Macroautophagy supports central CD4+ T cell tolerance by facilitating the direct presentation of endogenous self-antigens by medullary thymic epithelial cells.
Abstract
Macroautophagy serves cellular housekeeping and metabolic functions through delivery of cytoplasmic constituents for lysosomal degradation. In addition, it may mediate the unconventional presentation of intracellular antigens to CD4+ T cells; however, the physiological relevance of this endogenous MHC class II loading pathway remains poorly defined. Here, we characterize the role of macroautophagy in thymic epithelial cells (TECs) for negative selection. Direct presentation for clonal deletion of MHC class II–restricted thymocytes required macroautophagy for a mitochondrial version of a neo-antigen, but was autophagy-independent for a membrane-bound form. A model antigen specifically expressed in Aire+ medullary TECs (mTECs) induced efficient deletion via direct presentation when targeted to autophagosomes, whereas interference with autophagosomal routing of this antigen through exchange of a single amino acid or ablation of an essential autophagy gene abolished direct presentation for negative selection. Furthermore, when this autophagy substrate was expressed by mTECs in high amounts, endogenous presentation and indirect presentation by DCs operated in a redundant manner, whereas macroautophagy-dependent endogenous loading was essential for clonal deletion at limiting antigen doses. Our findings suggest that macroautophagy supports central CD4+ T cell tolerance through facilitating the direct presentation of endogenous self-antigens by mTECs.
Developing thymocytes test their TCR on peptide/MHC ligands presented by thymic stromal cells to ensure that a self-MHC–restricted and self-tolerant T cell repertoire is generated (Starr et al., 2003). They first interact with cortical thymic epithelial cells (cTECs) for positive selection and subsequently move into the thymic medulla, where they engage in tolerogenic contacts with medullary TECs (mTECs) and DCs (Petrie and Zuñiga-Pflucker, 2007).
Several unusual features of TECs have evolved to support distinct aspects of T cell differentiation (Klein et al., 2009). cTECs use unique proteases for the generation of MHC class I– or II–bound peptides that are essential for positive selection of a diverse T cell repertoire (Guerder et al., 2012). mTECs express the Autoimmune regulator (Aire) gene to make a broad spectrum of self-antigens visible to developing thymocytes for central tolerance induction through promiscuous expression of peripheral tissue antigens (PTAs; Derbinski et al., 2001; Anderson et al., 2002; Kyewski and Klein, 2006). A shared feature of cTECs and mTECs is their abundant and constitutive expression of MHC class II, rendering TECs unique among nonhematopoietic APCs. However, despite this commonality with professional APCs, both TEC subtypes display a conspicuously poor efficacy in presenting extracellular antigens along the classical MHC class II pathway, suggesting that they might instead use nonconventional, endogenous routes of MHC class II loading (Klein et al., 2001).
Two forms of autophagy have been implicated in endogenous MHC class II loading. Their common characteristic is the shuttling of cytoplasmic constituents into lysosomes (Strawbridge and Blum, 2007). In the case of chaperone-mediated autophagy, this occurs through Lamp-2a–dependent translocation of protein substrates across the lysosomal membrane (Kaushik and Cuervo, 2012). In the case of macroautophagy, portions of cytoplasm that can contain protein aggregates and organelles are sequestered into so-called autophagosomes, which subsequently fuse with lysosomal compartments for degradation of their cargo (Klionsky and Ohsumi, 1999). Both types of autophagy serve evolutionarily conserved housekeeping functions, but have been shown to potentially also intersect with the MHC class II–loading pathway (Zhou et al., 2005; Münz, 2009).
Evidence that macroautophagy can shuttle intracellular antigens onto MHC class II has been obtained in several cell culture models (Brazil et al., 1997; Nimmerjahn et al., 2003; Dörfel et al., 2005; Paludan et al., 2005; Riedel et al., 2008). In cell lines of diverse tissue origin, autophagosomal markers colocalized with MHC class II–loading compartments, and fusion of a model-antigen to light chain 3 (LC3), an autophagosomal membrane protein, enhanced recognition by specific CD4 T cells in vitro (Schmid et al., 2007). However, whether, where, and when macroautophagy-mediated endogenous MHC class II loading might be of physiological relevance in vivo remained unclear.
Macroautophagy occurs constitutively at low basal levels in essentially all tissues, reflecting its function in cellular homeostasis. Besides, macroautophagy can be strongly up-regulated upon nutrient deprivation and thereby provides metabolic substrates during starvation periods (Mizushima and Klionsky, 2007). As a marked exception from this dynamic adaptation to the metabolic status, the starvation-independent extent of constitutive macroautophagy in TECs is extraordinarily high (Mizushima et al., 2004). Specifically, roughly 70% of cTECs and 5–10% of mTECs contain unusually high numbers of autophagosomes (Nedjic et al., 2008). Of note, macroautophagy-positive mTECs without exception belong to the mature CD80hiMHCIIhi subset known to express the highest levels of Aire and the broadest spectrum of PTAs (Derbinski et al., 2005; Nedjic et al., 2008).
In line with a potential contribution of macroautophagy to the generation of MHC class II–bound peptides for T cell selection, the autophagosomal marker LC3 was found to co-localize with H2-DM–positive compartments in thymus cryosections (Kasai et al., 2009). Functional evidence for a critical role of macroautophagy in TECs for the generation of positively selecting peptide/MHC class II ligands was obtained in chimeric mice that harbored a thymus-graft carrying an autophagy gene 5 (Atg5)–null allele. This did not affect the principle capacity of TECs to support T cell development, but resulted in altered positive selection of several MHC class II–restricted anti–foreign TCRs (Nedjic et al., 2008). Consistent with this effect being caused by alterations in the composition of MHC class II–bound peptides on cTECs, positive selection was impaired for some CD4 T cell specificities but enhanced for others, whereas all MHC class I–restricted TCRs tested displayed normal selection (Nedjic et al., 2009).
It is much less clear whether macroautophagy-mediated antigen presentation by TECs, in particular those belonging to the medullary subset, might also be crucial for central CD4 T cell tolerance. Athymic nude mice grafted with Atg5−/− thymi developed chronic inflammation of the intestinal tract and inflammatory infiltrates in various other tissues (Nedjic et al., 2008). However, it remained open whether these autoimmune manifestations actually resulted from the escape of otherwise censored CD4 T cell specificities, be it through a lack of negative selection or a failure of deviation into the regulatory T (T reg) cell lineage. An alternative, but not mutually exclusive, scenario is that immune dysregulation and autoimmunity in these chimeras was driven by lymphopenia caused by impaired positive selection.
In this study, we specifically addressed the role of macroautophagy in TECs for central T cell tolerance. We tested (a) whether macroautophagy substrates are directly presented by TECs for negative selection of CD4 T cells, (b) whether interference with macroautophagy in TECs allows for the escape of forbidden CD4 T cell specificities, and (c) whether these two assumptions apply to an antigen that is expressed exclusively by Aire+ mTECs.
RESULTS
Direct presentation of mitochondrial or membrane-bound pigeon cytochrome c (PCC) by TECs
We performed a first series of experiments in a model consisting of two PCC-transgenic (tg) mouse lines: in the first (ePCC), PCC is expressed in its native form as a mitochondrial neo-self-antigen. In the second (mPCC), PCC is, through addition of a membrane anchor, expressed as a membrane protein on the cell surface (Oehen et al., 1996). In both cases, expression is driven by an MHC class I (H-2Kb) promoter construct. Previous work, primarily focusing on the fate of AD10 TCR-tg thymocytes, had established that both forms of PCC gain endogenous access to MHC class II of radioresistant thymic stromal cells, most likely TECs, resulting in negative selection of specific CD4 T cells (Oehen et al., 1996).
When studying the role of macroautophagy in this context, it was crucial to use a PCC-specific TCR whose positive selection was not affected by macroautophagy deficiency in TECs. Thus, we used the I-Ek–restricted AND TCR (Kaye et al., 1989).
