud SD, Mireles JR, Basu A. Proteolytic cleavage of p70 ribosomal S6 kinase by caspase-3 during DNA damage-induced apoptosis. Biochemistry. 2009;48:1474–1480. doi: 10.1021/bi801840s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou C, Elia A, Clemens MJ. Activation of p53 stimulates proteasome-dependent truncation of eIF4E-binding protein 1 (4E-BP1) Biol Cell. 2008;100:279–289. doi: 10.1042/BC20070121. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Yoneda K, Ueta E, Doi S, Osaki T. Enhanced apoptosis of squamous cell carcinoma cells by interleukin-2-activated cytotoxic lymphocytes combined with radiation and anticancer drugs. Eur J Cancer. 2000;36:2007–2017. doi: 10.1016/s0959-8049(00)00244-6. [DOI] [PubMed] [Google Scholar]
- Andrade F, Roy S, Nicholson D, Thornberry N, Rosen A, Casciola-Rosen L. Granzyme B directly and efficiently cleaves several downstream caspase substrates: implications for CTL-induced apoptosis. Immunity. 1998;8:451–460. doi: 10.1016/s1074-7613(00)80550-6. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Leo E, Stennicke HR, Welsh K, Salvesen GS, Reed JC. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher JA, Bertozzi CR. Chemical technologies for probing glycans. Cell. 2006;126:851–854. doi: 10.1016/j.cell.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Veugelers K, Motyka B, Goping IS, Shostak I, Sawchuk T, Bleackley RC. Granule-mediated killing by granzyme B and perforin requires a mannose 6-phosphate receptor and is augmented by cell surface heparan sulfate. Mol Biol Cell. 2006;17:623–633. doi: 10.1091/mbc.E05-07-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Leikina E, Frolov V, Bronk P, Zimmerberg J. An early stage of membrane fusion mediated by the low pH conformation of influenza hemagglutinin depends upon membrane lipids. J Cell Biol. 1997;136:81–93. doi: 10.1083/jcb.136.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais S, De Milito A, You H, Qin W. Targeting vacuolar H+-ATPases as a new strategy against cancer. Cancer Res. 2007;67:10627–10630. doi: 10.1158/0008-5472.CAN-07-1805. [DOI] [PubMed] [Google Scholar]
- Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, MacSwords J, Lathia JD, et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011;18:829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais S. Proton pump inhibitor-induced tumour cell death by inhibition of a detoxification mechanism. J Intern Med. 2010;267:515–525. doi: 10.1111/j.1365-2796.2010.02225.x. [DOI] [PubMed] [Google Scholar]
- Raaijmakers MH, Scadden DT. Divided within: heterogeneity within adult stem cell pools. Cell. 2008;135:1006–1008. doi: 10.1016/j.cell.2008.11.034. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Malik AK, Solar GP, Sherman D, Liang XH, Meng G, et al. VEGF regulates haematopoietic stem cell survival by an internal autocrine loop mechanism. Nature. 2002;417:954–958. doi: 10.1038/nature00821. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Shibuya M. The 230kDa mature form of KDR/Flk-1 (VEGF receptor-2) activates the PLC-gamma pathway and partially induces mitotic signals in NIH3T3 fibroblasts. Oncogene. 1997;14:2079–2089. doi: 10.1038/sj.onc.1201047. [DOI] [PubMed] [Google Scholar]
- You WK, Kasman I, Hu-Lowe DD, McDonald DM. Ricinus communis agglutinin I leads to rapid down-regulation of VEGFR-2 and endothelial cell apoptosis in tumor blood vessels. Am J Pathol. 2010;176:1927–1940. doi: 10.2353/ajpath.2010.090561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RD, Srinivasan S, Singh AJ, Mahoney JE, Gharahassanlou KR, Rahimi N. PEST motif serine and tyrosine phosphorylation controls vascular endothelial growth factor receptor 2 stability and downregulation. Mol Cell Biol. 2011;31:2010–2025. doi: 10.1128/MCB.01006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamerlik P, Lathia JD, Rasmussen R, Wu Q, Bartkova J, Lee M, et al. Autocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J Exp Med. 2012;209:507–520. doi: 10.1084/jem.20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Lang JY, Hung MC, Sengupta K, Banerjee SK, Baksi K, et al. Unfolded protein response is required in nu/nu mice microvasculature for treating breast tumor with tunicamycin. J Biol Chem. 2011;286:29127–29138. doi: 10.1074/jbc.M110.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougher AM, Wasserstrom H, Torley L, Shridaran L, Westdock P, Hileman RE, et al. Identification of a heparin binding peptide on the extracellular domain of the KDR VEGF receptor. Growth Factors. 1997;14:257–268. doi: 10.3109/08977199709021524. [DOI] [PubMed] [Google Scholar]
- Xu L, Fukumura D, Jain RK. Acidic extracellular pH induces vascular endothelial growth factor (VEGF) in human glioblastoma cells via ERK1/2 MAPK signaling pathway: mechanism of low pH-induced VEGF. J Biol Chem. 2002;277:11368–11374. doi: 10.1074/jbc.M108347200. [DOI] [PubMed] [Google Scholar]
- Taraboletti G, D'Ascenzo S, Giusti I, Marchetti D, Borsotti P, Millimaggi D, et al. Bioavailability of VEGF in tumor-shed vesicles depends on vesicle burst induced by acidic pH. Neoplasia. 2006;8:96–103. doi: 10.1593/neo.05583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Nat Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat AM, Karashima T, Davis DW, Lashinger L, Bar-Eli M, Millikan R, et al. The proteasome inhibitor bortezomib synergizes with gemcitabine to block the growth of human 253JB-V bladder tumors in vivo. Mol Cancer Ther. 2004;3:279–290. [PubMed] [Google Scholar]
- Kamat AM, Tharakan ST, Sung B, Aggarwal BB. Curcumin potentiates the antitumor effects of Bacillus Calmette-Guerin against bladder cancer through the downregulation of NF-kappaB and upregulation of TRAIL receptors. Cancer Res. 2009;69:8958–8966. doi: 10.1158/0008-5472.CAN-09-2045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.