Summary
Magnetotactic bacteria (MTB) use magnetosomes, membrane bound crystals of magnetite or greigite, for navigation along geomagnetic fields. In Magnetospirillum magneticum sp. AMB-1, and other MTB, a magnetosome gene island (MAI) is essential for every step of magnetosome formation. An 8-gene region of the MAI encodes several factors implicated in control of crystal size and morphology in previous genetic and proteomic studies. We show that these factors play a minor role in magnetite biomineralization in vivo. In contrast, MmsF, a previously uncharacterized magnetosome membrane protein encoded within the same region plays a dominant role in defining crystal size and morphology and is sufficient for restoring magnetite synthesis in the absence of the other major biomineralization candidates. In addition, we show that the 18 genes of the mamAB gene cluster of the MAI are sufficient for the formation of an immature magnetosome organelle. Addition of MmsF to these 18 genes leads to a significant enhancement of magnetite biomineralization and an increase in the cellular magnetic response. These results define a new biomineralization protein and lay down the foundation for the design of autonomous gene cassettes for the transfer of the magnetic phenotype in other bacteria.
Introduction
Biomineralization, the production of highly ordered inorganic minerals by living organisms, is a widespread process, examples of which are found in all major domains of life. Dozens of biologically synthesized minerals occur in nature, ranging from the calcium-based teeth and bones of animals to the silica tests of diatoms. The formation of these complex crystalline structures often relies on biomineralization proteins that control the nucleation of the crystals as well as their final geometries (Bartlett et al., 2006, Kröger & Poulsen, 2008). Despite its broad occurrence and clear implications for health (Kawasaki et al., 2009), the mechanisms and the evolutionary history of biomineralization processes are poorly understood.
The iron-oxide magnetite (Fe3O4) is a particularly interesting biomineral as it occurs in many animals displaying migratory behaviors, in the human brain (Kirschvink et al., 1992) as well as in magnetotactic bacteria (MTB) (Komeili, 2012). MTB are a phylogenetically and morphologically diverse group of aquatic microorganisms that share the ability to synthesize intracellular magnetic nano-crystals of magnetite or the iron sulfide, greigite (Fe3S4). The crystals are synthesized within a specialized membrane-bounded compartment that forms by invagination of the cell membrane. The crystal and the membrane that allows for its synthesis constitute an organelle called the magnetosome. Individual magnetosomes organize into one or several chains that allow the cells to passively align and navigate along geomagnetic field lines presumably facilitating the search for their favored low oxygen environments (Bazylinski & Frankel, 2004).
Magnetite crystals produced by MTB isolated in various environments are highly diverse in both size and geometry, and their number per cell can vary from less than a dozen to several hundred. Phylogenetic studies as well as the analysis of magnetofossils within ancient sediments imply that magnetite biomineralization by MTB might be the precursor to modern day biomineralization processes (Kirschvink & Hagadorn, 2000, Weiss et al., 2004). Thus, magnetosomes are often highlighted as potentially powerful models to explore the evolution and mechanistic basis of mineral formation by biological systems. In addition, the magnetic properties of magnetosomes have made them a source of inspiration for the development of various applications (Lang et al., 2007). Some of these applications rely on the simple principle of magnetic separation to detect and purify bio-molecules or specific cell types. Other proposed applications aim to develop magnetosomes into contrast agents for magnetic resonance imaging, use MTB for bioremediation purposes or even exploit their magnetic properties to locally heat and kill tumor cells (Alphandery et al., 2011, Lee et al., 2011, Martel et al., 2009, Mokrani et al., 2010, Tanaka et al., 2010). Despite the clear and persistent interest in MTB, the factors controlling the properties of magnetite crystals or their modes of action in vivo are only beginning to be understood. The motivation behind the study presented here was to identify proteins controlling mineral properties in vivo in order to shed light on the mechanisms of magnetite formation. In turn this will facilitate the development of more reliable and reproducible methods to synthesize nano-crystals of magnetite in vitro.
One of the primary paths for the discovery of magnetite biomineralization factors has been through the identification of proteins physically associated with magnetosomes (Arakaki et al., 2003, Grünberg et al., 2004, Grünberg et al., 2001, Tanaka et al., 2006). Four of these factors are small molecular mass proteins (MamC, MamD, Mms5 and Mms6) that are only found in magnetotactic bacteria, localize to magnetosomes and bind tightly to the magnetite crystals therefore suggesting a role in the control of biomineralization (Arakaki et al., 2003, Taoka et al., 2006). In Magnetospirillum gryphiswaldense MSR-1, mamC and mamD are part of a four-gene operon whose deletion leads to a modest decrease in magnetite crystal size, confirming an in vivo role for these proteins in biomineralization (Scheffel et al., 2008). The role of Mms6 in biomineralization was first investigated in vitro where a recombinant version of this protein improved the uniformity of magnetite crystals synthesized in vitro (Amemiya et al., 2007, Arakaki et al., 2010). Recently, mms6 was inactivated in Magnetospirillum magneticum AMB-1 and shown to also participate in the control of the size and geometry of the magnetite crystals in vivo (Tanaka et al., 2011).
In addition to these proteomic efforts, genetic analyses of magnetosome formation have also led to the discovery of a number of important biomineralization factors (Ding et al., 2010, Murat et al., 2010). Magnetotactic bacteria share a conserved genomic island, termed the Magnetosome Island (MAI), which is essential for magnetosome formation and encodes most of the proteins found to be physically associated with purified magnetosomes. In a recent comprehensive genetic survey of the 100-kilobase MAI of Magnetospirillum magneticum AMB-1, we demonstrated that the 18-gene mamAB cluster is essential for the stepwise assembly of the organelle and encodes for proteins participating at each step of its formation (Murat et al., 2010, Quinlan et al., 2011). Besides the mamAB region, an 8-gene region termed R3, encompassing the mamFDC and mms6 gene clusters, also plays an important role in magnetite biomineralization, as its deletion leads to the production of small and misshapen crystals (Murat et al., 2010). Intriguingly, the biomineralization phenotype of the ΔR3 mutant in AMB-1 (and that of the deletion of the equivalent region in MSR-1) is more severe than that of mms6 in AMB-1 or a mamGFDC deletion mutant in the closely related bacterium MSR-1 (Lohsse et al., 2011, Murat et al., 2010, Scheffel et al., 2008). Given the clear role of Mms6 in magnetite biomineralization in vivo and in vitro, we hypothesized that the more severe phenotype of the ΔR3 mutant may be due to a combined loss of mms6 and mamFDC. An additional possibility was that the other genes of R3, not previously characterized genetically, could play a dominant role in magnetite biomineralization.
These observations prompted us to investigate the role of the mamFDC and mms6 cluster (mms6cl) genes in biomineralization in AMB-1. By generating independent non-polar deletions of these genes, we show that the deletion of the previously uncharacterized gene mmsF, but not that of mms6 or mamFDC, is primarily responsible for the severe phenotype of the ΔR3 mutant. We show that initiation of magnetite synthesis does not seem to be delayed in absence of mmsF. Instead, crystal growth follows a pattern reminiscent of wild type AMB-1 until the crystals reach about 25 nm in length at which point crystal growth stalls in the ΔmmsF mutant while it proceeds in wild type cells. The key role of this protein in biomineralization is further highlighted in our surprising finding that in the 8-gene deletion mutant ΔR3, the expression of MmsF is sufficient to restore the synthesis of mature magnetite crystals. To investigate the capabilities of MmsF more broadly, we constructed a strain in which the entire MAI, save for the mamAB gene cluster, is deleted. Surprisingly, these 18 genes are sufficient for the early steps of magnetosome formation in AMB-1 but only lead to the biomineralization of small magnetite that do not facilitate the orientation of cells in a magnetic field. However, the expression of mmsF in this ‘minimal-island’ strain results in a significant improvement of magnetite synthesis that allowed the cell pellets to be attracted to a magnet. Taken together, these results demonstrate that MmsF, and to a lesser degree Mms6, are major players in the control of magnetite biomineralization in AMB-1. In addition, the work presented here lays out a potential blueprint for the development of an autonomous genetic unit for magnetic mineral production.