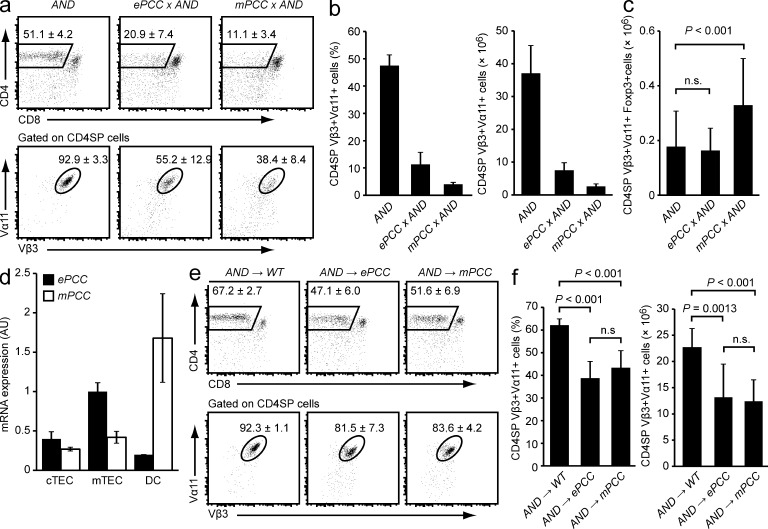
ePCC × AND and mPCC × AND double-tg mice, in which the neo-antigens were ubiquitously expressed in hematopoietic and nonhematopoietic tissues, displayed a similar extent of late negative selection of AND+ thymocytes at the CD4 single-positive (SP) stage (Fig. 1, a and b). In ePCC × AND double-tg mice, there was no increase in the absolute number of AND+ T reg cells, and in mPCC × AND double-tg mice, there was only a very modest, yet significant increase in the absolute number of AND+ T reg cells (P < 0.001), indicating that in both systems clonal deletion was the primary mechanism of tolerance (Fig. 1 c). Previous northern analyses had shown that the mPCC mRNA was much more abundant than the ePCC mRNA in total thymus tissue (Oehen et al., 1996). Quantitative (q)PCR with fractionated thymic cells indicated that this difference most likely stemmed from substantially higher expression of the mPCC transgene in DCs (Fig. 1 d). In contrast, in cTECs both mRNAs were expressed at identical levels (P = 0.088), whereas in mTECs the ePCC mRNA was actually about twofold more abundant (P = 0.002).
Figure 1.
Expression of ePCC or mPCC in nonhematopoietic thymic stromal cells is sufficient for partial negative selection of specific CD4SP cells. (a) Thymi of AND single-tg, ePCC × AND, and mPCC × AND mice were analyzed by flow cytometry. The mean frequency ± SD of CD4SP cells (top row) and of AND-TCR+ cells (Vα11+Vβ3+) among gated CD4SP thymocytes (bottom row) is indicated. (b) Frequencies and absolute numbers of CD4SP Vα11+Vβ3+ cells in mice of the indicated genotypes. (c) Absolute numbers of CD4SP Vα11+Vβ3+Foxp3+ T reg cells in mice of the indicated genotypes. Data in a–c are from at least 10 mice per genotype. (d) Relative abundance of the respective tg mRNA in purified cTECs, mTECs, and DCs from ePCC or mPCC tg mice as determined by qPCR. Thymi were enzymatically digested and enriched for low-density stromal cells, and mTECs, cTECs, and DCs were FACS-sorted for mRNA preparation. Values (arbitrary units, AU) are normalized to expression in mTECs from ePCC mice and indicate the mean ± SD from three independent biological replicates. (e) Thymocyte subsets in WT, ePCC, or mPCC recipients 4 to 5 wk after irradiation and reconstitution with AND bone marrow cells. Dot plots show the mean frequency ± SD of CD4SP cells (top row) and of AND-TCR+ cells (Va11+Vβ3+) among gated CD4SP thymocytes (bottom row). Data are representative of at least nine chimeras per group. (f) Frequencies and absolute numbers of CD4SP Va11+Vβ3+ cells in e.
BM chimeras were generated to eliminate concomitant neo-antigen expression by hematopoietic cells. This revealed an extent of partial deletion that was very similar between the two models when the transgenes were expressed exclusively by nonhematopoietic thymic stromal cells (Fig. 1, e and f).
Macroautophagy is necessary for direct presentation of mitochondrial, but not membrane–bound, PCC
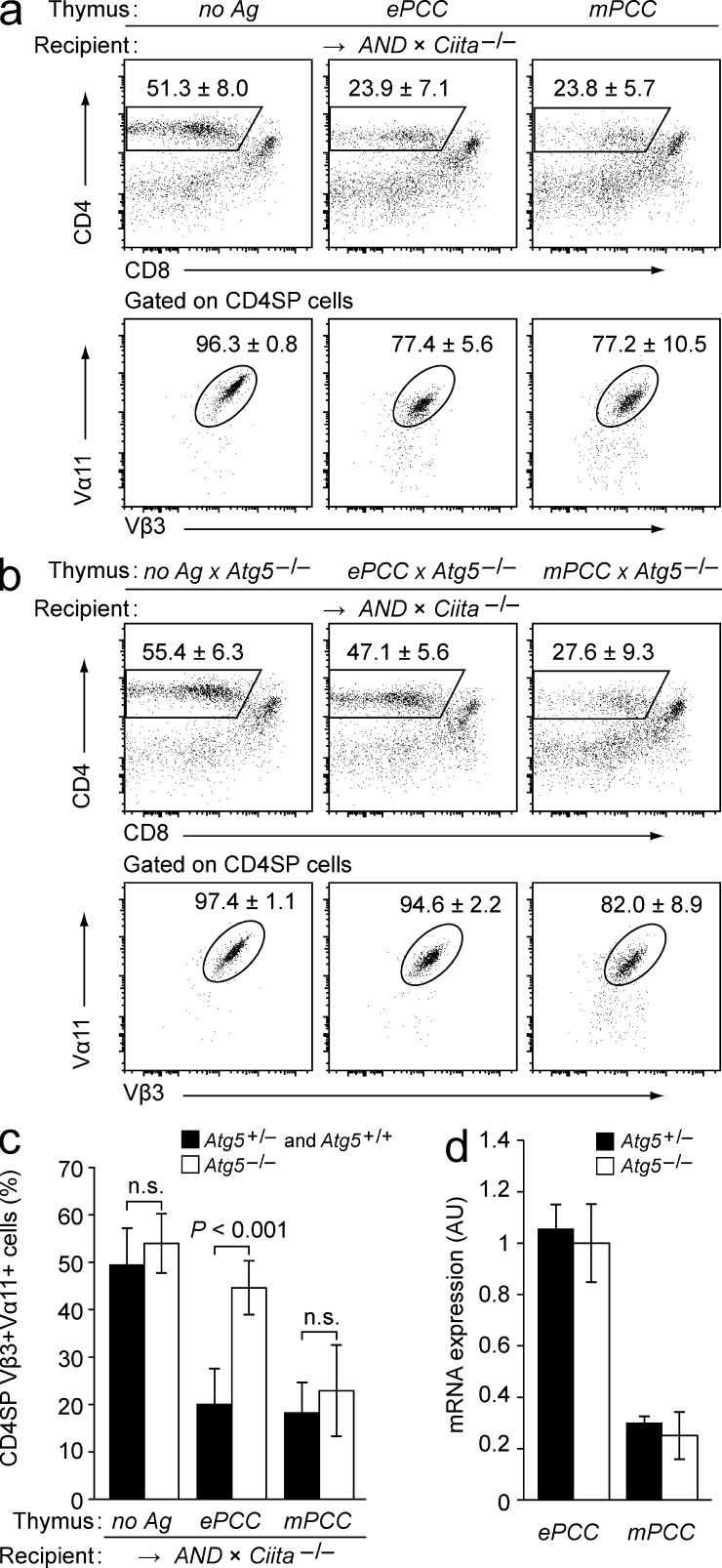
We then asked whether negative selection was mediated through direct endogenous presentation by nonhematopoietic thymic stromal cells or required indirect presentation by DCs. To generate chimeras lacking MHC class II on hematopoietic APCs, thymi from neo-antigen tg embryos were transplanted under the kidney capsule of AND-tg recipients on a class II transactivator (Ciita)–deficient background (Chang et al., 1996). Irrespective of whether PCC was expressed in the graft as mitochondrial or membrane-associated antigen, absence of MHC class II on hematopoietic APCs did not interfere with efficient negative selection (Fig. 2 a). Thus, both forms of PCC were directly presented by nonhematopoietic thymic stromal cells. Among these only cTECs and mTECs constitutively express MHC class II, so that negative selection with a high likelihood was a result of direct antigen presentation by TECs.
Figure 2.
Direct presentation of mitochondrial or membrane-bound PCC by TECs is sufficient for negative selection, but differs in its requirement for autophagy. (a) Subset composition of antigen non-tg (no Ag), ePCC, or mPCC thymi 4–6 wk after transplantation into AND × Ciita−/− recipients. The mean frequency ± SD of CD4SP cells (top row) and of AND-TCR+ cells (Vα11+Vβ3+) among gated CD4SP thymocytes (bottom row) is indicated. Data are representative of ≥11 chimeras per group. (b) Subset composition of WT × Atg5−/−, ePCC × Atg5−/−, or mPCC × Atg5−/− thymi 4–6 wk after transplantation into AND × Ciita−/− recipients. The mean frequency ± SD of CD4SP cells (top row) and of AND+ cells (Vα11+Vβ3+) among gated CD4SP thymocytes (bottom row) is indicated. Data are representative of ≥10 chimeras per group. (c) Frequencies of CD4SP Vα11+Vβ3+ cells among total thymocytes in a and b. (d) Relative abundance of the respective tg mRNA in purified total TECs from ePCC and mPCC tg thymi on Atg5+/− or Atg5−/− background as determined by qPCR. TECs were isolated from grafted E14-16 thymi 4–5 wk after transplantation into WT recipients. Values (arbitrary units; AU) are normalized to expression in TECs from ePCC Atg5−/− thymi and indicate the mean ± SD from three independent biological replicates.
To address whether direct presentation of PCC depended on macroautophagy (hereafter referred to as autophagy), we used a mouse model lacking Atg5. Atg5−/− mice die shortly after birth, presumably owing to incapacity to bridge a postnatal metabolic crisis associated with the switch from transplacental nutrient supply to milk nutrients (Kuma et al., 2004). However, Atg5−/− embryos do not show signs of developmental abnormalities (Kuma et al., 2004) and can be used as thymus-donors for transplantation experiments (Nedjic et al., 2008). Atg5−/− thymus grafts support bulk T cell differentiation, do not differ from Atg5+/+ controls in their stromal cell composition and show normal expression MHC class II on TECs (Nedjic et al., 2008).
Thymi from Atg5−/− embryos were transplanted under the kidney capsule of AND × Ciita−/− recipients (Fig. 2, b and c; compare with Fig. 2 a). As expected based on previous work, positive selection of AND thymocytes in the absence of cognate antigen was not affected by Atg5-deficiency in TECs (Nedjic et al., 2008). However, disruption of the Atg5 gene in ePCC thymi almost completely abolished negative selection. In contrast, negative selection in mPCC tg lobes was not affected by Atg5 deficiency. Importantly, disruption of Atg5 did not influence ePCC or mPCC mRNA-amounts in TECs, excluding that interference with autophagy might have differentially affected the expression of the two neo-antigens (Fig. 2 d). In sum, these observations indicated that autophagy was necessary for the direct tolerogenic presentation of mitochondrial PCC, whereas membrane-associated PCC was directly presented irrespective of whether or not nonhematopoietic thymic stromal cells, i.e., most likely TECs, harbored an intact Atg5 gene.
Targeting of a model antigen to autophagosomes
The PCC system was not suited to assess the role of autophagy for tolerance to an antigen that is expressed exclusively by mTECs. This is of particular interest given its implications for how promiscuously expressed PTAs induce central tolerance. Against this background, we set out to generate a novel model system where the same mTEC-specific antigen was deliberately targeted for autophagosomal degradation in one setting but not in a second setting.
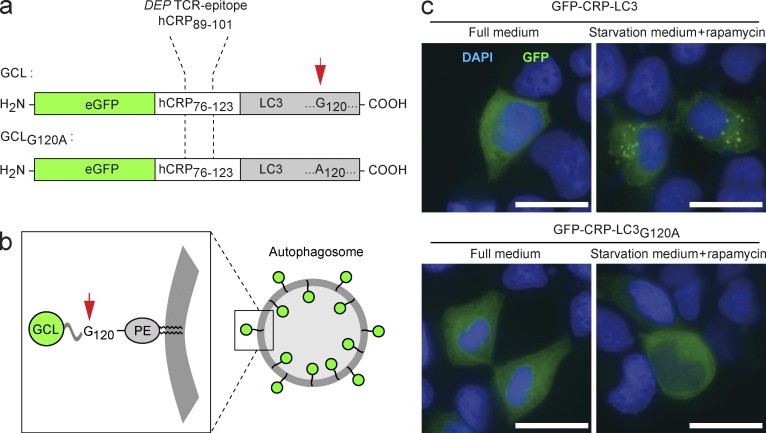
A tripartite hybrid protein was cloned, consisting of an N-terminal enhanced GFP as reporter component followed by a CD4 T cell epitope of human C-reactive protein (hCRP) as antigenic determinant and a C-terminal LC3 for autophagosomal targeting (Fig. 3 a). This fusion protein will be referred to as GCL (GFP-CRP-LC3). During autophagy, LC3 is linked to the inner and outer autophagosomal membrane, and antigen-LC3 fusion proteins have been shown to target the respective antigen to autophagosomes in cell culture models (Schmid et al., 2007).
Figure 3.
GCL is targeted to autophagosomes, whereas a difference by 1 aa in GCLG120A abolishes autophagosomal routing. (a) Schematic representation of the GCL and GCLG120A fusion proteins. (b) During autophagy, LC3 is covalently coupled to the inner and outer autophagosomal membrane. After fusion with lysosomal compartments, LC3 in the autophagosomal lumen is broken down proteolytically. (c) HEK 293T cells were transfected with expression vectors encoding for GCL or GCLG120A. Subsequently, cells were either cultured in full medium or transferred to starvation medium. Bars, 20 µm. Images are representative of two independent experiments.
We chose the CRP epitope as antigenic determinant to establish an independent model whose key components (antigen, TCR, and restricting MHC element) were different from the PCC and AND systems. Importantly, a CRP-specific tg TCR was available whose positive selection is independent of autophagy in TECs (Nedjic et al., 2008).
Anchoring of LC3 to the autophagosomal membrane requires the covalent attachment of phosphatidylethanolamine to glycine at position 120 (Fig. 3 b; Kabeya et al., 2000). To produce a variant of the GCL antigen that would not be attached to the autophagosomal membrane, we generated a second fusion protein in which position 120 of LC3 was changed to alanine (GCLG120A; Fig. 3 a).
Expression plasmids encoding for GCL or GCLG120A were transfected into HEK 293T cells. When cultured in full medium, both transfectants displayed a diffuse cytoplasmic GFP staining. Induction of autophagy through culturing in starvation medium led to the emergence of GFP+ punctae in cells expressing the GCL vector, indicating the incorporation of the GCL protein into nascent autophagosomes. In contrast, no such starvation-induced segregation of GFP into autophagosomes was seen with GCLG120A-expressing cells (Fig. 3 c). Thus, the variation of a single amino acid resulted in a differential propensity of GCL and GCLG120A to co-localize with autophagosomes.