Results
The deletion of the mms6 gene cluster is responsible for the biomineralization defect in the ΔR3 mutant
Previous work has implicated the MamGFDC and Mms6 proteins in magnetite biomineralization in MSR-1 and AMB-1 respectively (Arakaki et al., 2003, Scheffel et al., 2008, Tanaka et al., 2011). The deletions of the mamGFDC operon in MSR-1 and that of mms6 in AMB-1 cause magnetite crystals to be smaller than those of the wild type strain (69% and 56% of wild type crystal size respectively) (Scheffel et al., 2008, Tanaka et al., 2011). Recently, we showed that the simultaneous deletion of eight genes including the mamFDC and mms6 gene clusters in AMB-1, grouped in a chromosomal region named R3 (Figure 1a), leads to a phenotype that is significantly more severe than the reported phenotypes of either mamGFDC deletion or mms6 deletion mutants alone (Murat et al., 2010). Similarly severe phenotypes were also seen in a combined deletion of these gene clusters in MSR-1 (Lohsse et al., 2011). In the R3 deletion mutant, crystals are approximately 40% of wild type crystals in length (47.5 ± 15 nm; n= 248 and 18.9 ± 10 nm; n = 215 in AMB-1 and ΔR3 respectively, Table 1) and 28% of the wild type crystals in width. As a result, these crystals have a more elongated appearance with a width to length ratio of 0.662 ± .17 as compared to a ratio of 0.854 ± 0.165 for the cubooctahedral wild type crystals (see supplementary tables S4-6 for comparisons of statistical significance for all of the measurements presented in this manuscript). The severe phenotype of the R3 deletion could be due to the accumulated loss of all mms genes. Alternatively, one or more of the additional genes in this region (amb0950, amb0955 or mmsF) may be responsible for the bulk of the defect observed in this mutant. To distinguish between these possibilities, we generated various targeted deletions in this region and investigated the resulting defects in magnetite biomineralization.
Figure 1. Biomineralization phenotypes of the R3 gene mutants in AMB-1.
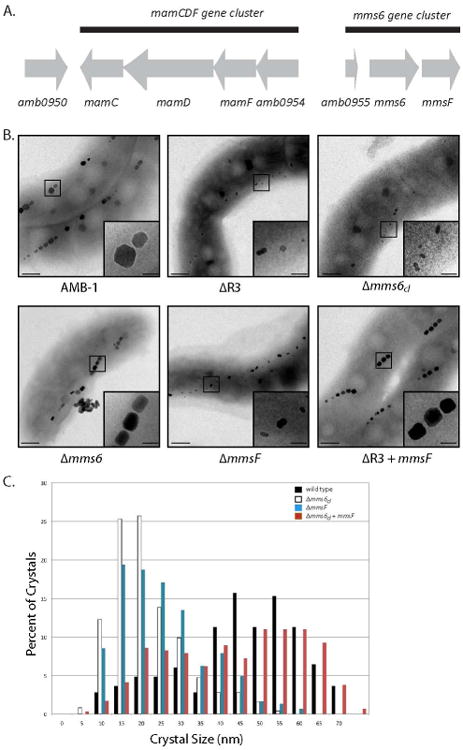
A. The genes and ORFs comprised in the region R3 are represented by grey arrows, and indicate the orientation of transcription. The gene name, or gene number, is indicated below each gene or ORF. The black lines show the groups of genes identified as gene clusters. B. Electron micrographs of (from top to bottom, left to right) wild type AMB-1, ΔR3, Δmms6cl, Δmms6, ΔmmsF mutants and the ΔR3 mutant complemented with mmsF. The insets are showing a higher magnification of the region boxed in the micrograph. Scale bars: 100 nm, insets 50 nm. C. The ΔmmsF mutant mimics the ΔR3 biomineralization phenotype. Crystal size distribution in wild type AMB-1 (black bars), Δmms6cl, (white bars), ΔmmsF (blue bars) and Δmms6cl + mmsF (red bars).
Table 1.
Measurements of magnetic response and crystal properties for various strains. Cmag, average crystal length and average crystal width of various strains were measured as detailed in Experimental Procedures. Shape factor is calculated as the width divided by the length (given in nanometers in the table). Number of crystals measured for each strain (n) is presented in the last column.
| Strain | Plasmid | Cmag | Average Length | Average Width | Average Shape Factor | n |
|---|---|---|---|---|---|---|
| AMB-1 | - | 1.97 ± .24 | 47.5 ±15 | 40 .4 ±14 | 0.854 ± 0.165 | 248 |
| ΔR3 | - | 1.03 ± .01 | 18.9 ± 10.0 | 11 .4 ± 4.5 | 0.662 ± .170 | 215 |
| mms6cl | 2.34 ± .2 | 46.0 ± 15.5 | 33.8 ± 13 | 0.734 ± .126 | 242 | |
| mmsF | 1.77 ± .16 | 40.8 ± 17 | 34.5 ± 15 | 0.840 ± .111 | 291 | |
| Δmms6cl | - | 1.04 ± .01 | 19.3 ± 9 | 13.1 ± 6 | 0.716 ±.177 | 253 |
| mms6cl | 2.1 ± .04 | 42.3 ± 17.5 | 33.9 ± 15 | 0.793 ± .110 | 274 | |
| amb0955 | 1.04 ± 03 | 22.4 ± 10 | 14.5 ± 6 | 0.671 ± .153 | 136 | |
| mms6 | 1.15 ± .08 | 27.8 ± 13.6 | 20.8 ± 11.8 | 0.757 ± .187 | 161 | |
| mmsF | 2.1 ± .04 | 36.7 ± 15 | 25.6 ± 12 | 0.706 ±.136 | 258 | |
| Δamb0955 | - | 1.88 ± .09 | 46.3 ± 17 | 39 ± 15 | 0.834 ± .121 | 78 |
| Δmms6 | - | 1.65 ± .11 | 38.8 ± 15.3 | 28.4 ± 12.5 | 0.731 ± .127 | 224 |
| mms6 | 1.67 ± .08 | 42.1 ± 15.5 | 35.7 ± 14.5 | 0.847 ±.122 | 116 | |
| ΔmmsF | - | 1.08 ± .05 | 23.0 ± 10.6 | 15.6 ± 7 | 0.705 ± .149 | 304 |
| mmsF | 1.59 ± .11 | 42.0 ± 16.8 | 33.1 ± 14 | 0.785 ± .113 | 309 | |
| ΔmamFDC | 1.66 ± .04 | 46.23 ± 13.6 | 34.9 ± 11.4 | 0.756 ± .116 | 171 | |
| miniMAI | - | 1 | 23.42 ± 8.23 | ND | ND | 165 |
| miniMAI_mmsF | mmsF | 1.01 ± .01 | 29.65 ± 12.46 | ND | ND | 351 |
We began by generating a mutant carrying a deletion of the mamFDC genes (amb0953, amb0952 and amb0951 respectively, Figure 1a) along with amb0954 a putative ORF with a position and orientation consistent with a possible co-transcription with mamF, mamD and mamC. The mutants generated in this study are listed in Table S1 (SI). The deletion of the mamFDC cluster has only a minor impact on the products of magnetite biomineralization and leads to the formation of crystals with a length similar to that of wild type and a slight reduction (86.4 %) in the average crystal width (width is 40.4 ± 14 nm and 34.9 ± 11.4 nm for wild type and ΔmamFDC crystals respectively). As a result, the ability of ΔmamFDC mutant cells to align in an applied magnetic field, as quantified by the coefficient of magnetism (Cmag (Schüler et al., 1995, Murat et al., 2010)) (summarized in Table 1, Figure S1C), is only slightly lower than that of the wild type cells.
In contrast to the mild phenotype seen with the deletion of the mamFDC cluster, the deletion of the mms6 cluster phenocopies the ΔR3 mutant in nearly every respect. The Δmms6cl mutant is weakly magnetic and produces smaller and elongated crystals (19.3 ± 9 nm in length and 13.1 ± 6 nm in width, Table 1) which are very similar to the crystals synthesized in the ΔR3 mutant (Figure 1). We introduced the mms6 cluster, placed under the control of the tac promoter, at a chromosomal locus neutral for magnetosome formation in the Δmms6cl mutant (plasmids are listed in Table S2, SI). The low Cmag of the Δmms6cl mutant was successfully complemented with this construct, crystal length was restored to 42.3 ± 17.5 nm but crystal width could only be complemented to about 84 % of wild type value (33.9 ± 15 nm; n = 274; Table 1). When introduced in the ΔR3 strain, this construct resulted in a full complementation of the mutant's magnetic properties (Cmag = 2.34) and crystal length (46 ± 15 nm). However, similarly to what we observed in the Δmms6cl mutant, crystal width could only be increased to 84 % of wild type values in the complemented strain. These results suggest that one or more genes in the mms6 gene cluster are responsible for the severe biomineralization defect in the ΔR3 mutant since the mms6 cluster can complement the ΔR3 phenotype in the absence of the mamFDC genes (Table 1).