Differential direct presentation of GCL and GCLG120A by mTECs
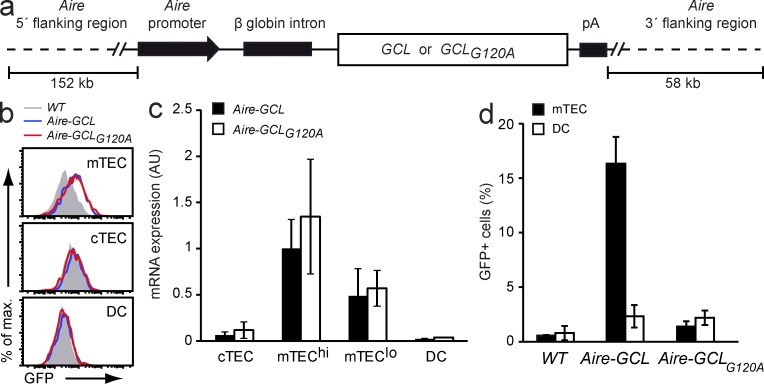
To express the fusion proteins in mTECs of tg mice, bacterial artificial chromosomes were generated bringing GCL or GCLG120A under the transcriptional control of the Aire gene-locus (Fig. 4 a). Microinjections yielded seven and nine founders for the Aire-GCL and the Aire-GCLG120A construct, respectively. Based on FACS analyses of GFP-fluorescence and qPCR data, a matched pair of tg lines was chosen for further analyses (Fig. 4, b and c). Compared with other founder lines, these two mouse strains (referred to as Aire-GCL and Aire-GCLG120A) displayed intermediate expression of GCL or GCLG120A, respectively.
Figure 4.
GCL and GCLG120A are equally expressed in mTECs of tg mice, yet differ in their direct presentation by mTECs. (a) Diagram of the Aire-GCL and Aire-GCLG120A BAC transgenes driving expression of the fusion proteins in mTECs. (b) Thymic stromal cells from WT (gray), Aire-GCL (blue), and Aire-GCLG120A (red) mice were analyzed by flow cytometry for GFP-fluorescence. Data are representative of two independent experiments, each with cells from three or more animals per genotype. (c) Abundance of tg mRNAs in thymic stromal cells from Aire-GCL or Aire-GCLG120A mice as determined by qPCR. Values are normalized to mTEChi from Aire-GCL mice and indicate the mean ± SD from three biological replicates. (d) NFAT-GFP reporter hybridoma cells expressing the hCRP89-101 specific DEP-TCR were co-cultured with mTECs (filled bars) or DCs (open bars) from Aire-GCL or Aire-GCLG120A mice. The mean frequency ± SD of GFP+ hybridoma cells is indicated. Data summarize three independent experiments with material from six or more thymi.
We next addressed whether neo-antigen expression in Aire-GCL and Aire-GCLG120A mTECs resulted in ex vivo measurable direct or indirect antigen presentation by mTECs or DCs, respectively. mTECs or DCs were purified and co-cultured with CRP89-101-specific T hybridoma cells carrying an NFAT-driven GFP reporter. With cells purified from Aire-GCL mice, mTECs induced a strong up-regulation of GFP in responder hybridoma cells, whereas DCs only lead to a marginal response (P = 0.048). In contrast, both mTECs and DCs from Aire-GCLG120A mice only elicited a very weak response as compared with WT control cells (P = 0.008 and P = 0.013, respectively; Fig. 4 d). Thus, direct presentation of an autophagy substrate by mTECs was substantially more efficient as compared with an antigen-variant that was expressed at identical levels yet differed by 1 aa and was therefore not targeted to autophagosomes.
mTECs directly present GCL, but not GCLG120A, for negative selection
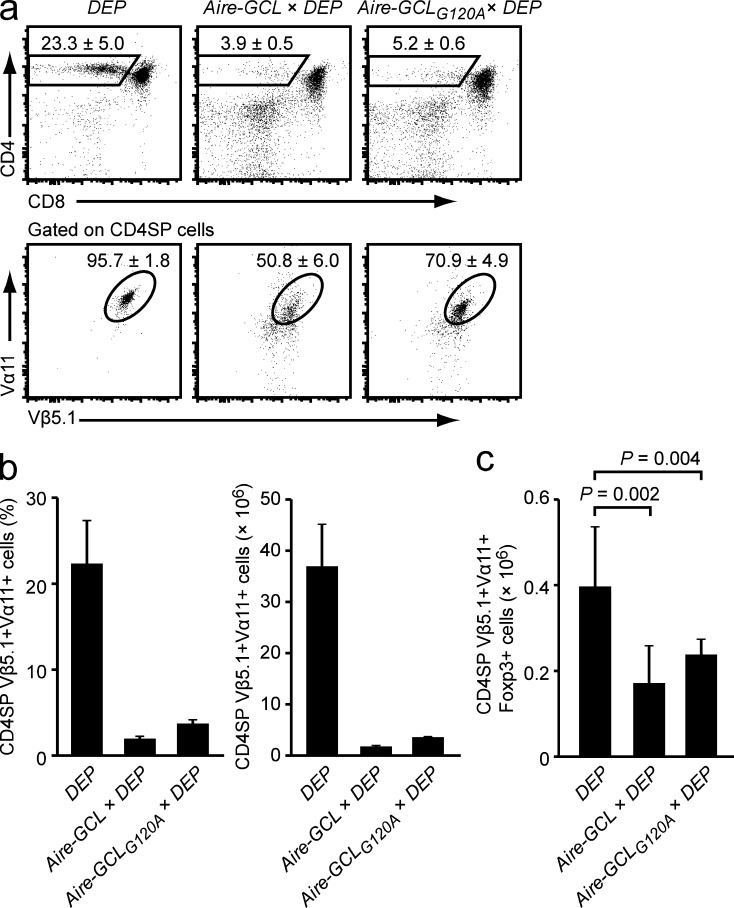
Aire-GCL and Aire-GCLG120A mice were bred to DEP TCR-tg mice that express a TCR that recognizes the epitope CRP89-101 (Klein et al., 1998). Both double tg strains displayed a similar extent of late negative selection at the CD4SP stage (Fig. 5, a and b). In both Aire-GCL × DEP and Aire-GCLG120A × Aire-GCL double-tg mice there was a similar decrease in the absolute number of CRP-specific (Vα11+Vβ5.1+) T reg cells as compared with DEP single-tg mice, indicating that in both systems central tolerance operated through clonal deletion rather than T reg induction (Fig. 5 c).
Figure 5.
Similar phenotype of negative selection in Aire-GCL × DEP and Aire-GCLG120A × DEP mice. (a) Thymi of DEP single tg, Aire-GCL × DEP, and Aire-GCLG120A × DEP mice were analyzed by flow cytometry. The mean frequency ± SD of CD4SP cells (top row) and of DEP-TCR+ cells (Vα11+Vβ5.1+) among gated CD4SP thymocytes (bottom row) is indicated. (b) Frequencies and absolute numbers of CD4SP Vα11+Vβ5.1+ cells in mice of the indicated genotypes. Data in a and b are from ≥10 mice per genotype. (c) Absolute numbers of CD4SP Vα11+Vβ5.1+Foxp3+ T reg cells in mice of the indicated genotypes. Data in c are from at least five mice per genotype.
Given the marked difference in antigen presentation by purified thymic APCs from Aire-GCL or Aire-GCLG120A mice (see Fig. 4 d), we were surprised to find a very similar extent of clonal deletion in both strains. We therefore hypothesized that distinct modalities of antigen presentation may operate in the two systems, yet elicit an essentially identical outcome. To exclude the formal possibility that ectopic GCL- or GCLG120A-expression in hematopoietic cells might contribute to negative selection, we first verified in BM chimeras whether expression exclusively in thymic epithelium was sufficient to mediate negative selection. Indeed, the thymi of DEP → Aire-GCL and DEP → Aire-GCLG120A BM chimeras recapitulated the phenotype of the respective double tg constellations (Fig. 6 a compared with Fig. 5 a).
Figure 6.
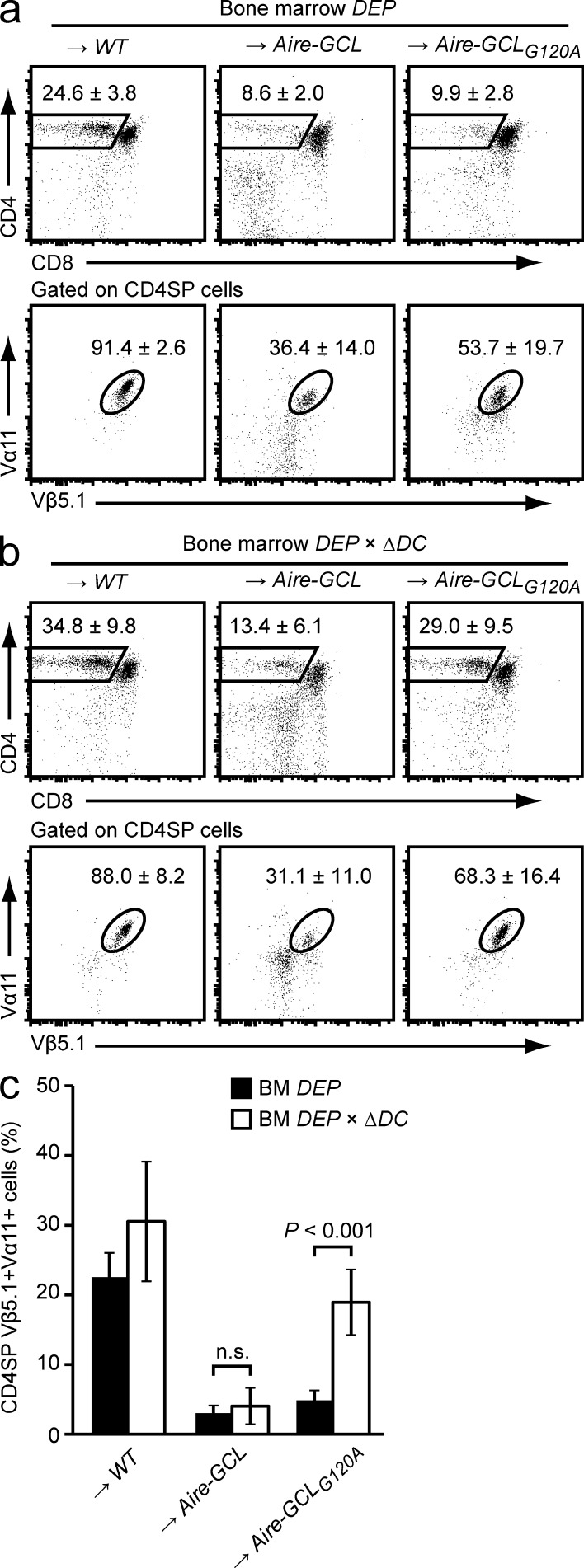
mTECs directly present GCL, but not GCLG120A, for negative selection. (a) Thymocyte subsets in WT, Aire-GCL, or Aire-GCLG120A recipients 4–5 wk after reconstitution with DEP BM cells. Dot plots show the mean frequency ± SD of CD4SP cells (top row) and of DEP-TCR+ cells (Vα11+Vβ5.1+) among gated CD4SP cells (bottom row). Data are representative of ≥10 chimeras per group. (b) WT, Aire-GCL, or Aire-GCLG120A recipients were reconstituted with DEP × ΔDC BM. 4–5 wk later, thymi were analyzed by flow cytometry. Plots show the mean frequency ± SD of CD4SP cells (top row) and of DEP-TCR+ cells (Vα11+Vβ5.1+) among gated CD4SP cells (bottom row). Data are representative of nine or more chimeras per group. (c) Frequencies of CD4SP Vα11+Vβ5.1+ cells among total thymocytes in a and b.
We next asked whether negative selection was mediated through direct antigen presentation by mTECs, or required antigen transfer to and indirect presentation by DCs. Aire-GCL or Aire-GCLG120A mice were reconstituted with DEP-tg BM harboring a diphtheria toxin–based suicide cassette controlled by CD11c (ΔDC mice; Ohnmacht et al., 2009). Irrespective of the lack of DCs, DEP × ΔDC → Aire-GCL chimeras recapitulated the deletion phenotype of DC-sufficient DEP → Aire-GCL chimeras (Fig. 6, b and c; compare Fig. 6 a). In contrast, DC-deficiency in DEP × ΔDC → Aire-GCLG120A chimeras resulted in a nearly complete rescue from negative selection. Together, these findings documented a differential capacity of GCL or GCLG120A mTECs to directly present the respective endogenously expressed neo-antigen for negative selection. Specifically, negative selection resulting from autophagosome-targeted GCL was mediated by directly presenting mTECs, whereas efficient clonal deletion by GCLG120A was dependent on indirect presentation by DCs.
Direct antigen presentation by Aire-GCL mTECs is autophagy dependent
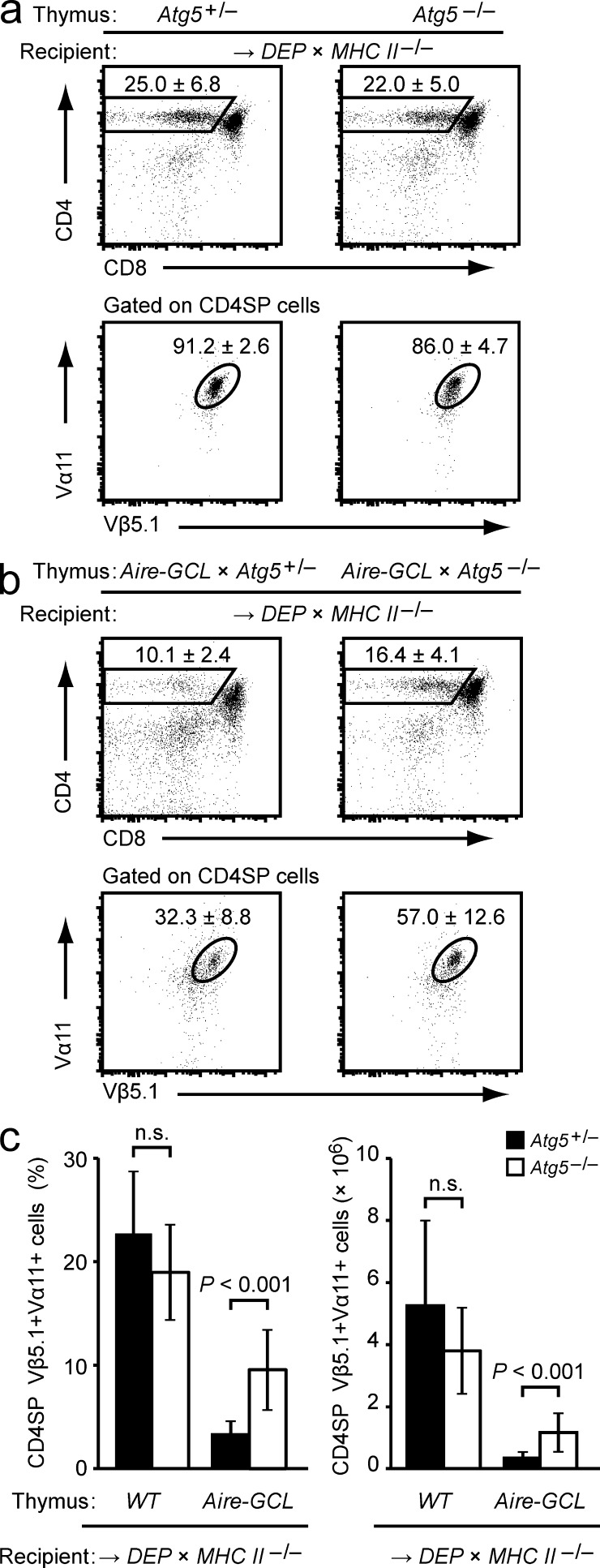
To test whether autophagy was responsible for the superior direct presentation of GCL, we generated a series of thymus chimeras to interfere with autophagy in TECs. As recipients of thymus grafts, we reconstituted WT mice with H2-Ab1–deficient DEP-tg BM. Because of the disrupted I-Ab β-chain gene in BM-derived cells, these mice (for clarity referred to as DEP × MHCII−/−) lacked MHC class II expression in DCs and were therefore indirect presentation–incompetent. Comparison of antigen nonexpressing chimeras confirmed that positive selection of the DEP TCR proceeded efficiently on both Atg5+/− and Atg5−/− thymic epithelium (Fig. 7 a; Nedjic et al., 2008). When the grafted epithelium expressed the Aire-GCL transgene and was autophagy-sufficient, negative selection of DEP T cells occurred (Figs. 7, b and c). Because this setting recapitulated the constellation in the DEP × ΔDC → Aire-GCL BM chimeras described in Fig. 6 (b and c), this was expected and confirmed that direct presentation by Aire-GCL mTECs was sufficient for efficient deletion. In contrast, when the grafted Aire-GCL epithelium was autophagy deficient, a significantly reduced extent of negative selection was seen (P < 0.001; Fig. 7, b and c). Thus, efficient endogenous loading of a GCL-derived epitope onto MHC class II of mTECs for negative selection required an intact autophagy pathway.