Mms6, but not Amb0955, plays a role in controlling magnetite crystal properties in AMB-1
We next investigated the role of each of the mms6 cluster genes in magnetite biomineralization. Mms6 appeared to be a good candidate to explain the ΔR3 mutant biomineralization phenotype as this protein is known to improve the properties of magnetite crystals synthesized in vitro (Amemiya et al., 2007) and was more recently shown to participate in the control of magnetite synthesis in vivo in AMB-1 (Tanaka et al., 2011). The mms6 gene cluster contains two additional genes: amb0955, a hypothetical protein, and mmsF, whose product was found to be associated with purified magnetosomes in MSR-1 and shares 64 % and 65 % sequence identity with the conserved magnetosome protein MamF from MSR-1 and AMB-1 respectively. Considering the established evidence regarding the role of Mms6 in magnetite formation, we hypothesized that this protein could be the primary or even sole biomineralization factor in the mms6 gene cluster. We generated a single non-polar deletion of mms6 and showed that the magnetic properties of the Δmms6 mutant were only slightly lower than that of wild type (Cmag values of 1.97 ± 0.24 and 1.65 ± 0.11 for wild type and Δmms6 respectively; Table 1 and Figure 1B). In the Δmms6 mutant, the crystals are smaller (length: 38.6 ± 15 nm, n= 225) as compared to wild type crystals (length: 47.5 ± 15 nm, n= 248), but significantly bigger than the crystals produced by the Δmms6cl mutant (length: 19.3 ± 9 nm, n= 253) (Table 1). However, in both the Δmms6 and the Δmms6cl mutants, the crystals are elongated, as evidenced by an increased shape factor (ratio width/length). In wild type AMB-1 cells, the shape factor is 0.854 ± 0.165, leading to roughly cubic crystals, whereas it is 0.716 ± 0.177 and 0.731 ± 0.127 in the Δmms6cl and the Δmms6 mutants respectively where the crystals are mostly parallelepipeds. To confirm this result, we assessed the ability of mms6 to complement the magnetite formation defect of the Δmms6cl mutant. A plasmid expressing mms6 from the tac promoter (listed in Table S1) was introduced in the Δmms6cl mutant. This construct allows for significant complementation of the Δmms6 mutant (80 % of wild type Cmag, crystal length and width, Table 1). However, when mms6 was expressed in the Δmms6cl mutant, the magnetic properties of the mutant were only partially improved as determined by measuring the Cmag (Table 1). This strongly suggested that the absence of mms6 is not entirely responsible for the severe biomineralization phenotype of the Δmms6cl mutant. It further implied that either the combination of the mms6 deletion with that of another gene in the cluster, or the deletion of amb0955 and/or mmsF, was responsible for the mutant phenotype. We deleted amb0955 in AMB-1 (Figure S1A) and observed that the Δamb0955 mutant displays magnetic properties and magnetite crystals indistinguishable from those of wild type (Table 1). In addition, the expression of amb0955 in the Δmms6cl mutant did not rescue the severe magnetic defects of the Δmms6cl mutant (Figure S1A), suggesting that Amb0955 does not play a role in biomineralization. Altogether, this analysis pointed towards mmsF, the third and last gene of the mms6 cluster, as playing a significant role in magnetite biomineralization in AMB-1.
MmsF regulates magnetite crystal size and geometry, even in the absence of Mms6 and the MamFDC proteins
Since the absence of neither amb0955 nor mms6 could on their own explain the severe defects of the Δmms6cl mutant, we investigated the biomineralization phenotype of a strain carrying a nonpolar deletion of the third gene of the cluster, mmsF. Interestingly, the Cmag of the ΔmmsF mutant, as well as the dimensions and appearance of its crystals were very similar to that of the Δmms6cl mutant (Table 1 and Figure 1). The ΔmmsF mutant synthesizes crystals that are on average 23 ± 11 nm in length (n= 304), essentially similar to the 19.3 ± 9 nm (n= 253) average crystal length seen in the Δmms6cl mutant (Table 1). The size distribution profiles of the crystals measured in both mutants are also strikingly similar (Figure 1C).
Since the phenotypes of the ΔmmsF and Δmms6cl mutants were almost indistinguishable, it suggested that the loss of mmsF alone could be responsible for the severe defect of the cluster mutant. When mmsF is expressed on the chromosome of the ΔmmsF mutant, it allows for the formation of large crystals (88 and 81% of wild type crystal length and width respectively) presenting an overall cubic symmetry (shape factor = 0.785 ± 0.113). When expressed in the Δmms6cl mutant, mmsF allows for the complementation of the mutant's magnetic properties to wild type values (Cmag = 1.97 and 2.1 for the wild type and the Δmms6cl strain expressing mmsF respectively, Table 1). The crystals measured remain smaller than those synthesized in wild type cells (length 36.7 ± 15 nm, n= 258) which is not surprising since this strain still lacks mms6. In fact, the Δmms6cl expressing mmsF produces crystals that are nearly identical in their dimensions to those formed in the Δmms6 mutant (See Table 1 and size distribution in Figure S1B).
To investigate whether the function of MmsF is dependent on the previously identified biomineralization proteins MamFDC, we tested its ability to restore magnetite formation in the ΔR3 mutant, that is, in the absence MamF, MamD, MamC and Mms6. We show that the expression of mmsF in the ΔR3 mutant leads to the formation magnetosomes that are wild type in their appearance as well as nearly wild type response to magnetic fields. (Table 1, Figure 1). This result shows that in the absence of MamFDC and Mms6, previously identified as abundant and important biomineralization proteins, MmsF is sufficient to synthesize large cubooctahedral magnetite crystals. It is important to note that MmsF can complement the ΔR3 mutant more efficiently than it does the Δmms6cl mutant, suggesting that in the absence of Mms6, the MamFDC proteins may hamper the activity of MmsF in vivo.
MmsF affects geometry of magnetite crystals in AMB-1
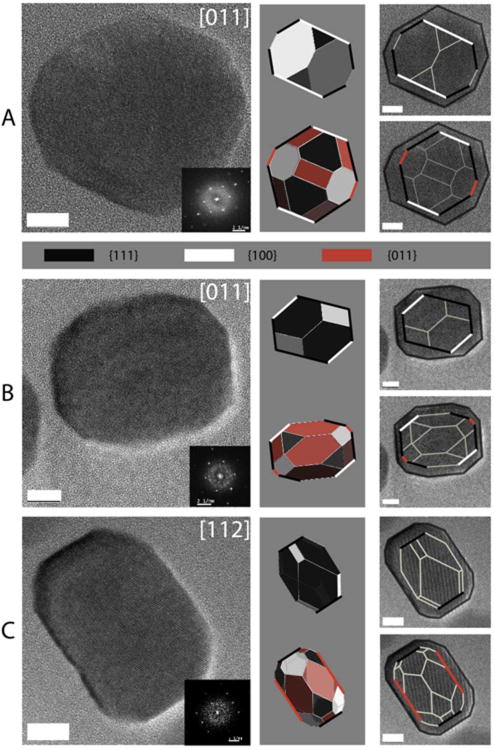
We used High Resolution X-ray diffraction (HRXRD) to determine the nature of the particles produced in the ΔmmsF mutant. We determined that in both the ΔmmsF and Δmms6cl mutants the size of the lattice parameter (a) is characteristic of magnetite (8.394 < a < 8.397 Å) (Faivre et al., 2010). Thus, MmsF neither functions in determining the chemical composition nor the mineral phase of the iron oxide minerals produced in AMB-1. Furthermore, using High Resolution Transmission Electron microscopy (HRTEM), we verified that the minerals produced in the ΔmmsF mutant were crystalline. As seen in Figure 2B and 2C (left panels), the particles in the mutant display the characteristic striations indicative of a crystalline structure and the FFT pattern are characteristics of the magnetite structure.
Figure 2. Magnetite nanocrystals with altered geometric properties are produced in the absence of mmsF.
Left, HRTEM images of representative magnetite crystals and corresponding Fast Fourier Transforms (FFT) (inset) in A. wild type AMB-1, B-C. ΔmmsF mutants. Right: Crystal models (gray background) and projected frame models (superimposed with the original HRTEM images) oriented as in the HRTEM image. Face forms {100}, {110} and {111} are colored according to the scheme indicated in the gray band between A and B. Faces parallel to lattice planes belonging to the zone axis of each micrograph are outlined with the corresponding color. Scalebar: 10nm.