Figure 7.
Direct antigen presentation by Aire-GCL mTECs is autophagy-dependent. (a) Thymocyte subsets in Atg5+/− or Atg5−/− thymi 4–6 wk after transplantation into DEP × MHC Class II−/− recipients. Plots show the mean frequency ± SD of CD4SP cells (top row) and of DEP-TCR+ cells (Vα11+Vβ5.1+) among gated CD4SP thymocytes (bottom row). Data are representative of ≥11 chimeras per group. (b) Thymocyte subsets in Aire-GCL × Atg5+/− or Aire-GCL × Atg5−/− thymic lobes 4–6 wk after transplantation into DEP × MHC Class II−/− recipients. Plots show the mean frequency ± SD of CD4SP cells (top row) and of DEP-TCR+ cells (Vα11+Vβ5.1+) among gated CD4SP thymocytes (bottom row). Data are representative of ≥13 chimeras per group. (c) Frequencies and absolute numbers of CD4SP Vα11+Vβ5.1+ cells in a and b.
Direct and indirect presentation of GCL operate in parallel
The data depicted in Fig. 6 (b and c) had shown that clonal deletion by the GCLG120A antigen-variant that was not targeted to autophagosomes depended on indirect presentation by DCs. In conjunction with our findings from Fig. 4 d, this indicated that antigen presentation by DCs close to the detection-limits of our in vitro assay could be sufficient for negative selection in vivo. By inference, it therefore remained possible that indirect presentation by DCs in the Aire-GCL system, despite being hardly above the limit of detection in vitro (Fig. 4 d) and not being essential for negative selection in vivo (Fig. 6, b and c), operated in parallel with direct presentation by mTECs. Of note, the experimental approach depicted in Fig. 7 by design excluded such a potential contribution of DC-mediated indirect presentation.
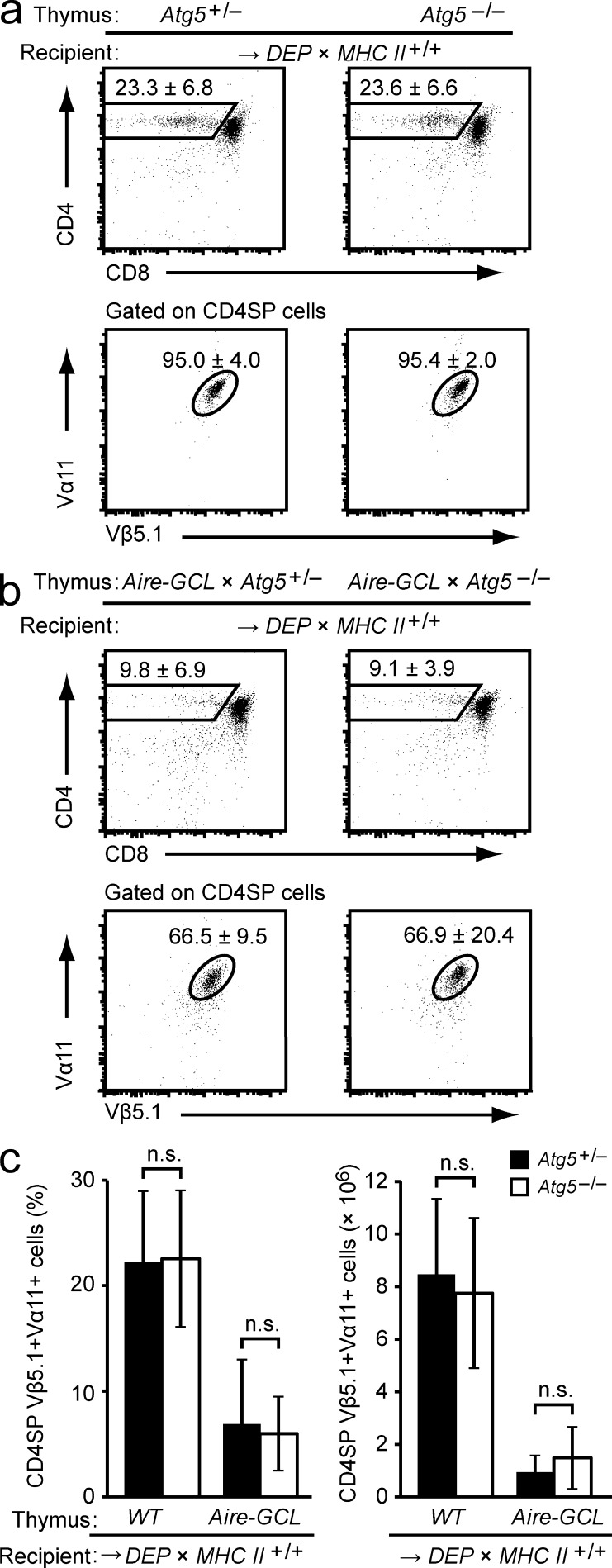
To establish a constellation in which indirect presentation by DCs—if occurring and being sufficiently strong—would compensate for the diminished direct presentation by Atg5-deficient mTECs, we transplanted Atg5−/− Aire-GCL thymi into MHC class II-sufficient DEP-tg recipients. Contrary to the observations with MHC class II-deficient hematopoietic APCs, an identical extent of negative selection was seen irrespective of whether the autophagy pathway in the grafted Aire-GCL epithelium was intact or disrupted (Fig. 8, a–c, compared Fig. 7). Thus, autophagy-dependent direct presentation by mTECs and autophagy-independent indirect presentation by DCs operate in parallel in the Aire-GCL thymus and can compensate for each other.
Figure 8.
Indirect presentation by DCs can compensate for impaired direct presentation by mTECs in Aire-GCL thymi. (a) Subsets in Atg5+/− or Atg5−/− thymi 4–6 wk after transplantation into DEP × MHC Class II+/+ recipients. Plots show the mean frequency ± SD of CD4SP cells (top row) and of DEP-TCR+ cells (Vα11+Vβ5.1+) among gated CD4SP thymocytes (bottom row). Data are representative of ≥19 chimeras per group. (b) Thymocyte subsets in Aire-GCL × Atg5+/− or Aire-GCL × Atg5−/− thymic lobes 4 to 6 wk after transplantation into DEP × MHC Class II+/+ recipients. Plots show the mean frequency ± SD of CD4SP cells (top row) and of DEP-TCR+ cells (Vα11+Vβ5.1+) among gated CD4SP thymocytes (bottom row). Data are representative of ≥10 chimeras per group. (c) Frequencies and absolute numbers of CD4SP Vα11+Vβ5.1+ cells in a and b.
Crucial role of autophagy-dependent direct presentation at low antigen doses
The redundancy of direct and indirect presentation in the Aire-GCL system was intriguing, particularly considering that CRP89-101 presentation as determined in vitro was considerably higher with mTECs as compared with DCs (Fig. 4 d). Given that indirect presentation by DCs, despite barely being measurable in vitro, was sufficient for negative selection, we reasoned that the level of direct presentation by Aire-GCL mTECs might substantially exceed the minimal requirement for saturating negative selection. By inference, the capacity of DCs to promote negative selection through the disfavored indirect pathway would be limited by the amount of GCL antigen that was expressed by mTECs.
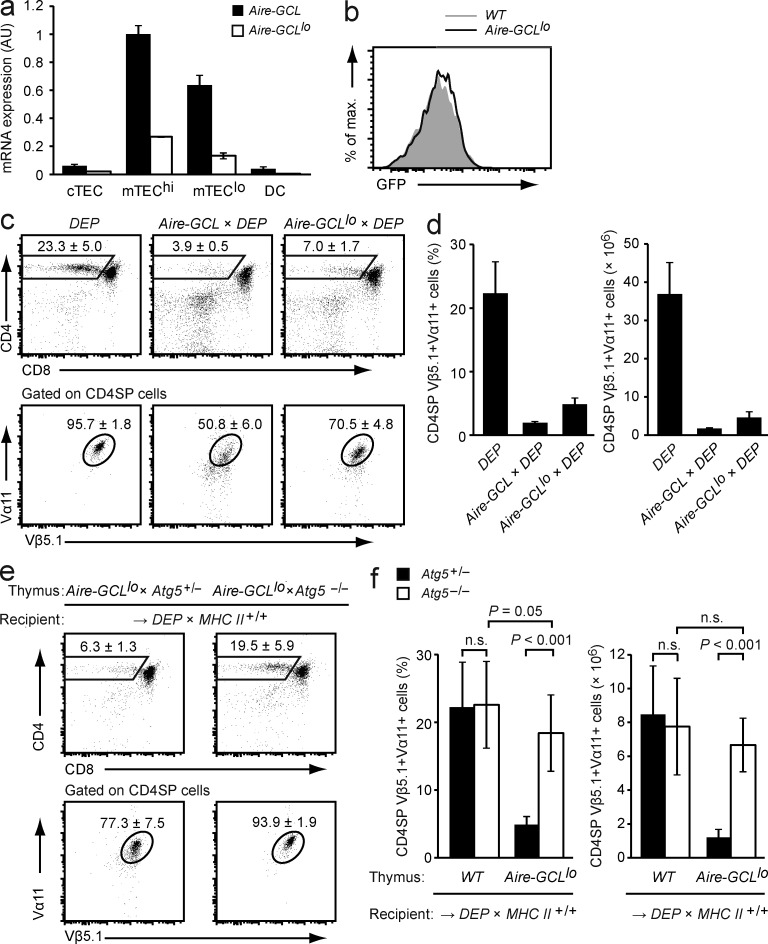
To test this hypothesis, we extended our analyses to a second Aire-GCL founder strain, hereafter referred to as Aire-GCLlo, which expressed the transgene at roughly fourfold lower mRNA levels compared with Aire-GCL mice (Fig. 9 a). GFP fluorescence was not detectable in Aire-GCLlo mTECs, indicating substantially lower protein expression (Fig. 9 b). Consistent with this, no direct presentation of the CRP-epitope by Aire-GCLlo mTECs was measurable in vitro using the reporter-hybridoma assay (unpublished data). However, regardless of these considerable quantitative differences in antigen expression, Aire-GCLlo × DEP double-tg mice displayed almost the same extent of deletion of DEP+ thymocytes as compared with Aire-GCL × DEP mice (Fig. 9, c and d).
Figure 9.
Autophagy-dependent direct presentation by mTECs is essential for negative selection at limiting antigen expression. (a) Expression of GCL mRNA in thymic stromal cells from Aire-GCL or Aire-GCLlo mice as determined by qPCR. Values (mean ± SD from three independent biological replicates) are normalized to expression in Aire-GCL mTEChi. (b) GFP fluorescence in mTECs from WT (gray) and Aire-GCLlo (black line) mice. Data are representative of three independent experiments with pooled cells from four or more animals per genotype. (c) Thymi of DEP single-tg, Aire-GCL × DEP, and Aire-GCLlo × DEP mice were analyzed by flow cytometry. The mean frequency ± SD of CD4SP cells (top row) and of DEP-TCR+ cells (Vα11+Vβ5.1+) among gated CD4SP thymocytes (bottom row) is indicated. (d) Frequencies and absolute numbers of CD4SPVα11+Vβ5.1+ cells in mice of the indicated genotypes. Data in c and d are from ≥8 mice per genotype. (e) Thymocyte subsets in Aire-GCLlo × Atg5+/− or Aire-GCLlo × Atg5−/− thymic lobes 4–6 wk after transplantation into DEP × MHC Class II+/+ recipients. The mean frequency ± SD of CD4SP cells (top row) and of DEP-TCR+ cells (Vα11+Vβ5.1+) among gated CD4SP thymocytes (bottom row) is indicated. Data are representative of at least 17 chimeras per group. (f) Frequencies and absolute numbers of CD4SP Vα11+Vβ5.1+ cells among total thymocytes in (e) as compared with antigen nonexpressing (WT) Atg5+/− (n = 19) or Atg5−/− (n = 20) thymic lobes. Data from antigen nonexpressing thymi are also shown in Fig. 8 a.
Transplantation of autophagy-sufficient Aire-GCLlo thymi into DEP recipients recapitulated the extent of negative selection seen in Aire-GCLlo × DEP double tg mice, confirming that antigen expression in mTECs was sufficient for negative selection (Fig. 9, e and f). However, negative selection was essentially abolished when these grafts harbored a disrupted Atg5 gene. Thus, despite seeding of the graft with indirect presentation–competent DCs, these cells did not compensate for impaired direct presentation by Atg5-deficient mTECs. Collectively, these observations indicated that at limiting amounts of antigen expression in mTECs, autophagy-dependent shuttling of an endogenous antigen onto MHC class II was an essential and nonredundant mechanism of direct antigen presentation for central tolerance.
DISCUSSION
Our results underscore the autonomous capacity of TECs to induce CD4 T cell tolerance through direct presentation of endogenously expressed self-antigens (Oukka et al., 1996; Klein et al., 1998; Aschenbrenner et al., 2007; Hinterberger et al., 2010; Hubert et al., 2011). In two independent model systems, we found that macroautophagy was essential for endogenous antigen-loading onto MHC class II of TECs for negative selection. Interference with autophagy resulted in the escape of forbidden CD4 T cell–specificities, whereby the requirement for autophagy varied with (a) the subcellular distribution, (b) the inherent propensity for autophagosomal routing, and (c) the expression level of the respective antigen.
In the particular model systems studied here, clonal deletion was the predominant/exclusive mode of CD4 T tolerance associated with autophagy-mediated antigen presentation by TECs. Nevertheless, it appears unlikely that distinct modalities of how TECs or other thymic APCs generate MHC II–bound epitopes are critical qualitative determinants of the ensuing fate of autoreactive CD4 T cells. Given that antigen presentation by mTECs can lead to both clonal deletion and T reg differentiation (Hinterberger et al., 2010), and considering the at least partial redundancy of TECs and DCs for T reg induction (Wirnsberger et al., 2009), we predict that for other combinations of TCR and/or self-antigen, T reg-induction may also depend upon autophagy-mediated direct antigen presentation by TECs. However, future experimentation is needed to test this hypothesis.