HRTEM was also used to further investigate the effects of the loss of MmsF on crystal geometry. In Figure 2, representative HRTEM images of magnetite nanocrystals formed in the wild type (Fig 2A, left) and the ΔmmsF mutant (Fig 3B-C, left) are shown. In the wild type, particles mostly exhibit a shape factor close to 1 (See Fig 2A, left). Their projected shape can be modeled, as described in previous works (Faivre et al., 2005, Faivre et al., 2008), by different morphologies, namely cubooctahedral with the {100} form more extended than the {111} (Fig 2A, right, upper model) or cubooctahedral-like with equally expressed {100} and {111} faces and small {110} faces. From the recorded set of HRTEM images it was not possible to unambiguously discriminate between these two cases but based on the observed intensity of the phase contrast at the edges of the particle and their aspect ratios, the second model seems to be more likely. Moreover, the ΔmmsF mutant grows particles with a lower aspect ratio (0.705 ± .149 as compared to 0.854 ± 0.165 in the wild type), as can be appreciated by comparing Fig 2A and B-C. The elongation direction coincides with the direction of the magnetosome chain and corresponds to the <111> direction of the magnetite crystal lattice, as indicated by high-resolution images of the particles (Fig 2B and C). However, although the particles exhibit a different aspect ratio their projected shape is not compatible with cubic crystals delimited by extended {100} form faces and is therefore distinct from the geometry of the wild type crystals. These results suggest that MmsF would participate in controlling the size and shape of the crystals, perhaps by controlling crystal growth and/or nucleation in AMB-1.
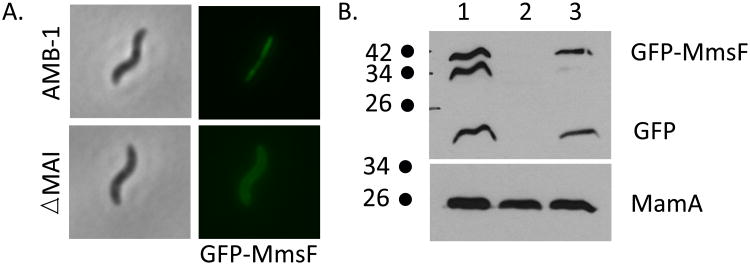
Figure 3. MmsF is a magnetosome membrane protein.
A. GFP-MmsF has a magnetosome-dependent localization in AMB-1. The localization of GFP-MmsF was determined in AMB-1 and a ΔMAI mutant that no longer makes magnetosomes. In wild type cells, GFP-MmsF localizes as a line reminiscent of the position of the magnetosome chain. In the absence of magnetosomes, GFP-MmsF localizes around the cell membrane. B. GFP-MmsF co-purifies with the magnetosomes in AMB-1. Western blot of whole cell lysates (lane 1) of wild type cells expressing GFP-MmsF on a plasmid were adsorbed on a magnetized column: non-magnetic material in the flow through (lane 2) and magnetosomes and its constituents eluted from the column (lane 3) revealed using an anti-GFP antibody. The peripherally associated magnetosome protein MamA is used as an internal control and revealed by anti-MamA polyclonal antibodies (bottom panel). The size of the molecular weight markers are indicated on the left. The anti-GFP antibody also revealed an additional band of an intermediate size between GFP-MmsF (42 kDa) and GFP (22 kDa) that may correspond to a partially degraded GFP-MmsF fusion protein potentially cleaved in a protease-accessible region of MmsF. This band was seen primarily in the lysate but is also found in the purified magnetosome fraction.
MmsF localizes to the magnetosomes in AMB-1
MmsF is a small protein (107 residues in AMB-1) that was found to be associated with the purified magnetosome fraction in MSR-1 (Grünberg et al., 2004). Considering its role in magnetite biomineralization in AMB-1, we hypothesized that MmsF would also be associated with the magnetosomes in AMB-1. We determined the localization of a version of MmsF tagged on its N-terminus with the green fluorescent protein (GFP) in wild type cells. This fusion protein is functional in vivo as it can complement the ΔmmsF biomineralization phenotype. As shown in Figure 3A, GFP-MmsF localizes as a rather straight line, continuous or dotted, extending across the longer axis of the cells, only rarely spanning the entire length of the cell. This localization is characteristic of other magnetosome-associated proteins such as MamA, MamJ, MamI and MamC (Komeili et al., 2004, Lang & Schüler, 2008, Murat et al., 2010, Quinlan et al., 2011, Scheffel et al., 2006). In a strain where the MAI region is deleted (ΔMAI, Table S2), which does not make magnetosome organelles, GFP-MmsF localizes homogeneously around the cell membrane, which is in agreement with its predicted membrane localization. The magnetosome-dependent localization of MmsF suggests that it is primarily associated with magnetosomes in vivo. To confirm these results we used a biochemical fractionation assay to assess the distribution of MmsF within the cell. Whole cell lysates of wild type AMB-1 expressing GFP-MmsF were passed over a magnetic column, enriching for magnetosomes and their associated proteins. Western blotting against GFP and other known magnetosome proteins was then used to assay the localization of GFP-MmsF within the cell. As can be seen in figure 3B, nearly all of the GFP-MmsF is found in the magnetosome fraction. In contrast, the magnetosome-associated protein MamA is found in both the nonmagnetic as well as the magnetosome fraction due to its cytoplasmic localization. The western blot analysis of GFP-MmsF also reveals the presence of cleavage products that are smaller than the full-length protein. This cleavage may be an artifact of the extract preparation process or it may reflect the production of smaller protein products in AMB-1. Given that a background of cytoplasmic fluorescence was not observed and the protein's localization varies depending on specific strain genotypes, we believe that these results are not influenced by free GFP that may have resulted from proteolytic cleavage. The combination of fluorescence microscopy experiments and biochemical fraction studies demonstrate that MmsF is associated with the magnetosome membrane in vivo.
The deletion of mmsF halts the maturation of magnetite crystals in AMB-1
Magnetite crystals are overall smaller in the ΔmmsF mutant as compared to wild type crystals, yet a percentage (22.7 %) of the crystals can reach lengths above 35 nm (Figure 1C). Crystals above 35 nm are able to stably hold a magnetic moment, therefore contributing to the cells overall magnetic response. These observations suggest that the protein is required for the proper timing of biomineralization initiation or that it is important for maturation of magnetite crystals. To distinguish between these two hypotheses, we studied the timing of the initiation of crystallization and growth of magnetite in wild type AMB-1 and in the ΔmmsF deletion strain. Cells that were passaged three times in media lacking iron (see Material and Methods and (Quinlan et al., 2011)) were used to inoculate iron-containing media at 30°C in microaerobic conditions, allowing for optimal magnetite synthesis. Cell growth, Cmag and magnetite formation were monitored over time between 30 minutes and 24 hours post-inoculation in the iron-containing medium. No electron-dense particles were observed in the cells by TEM prior to their transfer to the iron-replete media (t0). After 30 minutes, both wild type and mutant cells contained electron dense particles (smaller than 20 nm), aligned in chains, and located at the inner curvature of the cells (shown in Figure 4, right panels). At this stage, most particles are oddly shaped, in both strains, and show heterogeneous geometries. Their localization and density suggests these particles could represent the early stages of magnetite formation as previously shown (Quinlan et al., 2011). This observation indicates that there is no delay in the initiation of magnetite biomineralization in the ΔmmsF mutant. The average crystal size in the mutant and wild type cells are seemingly identical 30 minutes post transfer into iron-replete media (13.78 ± 5.75 nm and 16.96 ± 5.91 nm respectively). By compiling the data from three independent experiments, we find that crystal distribution in the wild type and in the ΔmmsF mutant look similar up to about 2-4 hours post transfer in iron-replete media. Beyond that time, crystal size distribution does not change in the mutant, while becoming broader in the wild type as more crystals occupy the larger size range (Figure 4, left panels). Yet, in the ΔmmsF mutant, the proportion of oddly shaped crystals (see insets in Figure 4, right panels) remains high until 5 hours post transfer, while a vast majority of the wild type crystals have a normal cubooctahedral geometry starting as early as 2.5 hours post transfer. These results indicate that the activity of MmsF is most likely required during the maturation of the crystal after the initiation of biomineralization.
Figure 4. Biomineralization is slowed down and stalls in the ΔmmsF mutant after initial nucleation events.
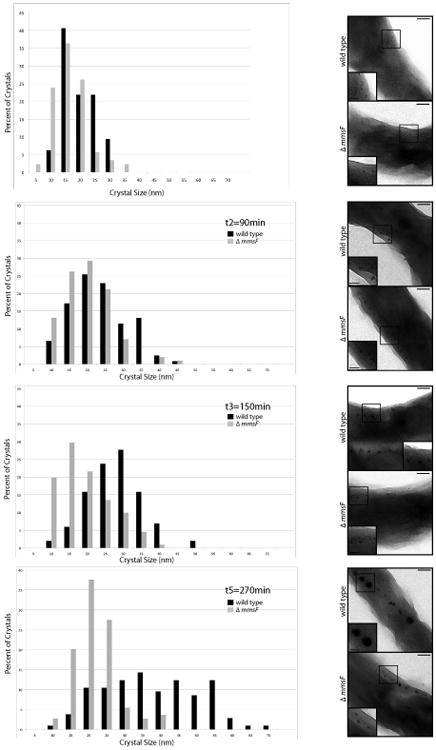
A. Crystal size distribution in wild type AMB-1 (black bars) and the ΔmmsF (grey bars) strains 30, 90, 150 and 270 minutes post transfer of the cultures in an iron-replete media. The data shown comes from one data set representative of three independent experiments. B. Early stages of magnetite formation in AMB-1 and ΔmmsF. Electron micrographs of glutaraldehyde-fixed cells at the time points indicated in A. Insets are higher magnification of the region boxed in the micrograph. Scale bars: 100 nm, insets 50 nm.
The mamAB operon is sufficient for the synthesis of mineral-containing magnetosome organelles
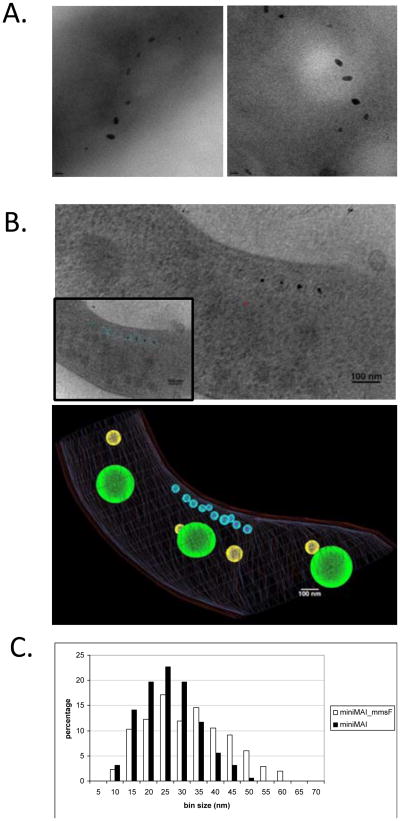
Our results thus far indicate that amongst the proteins encoded by the R3 region, MmsF performs a prominent and critical role in magnetite biomineralization. To investigate the role of MmsF more broadly, we sought to investigate its effect on crystal formation in a strain where the 100-gene magnetosome island is reduced to a minimal set of genes essential for magnetosome formation. In a previous global genetic dissection of the magnetosome island of AMB-1, we showed that the 18 genes of the mamAB cluster (R5) are essential for various steps of magnetosome formation such as membrane biogenesis, magnetosome protein sorting, organization of magnetosomes into chains, as well as biomineralization. Additionally, a recent genetic analysis of the magnetosome island has shown that the mamAB cluster is indeed sufficient for the production of magnetosomes in MSR-1 (Lohsse et al., 2011). Thus, we hypothesized that the mamAB cluster may also encode for all the factors required for the establishment of the magnetosome chain in AMB-1. To test this hypothesis, we deleted the regions surrounding the mamAB cluster (R1 to R4 and R6 to R14) in order to create a minimal 18-gene MAI. First, the ∼25 kilobase region upstream of R5, which includes R3, was deleted in AMB-1. As expected, this mutant displays a severe defect in magnetite formation, reminiscent of the ΔR3 mutant phenotype (Figure S2). Second, the MAI regions downstream of R5 (∼60 kilobases) were simultaneously deleted, so that only the mamAB cluster would remain from the MAI. The cell pellets of the mutant, termed miniMAI, were light in color and could not be pulled using a bar magnet, initially suggesting the absence of any magnetosome organelles in this mutant. However, TEM analysis of the miniMAI strain revealed the presence of small electron dense particles that in most cells were aligned in a chain, and where located near the inner curvature of the cells (Figure 5A). HRTEM analysis of these particles showed that they are indeed crystalline with crystal fringe dimensions of magnetite. Electron Cryo-Tomography (ECT) of the miniMAI strain (17 cells were analyzed) showed that these small magnetite particles are indeed found within compartments that resemble the magnetosome membrane of wild type cells in their appearance and dimensions (mostly within the 60-75 nm range), (Figure 5B). One clear defect, however, is that the miniMAI strain forms short magnetosome chains containing only 3 to 11 individual compartments that, in contrast to wild type cells, do not span the length of the cell (modeled in the inset Figure 5B). In these cells, 3 to 6 compartments contained magnetite crystals. These results suggest that the biomineralization proteins previously identified in the mamAB region (mamE, mamM, mamN, mamO, mamS, mamP, mamR, mamT) are sufficient to trigger the nucleation of small magnetite crystals, in the magnetosome compartments. They further demonstrate that the biomineralization proteins in R3, including MmsF, are not essential for the nucleation of magnetite and would most likely participate in the control of later stages of crystal growth.
Figure 5. MmsF can significantly enhance the magnetic properties of a miniMAI AMB-1 strain.
A. AMB-1 carrying a miniMAI still produces magnetic particles. Electron micrographs of the miniMAI strain showing aligned magnetite crystals in two representative cells. B. The miniMAI strain of AMB-1 produces membrane-enclosed magnetite crystals. Top panel: Tomographic slice of a miniMAI cell analyzed by Electron Cryo-Tomography. The electron dense magnetite crystals are enclosed within membranes similar to wild type magnetosome compartments. In the inset, the rest of the magnetosome compartments seen in different tomographic slices of the same cell are shown in light blue. Bottom panel: segmentation analysis of the internal structures seen above. The magnetosomes are represented in blue, the cell membrane in red, presumed polyhydroxybutyrate (PHB) granules in green, and putative storage granules in yellow. Scale bars: 100 nm. C. MmsF improves magnetic particle synthesis in the miniMAI strain. Crystal size distribution in the miniMAI (black bars) and the miniMAI strain expressing mmsF in cis (miniMAI_mmsF, white bars). Δmms6cl.
mmsF can enhance the magnetic properties of a ‘minimal-island’ strain
Since the loss of mmsF is responsible for the biomineralization defect of the ΔR3 mutant in AMB-1, we tested whether it could also enhance the biomineralization of magnetic particles in the miniMAI strain. When mmsF was expressed in the miniMAI strain, the cell pellets were significantly darker than those of the miniMAI strain alone and could be easily pulled towards a bar magnet, suggesting improved magnetite synthesis. TEM analysis of the strains showed that the addition of mmsF to the miniMAI strain led to a slight increase in the average length of the crystals (Table 1), and an overall increase in the number of crystals above 35 nm in length. These larger crystals represent 46.15 % of the total number of crystals in miniMAI strain expressing mmsF as compared to 20.85% in the miniMAI strain alone. In addition, we found only one crystal of average wild type dimensions (about 50 nm in length) in the miniMAI strain (1 out of 163 measured) whereas these larger particles represented 11.97 % of the crystals in the miniMAI_mmsF strain (42 out of 351 crystals), with the largest measured at 69 nm in length (Figure 5C). Accordingly, while the ΔMAI or miniMAI strains consistently failed to orient in an applied magnetic field in the quantitative Cmag assay (Cmag= 1 ± 0), the miniMAI_mmsF strain displayed a measurable, albeit poor, cellular response to external magnetic fields (Table 1) which may be the consequence of production of a larger number of magnetite crystals by this strain. These results further demonstrate the importance of MmsF in promoting the biomineralization of mature magnetite crystals. Additionally, they are a major step towards the development of a genetic cassette for the heterologous expression of magnetic organelles in other microorganisms.
Discussion
In the past decade, several approaches have been used to identify proteins involved in the control of magnetite biomineralization in magnetotactic bacteria (Arakaki et al., 2003, Grünberg et al., 2004, Grünberg et al., 2001, Komeili et al., 2004, Richter et al., 2007, Wahyudi et al., 2001). In particular, a proteomics approach allowed for the identification of four proteins (MamD, MamC, Mms5 and Mms6 (Arakaki et al., 2003), which are tightly associated with purified magnetosomes. Eight more factors influencing magnetite crystal formation were identified through targeted genetic approaches (MamG, MamF, MamP, MamR, MamS, MamT, FtsZ-like and MamE (Ding et al., 2010, Murat et al., 2010, Quinlan et al., 2011, Scheffel et al., 2008). Thus far, only Mms6 has been shown to influence magnetite synthesis both in vivo and in vitro (Amemiya et al., 2007, Prozorov et al., 2007, Tanaka et al., 2011). As a recombinant version of Mms6 was found to bind iron, and it is associated with magnetite in vivo, it was proposed that it could specifically bind to certain crystal faces where it would locally inhibit crystal growth. Yet, the mechanisms of initiation of magnetite formation or deposition and growth of the crystal lattices are unknown.
In a recent study, we performed a preliminary characterization of an AMB-1 mutant carrying an eight-gene deletion including four of the known biomineralization factors (mms6 and mamFDC) that results in a severe biomineralization defect (Murat et al., 2010). When we began this study, the homologs of the mamFDC genes (mamGFDC) had been characterized in M. gryphiswaldense MSR-1 where their deletion leads to a modest decrease in crystal size (Scheffel et al., 2008). In AMB-1, a mutant lacking the mamFDC cluster produces crystals similar in length to wild type crystals but the average crystal width is smaller. Overall, this translates into slightly elongated crystals and lower magnetic properties for the mutant. In MSR-1, while the crystals are smaller (in average 69 % of wild type) the proportions of the crystals remain comparable to those of wild type crystals. This observation suggests that the MamFDC proteins might play slightly different roles in biomineralization in AMB-1 and MSR-1.
From previous work, Mms6 was a clear candidate for being responsible for the biomineralization defect of the ΔR3 mutant. It is, thus far, the only MTB protein for which an in vitro role in biomineralization has been demonstrated (Arakaki et al., 2010, Amemiya et al., 2007). Recently, mms6 was inactivated in AMB-1 and the authors reported that the resulting crystals are smaller (56 % of wild type crystal length) and have an elongated morphology as compared to wild type crystals. Here we also made a deletion of mms6 and observed a milder defect in magnetite crystal properties (81 % of wild type crystal length) than what was reported by Tanaka et al. (Tanaka et al., 2011). It is possible that differences in growth conditions, such as the iron source used or oxygen levels, account for the varying phenotypes observed in these two mms6 mutant strains. However, we believe it is more likely that the mutational strategies used may be the main reason for the differences in the two reports. In this study, we used a two-step recombination strategy to isolate unmarked nonpolar deletions of mms6, whereas Tanaka et al. disrupted the gene through the insertion of an antibiotic resistance cassette. mms6 is directly upstream of mmsF, the two genes are separated by only 17 nucleotides and no promoter can be readily identified upstream of mmsF. These features strongly suggest that the two genes are co-transcribed. Considering the location of mmsF, it is likely that its expression would have been affected in a strain carrying a several-kilobase insertion in mms6 (Tanaka et al., 2011), and ultimately magnetite biomineralization. Nonetheless, it is clear that Mms6 plays a role in magnetite synthesis in AMB-1 that ensures the production of crystals with a specific aspect ratio.
Our work demonstrates that MmsF, which was first identified in purified magnetosome fractions extracted from MSR-1 cells, is essential for biomineralization of mature magnetite crystals. It is conserved in Magnetospirillum magnetotacticum MS-1, MSR-1, Magnetococcus sp. MC-1, and the magnetite-containing vibrio MV-1. MmsF is predicted to encompass three membrane-spanning helices but does not contain conserved domains or motifs that could help predict its function in vivo. Our functional GFP fusions to the N-terminus of MmsF indicate that its N-terminus is in the cytoplasm while the C-terminus faces the magnetosome compartment. The regions predicted to reside in the cytoplasm are short and poorly conserved: only one to three residues that would separate the second and third helices are conserved in all 5 MmsF homologues. However, the loops within the magnetosome compartment (the C-terminus and the loop between the first and second helices) contain highly conserved residues that could mediate MmsF's interactions with the mineral or other biomineralization protein. Although we were not able to assign a function to MmsF in biomineralization, the effects of its deletion suggest it contributes to both the overall growth of the crystal and its morphology, two features that may be related to one another. An in vitro characterization of MmsF is required to assess its role in magnetite synthesis as well as its ability to bind iron.
Interestingly, in AMB-1 there are three predicted MamF paralogs encoded by the amb4012, amb0953 and amb1026 genes, all of which share 64-65% sequence identity with MmsF. Despite this high level of homology there does not appear to be a functional redundancy between MmsF and MamF in AMB-1. All three versions of mamF are expressed at 2 to 4 times higher levels than mmsF in wildtype cells (see transcriptome data associated with Greene at al., 2012). Yet, they do not appear to contribute significantly to biomineralization when mmsF is deleted. Perhaps, MmsF and MamF arose in an ancient duplication event and have since diverged in function. Alternatively, it is possible that there is post-transcriptional modulation of MamF levels or activity such that it can perform a redundant function with MmsF in a conditional manner. A more focused study of MamF is needed to fully elucidate its function and its relationship with MmsF.
In exploring the central role of MmsF in biomineralization, we have also discovered a curious and potentially illuminating relationship between Mms6 and MamFDC proteins. When mmsF is introduced into the Δmms6cl mutant, a strain that still contains the mamFDC genes, the resulting magnetite crystals are similar in their dimensions and aspect ratio to those formed in the Δmms6 strain. In contrast, when mmsF is introduced into the ΔR3 mutant, a strain missing mms6 and mamFDC, the resulting crystals are essentially wild type in their characteristics. Thus, the biomineralization defect that can be attributed to the loss of mms6 is eliminated once the mamFDC genes are also deleted. These results suggest that one or several of the MamFDC proteins negatively regulate MmsF in the absence of Mms6. This observation, along with the wide range of biomineralization phenotypes described thus far in the literature, suggest that beyond the mere nucleation of magnetite, magnetite synthesis could rely on the precise orchestration of many discrete steps, where factors could influence each other's function, potentially in response to specific environmental stimuli.
To further highlight the importance of MmsF and explore its potential as a biomineralization factor in future applications, we sought to create a minimized magnetosome island strain in AMB-1. We find that the 18 genes of the mamAB region are sufficient for the formation of chains of small electron-dense particles in AMB-1. These particles are about 20 nm in length and are formed within membrane-bounded compartments that have wild type dimensions. This shows that the mamAB region contains all the factors required to invaginate and shape the magnetosome membranes. This is a similar approach to that used in a recent work by Schüler and colleagues where, amongst the core magnetotaxis components of the MAI of MSR-1, the mamAB region is sufficient for the production of small electron-dense particles, aligned in a chain (Lohsse et al., 2011). In both cases, the chain formed is not comparable to wild type chains in most respects: there are fewer magnetosomes (a third to one half) and the crystals remain small (Lohsse et al., 2011). Both MamK and MamA are expressed in miniMAI cells of AMB-1 at levels comparable to those seen in wild type (Figure S3) suggesting that some combination of other MAI genes is required for the production of wild type magnetosomes. However, we cannot exclude the possibility that mamAB genes other than mamA or mamK could be misregulated in the miniMAI strain. It is noteworthy that the chain is found at the inner curvature of the cell in the miniMAI strain, suggesting that the positioning of the chain depends on factors present in the mamAB region, or that the shape of the cell intrinsically pre-determines the location of the organelle in the cell. Furthermore, we show that the particles produced in the miniMAI strain of AMB-1 are indeed magnetite, indicating that the initial events that nucleate and determine the chemical identity of the mineral are also encoded by the mamAB gene cluster.
Finally, we show that when expressed in the miniMAI strain, mmsF can significantly improve magnetite production and allow the cells to react to an applied magnetic field. However, these minerals are still smaller than those of the wild type cells. Perhaps, as suggested above, the expression of some of the miniMAI genes is lower in this strain than that of wild type, thus preventing MmsF from performing its normal function. Alternatively, additional genes contributing to biomineralization may exist in other parts of the MAI. For instance, in our previous work we showed that the R2 region might also contain biomineralization factors (Murat et al., 2010). Additionally, while none of the individual deletions of regions R6-R14 have a significant effect on biomineralization the combined loss of R6-R14 has a noticeable biomineralization defect, implying that combinations of MAI genes outside of R3 and R5 may participate in mineral formation. Despite these concerns, the work presented here sets the foundation for the creation of a mobile genetic cassette for the heterologous expression of magnetosomes. Future engineering of the levels of expression of the mamAB and mmsF genes in the minimal strain, as well as a more expanded search for biomineralization factors may lead to higher levels of magnetite production and therefore optimize the overall magnetic capabilities of such a mobile cassette.
Experimental Procedures
Strains and Growth Conditions
The DH5αλpir strain of Escherichia coli was used for all clonings. In E. coli, the antibiotics were used as follows: kanamycin 50 μg/mL, and carbenicillin 100 μg/mL. The WM3064 strain of E. coli was used as a donor strain in the conjugations and grown in the presence of 300 μM diaminopimelic acid (DAP). Magnetospirillum magneticum strain AMB-1 was grown in MG medium completed with Wolfe's Vitamin Solution as previously described (Komeili et al., 2006). When needed, iron malate (3 mM ferric chloride, 9 mM malate) was added at a final concentration of 30 μM. Conjugations were performed as described previously (Murat et al., 2010) using the WM 3064 E. coli strain as a donor. For Cmag measurements and TEM analysis, cells were grown in sealed Balch tubes filled with 10 mL of MG medium and 20 mL headspace in a 30° incubator without shaking. Balch tubes were flushed with N2 for 2 minutes immediately after autoclaving. In AMB-1, antibiotics were used at the following concentrations: kanamycin at 15 μg/mL in solid media and at 7-10 μg/mL in liquid media, and carbenicillin at 20 μg/mL on solid and liquid media.
Molecular Biology and Gene Deletion in AMB-1
All PCRs were carried out using the Promega GoTaq Green Master Mix according to supplier using a Biorad MyCycler thermocycler. Enzymes (restriction enzymes, T4 DNA ligase) were purchased from New England Biolabs. Primers were ordered from Integrated DNA Technologies, Inc, Iowa, USA (listed in Table S3, SI). All deletions were created using a two-step recombination method previously described (Komeili et al., 2004, Murat et al., 2010). Briefly, flanking regions upstream and downstream of the region to be deleted were amplified and fused by PCR using a 21-nucleotide linker. The resulting PCR fragment was cloned between the SpeI sites of the pAK0 suicide plasmid, which contains a kanamycin resistance cassette and the sacB gene (Komeili et al., 2004). The plasmids were transferred into AMB-1 cells using conjugation and the transconjugants were selected on MG media plates containing kanamycin. The deletion mutants were selected on media containing 2 % sucrose and individually characterized by PCR as described previously (Murat et al., 2010). For complementation experiments, the genes of interest were cloned between the SpeI and NotI sites in pAK253, a plasmid which allows for the integration of the construct on the chromosome of AMB-1 (Murat et al., 2010). In the case of the Δmms6 mutant, we found that complementation efficiency was improved when mms6 was introduced on a replicative plasmid. Mms6 was cloned under the control of a tac promoter in a pAK22-derived vector carrying an ampicillin resistance cassette. Except for that of mms6, all complementation experiments were performed in the absence of antibiotics
Time-Course Experiments
To follow the early stages of magnetite synthesis, AMB-1 and the ΔmmsF mutant cells were passaged twice in 10 mL MG medium lacking iron in 20 mL Balch tubes and incubated at 30° in a microaerobic chamber, in which the oxygen concentration was maintained below 10% (Quinlan et al., 2011). To prevent magnetite synthesis, all glassware was soaked in oxalic acid for 24 hours prior to use and remove trace iron. The absence of crystals was verified using TEM on whole cells. Non-magnetic cells in exponential phase were then passaged into 15 mL MG medium containing iron malate, in Balch tubes which had been flushed with N2 for two minutes before and after the addition of vitamins and iron (t0). At each time point (starting 30 minutes post the addition of iron) absorbance and Cmag were monitored. Up to 24 hours post transfer into iron-replete media, two 15 mL cultures were pooled at each time point and cells were collected by filtration and fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer for visualization and analysis by TEM.
TEM and Crystal Analysis
For TEM characterization, mutants were grown in microarerobic conditions up to an OD 400 nm of 0.2-0.3. 500 μl of cells were spun down and resuspended in 10-20 μL of MG medium and adsorbed onto 400-mesh copper grid (Ted Pella Inc) coated with Formvar film. Samples were then analyzed with an FEI Tecnai 12 TEM equipped with Gatan Bioscan (1k × 1k) CCD Camera Model 792 at an accelerating voltage of 120 kV. For each mutant, at least 30 cells with 1 to 20 crystals were imaged and crystal size was measured by hand using the measuring tool in the GNU Image Manipulation Program 2.6. A statistical analysis of all the values obtained was carried out and are reported in the Supplemental Information (SI). The longest axis between two parallel crystal faces is reported as crystal size or the length and the axis perpendicular to that was considered the width. The shape factor represents the ratio of the short axis over the long axis.
XRD and HRTEM
XRD was measured at the μ-Spot synchrotron beamline (BESSY II, Helmholtz Zentrum Berlin) with a 100 μm beam of 15 keV (Fischer et al., 2011). High resolution images were acquired on a transmission electron microscope from Zeiss (LIBRA 200) operated at 200 kV at the Helmholtz Center for Energy (Berlin, Germany).
Fluorescence Microscopy
The GFP fusions were derived from the pAK22 plasmid and analyzed as described previously (Murat et al., 2010). For each construct, more than 200 cells were photographed under a 100× magnification and analyzed.
Magnetosome Purification
To study the association of GFP-MmsF with the magnetosomes, 1L of AMB-1 expressing this protein on a plasmid was grown to exponential phase in complete MG media containing kanamycin, and then divided into three fractions, from which cells were collected by centrifugation at 4°, 6000 rpm for 15 minutes. Pellets were resuspended in 5 mL of lysis buffer (20mM HEPES pH7.5; 25 mM NaCl; 0.5 mM EDTA; complete EDTA-free protease inhibitor) and exposed to three cycles of freeze-thaw using liquid nitrogen. Subsequently, cells were treated with 20 μg/mL lysozyme, 25 μg/mL DNaseI, 14.6 mM β-mercapto ethanol, and 1 mM PMSF on ice for 1.5 hours. The lysate was then sonicated on ice for four 5 second intervals using a microtip at output setting 2 of a Branson Sonifier 250, before being passed over a magnetized MACS® separation column equilibrated in 10 mL of elution buffer (20 mM HEPES pH 7.5; 25 mM NaCl, 14.1 mM β-mercapto ethanol; 1mM PMSF). After 10 washes of 1 mL elution buffer, magnetization of the column was removed and the remaining material was eluted in 5 mL of elution buffer using the plunger supplied with the columns.
Whole Cell sample preparation for Electron Cryo Tomography
Lacey formvar coated 3 mm copper TEM grids purchased from Ted Pella Inc., were glow discharged for 20 seconds to make the carbon hydrophilic. A 3 μL drop of 10 nm fiducial gold suspension was applied to the grids and allowed to dry. Using the FEI Vitrobot model Mark II, a 3 μL drop of a freshly prepared whole cell suspension was applied to the grid; the grid was blotted for 6 seconds and plunged into liquid ethane, temperature <-175 °C. The resulting specimen consisted of amorphous ice with cells frozen in near native state, thin enough to allow imaging with 300kV electrons.
Cryo TEM data collection
TEM specimens were transferred under liquid nitrogen into the JEOL JEM 3100FSC field emission cryo-TEM, equipped with an in-column Omega-type energy filter spectrometer. The microscope was operated at 300 kV with a energy slit width of 30 eV shifted to record zero loss images. Tomographic tilt series data sets were collected with the specimen tilted from −60° to +60° in increments of 1.0°. Images were recorded with a Gatan 2K charge-coupled device digital camera at underfocus values between −12 and −16 microns. The CCD pixel size was 24 microns. The image pixel size was usually 1.2 nm. SerialEM software (Mastronarde, 2005) was used to help automate the collection of low dose whole cell cryo-electron tomograms.
Tomographic data alignment and reconstruction
The images were pre-aligned in Etomo/IMOD software (Kremer et al., 1996). Final alignments were either done with RAPTOR (Amat et al., 2008) software or within IMOD and then combined using either weighted back-projection methods, using Etomophile/IMOD, or SIRT, using Tomo3d (Agulleiro & Fernandez, 2011, Agulleiro et al., 2010) to re-construct the three-dimensional structure.
Supplementary Material
Acknowledgments
We would like to thank the members of the Komeili lab for their helpful discussions and critical reading of the manuscript. A.K. was supported by a David and Lucille Packard Foundation Fellowship in Science and Engineering and the National Institute of Health (R01GM084122). We would also like to thank M. Wollgarten for access to the HRTEM. K.H.D and R.C. were supported by a U.S. Department of Energy Contract.
References
- Agulleiro JI, Fernandez JJ. Fast tomographic reconstruction on multicore computers. Bioinformatics. 2011;27:582–583. doi: 10.1093/bioinformatics/btq692. [DOI] [PubMed] [Google Scholar]
- Agulleiro JI, Garzon EM, Garcia I, Fernandez JJ. Vectorization with SIMD extensions speeds up reconstruction in electron tomography. J Struct Biol. 2010;170:570–575. doi: 10.1016/j.jsb.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Alphandery E, Faure S, Seksek O, Guyot F, Chebbi I. Chains of magnetosomes extracted from AMB-1 magnetotactic bacteria for application in alternative magnetic field cancer therapy. ACS Nano. 2011;5:6279–6296. doi: 10.1021/nn201290k. [DOI] [PubMed] [Google Scholar]
- Amat F, Moussavi F, Comolli LR, Elidan G, Downing KH, Horowitz M. Markov random field based automatic image alignment for electron tomography. J Struct Biol. 2008;161:260–275. doi: 10.1016/j.jsb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Amemiya Y, Arakaki A, Staniland SS, Tanaka T, Matsunaga T. Controlled formation of magnetite crystal by partial oxidation of ferrous hydroxide in the presence of recombinant magnetotactic bacterial protein Mms6. Biomaterials. 2007;28:5381–5389. doi: 10.1016/j.biomaterials.2007.07.051. [DOI] [PubMed] [Google Scholar]
- Arakaki A, Masuda F, Amemiya Y, Tanaka T, Matsunaga T. Control of the morphology and size of magnetite particles with peptides mimicking the Mms6 protein from magnetotactic bacteria. J Colloid Interface Sci. 2010;343:65–70. doi: 10.1016/j.jcis.2009.11.043. [DOI] [PubMed] [Google Scholar]
- Arakaki A, Webb J, Matsunaga T. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. Journal of Biological Chemistry. 2003;278:8745–8750. doi: 10.1074/jbc.M211729200. [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Ganss B, Goldberg M, Moradian-Oldak J, Paine ML, Snead ML, Wen X, White SN, Zhou YL. 3. Protein-protein interactions of the developing enamel matrix. Curr Top Dev Biol. 2006;74:57–115. doi: 10.1016/S0070-2153(06)74003-0. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- Ding Y, Li J, Liu J, Yang J, Jiang W, Tian J, Li Y, Pan Y. Deletion of the ftsZ-like gene results in the production of superparamagnetic magnetite magnetosomes in Magnetospirillum gryphiswaldense. J Bacteriol. 2010;192:1097–1105. doi: 10.1128/JB.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre D, Fischer A, Garcia-Rubio Is, Mastrogiacomo G, Gehring AU. Development of Cellular Magnetic Dipoles in Magnetotactic Bacteria. Biophysical Journal. 2010;99:1268–1273. doi: 10.1016/j.bpj.2010.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre D, Menguy N, Guyot F, Lopez O, Zuddas P. Morphology of nanomagnetite crystals: Implications for formation conditions. American Mineralogist. 2005;90:1793–1800. [Google Scholar]
- Faivre D, Menguy N, Posfai M, Schueler D. Environmental parameters affect the physical properties of fast-growing magnetosomes. American Mineralogist. 2008;93:463–469. [Google Scholar]
- Fischer A, Schmitz M, Aichmayer B, Fratzl P, Faivre D. Structural purity of magnetite nanoparticles in magnetotactic bacteria. J R Soc Interface. 2011;8:1011–1018. doi: 10.1098/rsif.2010.0576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene SE, Brilli M, Biondi E, Komeili A. Analysis of the CtrA Pathway in Magnetospirillum Reveals an Ancestral Role in Motility in Alphaproteobacteria. J Bacteriol. 2012 doi: 10.1128/JB.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg K, Müller EC, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol. 2004;70:1040–1050. doi: 10.1128/AEM.70.2.1040-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg K, Wawer C, Tebo BM, Schüler D. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl Environ Microbiol. 2001;67:4573–4582. doi: 10.1128/AEM.67.10.4573-4582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Buchanan AV, Weiss KM. Biomineralization in humans: making the hard choices in life. Annu Rev Genet. 2009;43:119–142. doi: 10.1146/annurev-genet-102108-134242. [DOI] [PubMed] [Google Scholar]
- Kirschvink JL, Hagadorn JW. A grand unified theory of biomineralization. Wiley-VCH; 2000. pp. 139–149. [Google Scholar]
- Kirschvink JL, Kobayashi-Kirschvink A, Woodford BJ. Magnetite biomineralization in the human brain. Proc Natl Acad Sci U S A. 1992;89:7683–7687. doi: 10.1073/pnas.89.16.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol Rev. 2012;36:232–255. doi: 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Komeili A, Vali H, Beveridge TJ, Newman DK. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci USA. 2004;101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kröger N, Poulsen N. Diatoms-from cell wall biogenesis to nanotechnology. Annu Rev Genet. 2008;42:83–107. doi: 10.1146/annurev.genet.41.110306.130109. [DOI] [PubMed] [Google Scholar]
- Lang C, Schüler D. Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl Environ Microbiol. 2008;74:4944–4953. doi: 10.1128/AEM.00231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C, Schüler D, Faivre D. Synthesis of magnetite nanoparticles for bio- and nanotechnology: genetic engineering and biomimetics of bacterial magnetosomes. Macromolecular bioscience. 2007;7:144–151. doi: 10.1002/mabi.200600235. [DOI] [PubMed] [Google Scholar]
- Lee N, Kim H, Choi SH, Park M, Kim D, Kim HC, Choi Y, Lin S, Kim BH, Jung HS, Park KS, Moon WK, Hyeon T. Magnetosome-like ferrimagnetic iron oxide nanocubes for highly sensitive MRI of single cells and transplanted pancreatic islets. Proc Natl Acad Sci U S A. 2011;108:2662–2667. doi: 10.1073/pnas.1016409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohsse A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One. 2011;6:e25561. doi: 10.1371/journal.pone.0025561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel S, Mohammadi M, Felfoul O, Lu Z, Pouponneau P. Flagellated Magnetotactic Bacteria as Controlled MRI-trackable Propulsion and Steering Systems for Medical Nanorobots Operating in the Human Microvasculature. Int J Rob Res. 2009;28:571–582. doi: 10.1177/0278364908100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mokrani N, Felfoul O, Afkhami Zarreh F, Mohammadi M, Aloyz R, Batist G, Martel S. Magnetotactic bacteria penetration into multicellular tumor spheroids for targeted therapy. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:4371–4374. doi: 10.1109/IEMBS.2010.5627105. [DOI] [PubMed] [Google Scholar]
- Murat D, Quinlan A, Vali H, Komeili A. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci U S A. 2010;107:5593–5598. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prozorov T, Mallapragada SK, Narasimhan B, Wang LJ, Palo P, Nilsen-Hamilton M, Williams TJ, Bazylinski DA, Prozorov R, Canfield PC. Protein-mediated synthesis of uniform superparamagnetic magnetite nanocrystals. Advanced Functional Materials. 2007;17:951–957. [Google Scholar]
- Quinlan A, Murat D, Vali H, Komeili A. The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol Microbiol. 2011;80:1075–1087. doi: 10.1111/j.1365-2958.2011.07631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Kube M, Bazylinski DA, Lombardot T, Glöckner FO, Reinhardt R, Schüler D. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. Journal of Bacteriology. 2007;189:4899–4910. doi: 10.1128/JB.00119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffel A, Gärdes A, Grünberg K, Wanner G, Schüler D. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. Journal of Bacteriology. 2008;190:377–386. doi: 10.1128/JB.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- Schüler D, Uhl R, Bäuerlein E. A simple light scattering method to assay magnetism in Magnetospirillum gryphiswaldense. Fems Microbiology Letters. 1995;132:139–145. [Google Scholar]
- Tanaka M, Arakaki A, Staniland SS, Matsunaga T. Simultaneously discrete biomineralization of magnetite and tellurium nanocrystals in magnetotactic bacteria. Appl Environ Microbiol. 2010;76:5526–5532. doi: 10.1128/AEM.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Mazuyama E, Arakaki A, Matsunaga T. MMS6 protein regulates crystal morphology during nano-sized magnetite biomineralization in vivo. J Biol Chem. 2011;286:6386–6392. doi: 10.1074/jbc.M110.183434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Okamura Y, Arakaki A, Tanaka T, Takeyama H, Matsunaga T. Origin of magnetosome membrane: proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics. 2006;6:5234–5247. doi: 10.1002/pmic.200500887. [DOI] [PubMed] [Google Scholar]
- Taoka A, Asada R, Sasaki H, Anzawa K, Wu LF, Fukumori Y. Spatial localizations of Mam22 and Mam12 in the magnetosomes of Magnetospirillum magnetotacticum. Journal of Bacteriology. 2006;188:3805–3812. doi: 10.1128/JB.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahyudi AT, Takeyama H, Matsunaga T. Isolation of Magnetospirillum magneticum AMB-1 mutants defective in bacterial magnetic particle synthesis by transposon mutagenesis. Appl Biochem Biotechnol. 2001;91-93:147–154. doi: 10.1385/abab:91-93:1-9:147. [DOI] [PubMed] [Google Scholar]
- Weiss BP, Kim SS, Kirschvink JL, Kopp RE, Sankaran M, Kobayashi A, Komeili A. Magnetic tests for magnetosome chains in Martian meteorite ALH84001. Proc Natl Acad Sci U S A. 2004;101:8281–8284. doi: 10.1073/pnas.0402292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.