The introduction of a disrupted Atg5 gene into TECs abolished the direct presentation of mitochondrial ePCC, but not of plasma membrane-located mPCC. The significance of this is twofold: first, the robust Atg5-independent negative selection by mPCC TECs indicates that possible Atg5 knockout–connected collateral effects caused by subtle alterations in TEC physiology unrelated to MHC class II–loading do not impede their capacity to directly tolerize CD4 T cells. Second, these findings suggest that the subcellular distribution of a given antigen is a critical determinant of whether or not its direct presentation depends on autophagy.
How may topological parameters specify that both ePCC and mPCC are directly presented by TECs, yet differ in their respective dependence on autophagy? Contrary to the traditionally held view that the autophagosomal content indiscriminately represents the cytoplasmic content, there is mounting evidence for a substantial degree of substrate selectivity in autophagy. For instance, ubiquitinated proteins or inclusion bodies may be earmarked for autophagosomal degradation through selective recognition by p62/SQSTM1 (sequestosome 1) and Alfy (autophagy-linked FYVE protein; Simonsen et al., 2004; Bjørkøy et al., 2005; Pankiv et al., 2007). In the same vein, organelles such as peroxisomes, parts of the nucleus, and mitochondria can be engulfed by autophagosomes, whereby the latter process is referred to as mitophagy (Kim et al., 2007; Mizushima, 2011). We deem it likely that mitophagy is required for the direct presentation of mitochondrial ePCC. In contrast, the type II membrane protein mPCC may gain access to MHC class II in an autophagy-independent manner because its biosynthetic pathway possibly intersects with nascent MHC class II compartments and/or through reentry of plasma membrane proteins into endosomes. In line with these topological considerations, epitopes derived from membrane proteins make up a disproportionally high fraction of the MHC class II ligandome (Chicz et al., 1993; Dongre et al., 2001). Direct support for a differential role of autophagy in the routing of organelle-derived versus membrane-bound antigens to MHC class II comes from a proteomic survey of autophagy-associated changes in the composition of MHC class II–bound peptides in a cultured cell line: peptides derived from various true intracellular sources, including organelles, were significantly enriched upon induction of autophagy, whereas the abundance of membrane-derived epitopes remained constant (Dengjel et al., 2005).
The different molecular composition of the two antigen-variants, their ubiquitous expression in both cTECs and mTECs, and the potential effects of unequal expression levels limited the conclusive interpretation of some of the findings in the ePCC/mPCC system. We therefore decided to generate the novel Aire-GCL/Aire-GCLG120A model in which, through expression of two closely related protein variants at identical levels and specifically in mTECs, essentially all of these variables could be controlled experimentally. Consistent with favored access of autophagy substrates to MHC class II, only the GCL variant was directly presented by mTECs to an extent that was quantifiable in vitro and that was sufficient for clonal deletion in vivo. Importantly, this gain-of-function read-out provides evidence for autophagy-mediated direct antigen presentation by mTECs that does not bear the caveats of loss-of-function assays that solely rely on the disruption of autophagy genes.
Our findings have implications for how PTAs that are expressed by Aire+ mTECs might be presented for central CD4 T cell tolerance. There is convincing evidence that both a direct pathway (Oukka et al., 1996; Klein et al., 1998; Aschenbrenner et al., 2007; Hinterberger et al., 2010; Hubert et al., 2011) and an indirect pathway via DCs (Gallegos and Bevan, 2004; Koble and Kyewski, 2009; Hubert et al., 2011) can operate in this context. The parameters governing the requirement for one or the other mechanism remain to be established (Klein et al., 2011). We suggest that membrane associated self-antigens as well as substrates of autophagy (as for instance specified by partitioning of the respective antigen into particular organelles such as mitochondria or the nucleus) may preferentially be presented via the direct pathway. In line with this, membrane-bound hemagglutinin or ovalbumin (OVA), as well as nuclear β-galactosidase, when expressed exclusively by mTECs, were all found to promote CD4 T cell tolerance through direct presentation by mTECs (Oukka et al., 1996; Aschenbrenner et al., 2007; Hubert et al., 2011). The other way around, unless the antigenic determinant in question is a bona fide secreted protein, as in the case of mTEC-derived soluble OVA, which indeed requires presentation by DCs (Hubert et al., 2011), it is less obvious why tolerance to certain antigens may depend upon antigen transfer to and presentation by DCs. In the same vein, it remains unclear why negative selection in the Aire-GCLG120A system is at least in part dependent on indirect presentation by DCs.
We deem it unlikely that there is a strict demarcation between the direct and the indirect pathway of antigen presentation, and for a given antigen, the two routes of antigen display are certainly not mutually exclusive. For instance, negative selection in the Aire-GCL system could be promoted by both mTECs or DCs. Conceivably, a certain degree of redundancy between the two mechanisms may afford robustness to central tolerance. However, the lower-expressing Aire-GCLlo system revealed that below a certain threshold of antigen abundance, the autophagy-dependent mode of direct presentation can be essential. Whether one of the two Aire-GCL systems more closely reflects physiological expression of self-antigens in mTECs may vary from case to case. In particular, for promiscuously expressed PTAs, the available amount of antigen in individual mTECs is likely to be limited. In addition, the expression of a given PTA frequently seems to be confined to a minute fraction (1–3%) of mTECs (Smith et al., 1997; Klein et al., 1998; Klein et al., 2001). Therefore, constraints in the amount and spatial distribution of self antigens within the thymic microenvironment may have been a driving force for TECs to co-opt an evolutionary ancient catabolic process to focus the presentation of limited supplies of self-determinants onto their own MHC class II molecules.
MATERIALS AND METHODS
Mice.
Atg5−/− (Kuma et al., 2004), MHC class II−/− (H2-Ab1−/−; Cosgrove et al., 1991), Ciita−/− (Chang et al., 1996), ePCC (provided by S. Hedrick, University of California San Diego, La Jolla, CA), mPCC (provided by R. Schwartz, National Institutes of Health, Bethesda, MD; Oehen et al., 1996), AND (Kaye et al., 1989), DEP (Klein et al., 1998), and ΔDC mice (Ohnmacht et al., 2009) have been described previously. C57BL/6 and B10.A mice were purchased from Charles River. Mice were maintained under specific pathogen-free conditions in individually ventilated cages. All animal studies and procedures were approved by local authorities (Regierung von Oberbayern; Az 55.2–1-54-2531-7-08).
LC3 fusion proteins.
The LC3 and LC3G120A cDNAs (gift from N. Mizushima [Tokyo Medical and Dental University, Tokyo, Japan] and T. Yoshimori [Osaka University, Osaka, Japan]) were subcloned into the pBluescript II vector (Stratagene). Using primers designed to introduce the appropriate restriction sites, the following elements were amplified, subcloned, and inserted 5′ of LC3 or LC3G120A in the listed order (5′ → 3′): a β-globin intron from pSG5 (Stratagene), a cDNA encoding for eGFP from pTagGFP-C (Evrogen) and a cDNA fragment encoding for aa 76–123 of hCRP. For in vitro expression, the constructs were subcloned into the pMSCVneo vector (Takara Bio Inc.).
Aire-GCL and Aire-GCLG120A mice.
Constructs containing a β-globin intron followed by cDNAs encoding for GCL or GCLG120A, respectively, and a poly-A signal were inserted by homologous recombination into the mouse BAC RP23-77011 (BACPAC; Hinterberger et al., 2010). The targeting vectors contained homology boxes spanning nucleotides −379 to −2 (5′) and 17 to 399 (3′) of the Aire gene. A neomycin resistance gene flanked by FRT sites was used to select successfully recombined clones and was subsequently removed. Tg mice were generated by injection of linearized BAC DNA into pronuclei of C57BL/6 zygotes. For genotyping, the following primers were used: forward 5′-GATCCTGCAGTCTAAGGATATAGGATAC-3′ and reverse 5′-GCACCATAGTTATAAACCAG-3′.
Antibodies and flow cytometry.
FITC-conjugated mAbs to Vβ3 (KJ25), Vβ5.1/5.2 (MR9-4), Alexa Fluor 488 (A488)–conjugated mAb to Ly51 (6C3), phycoerythrin (PE)-conjugated mAbs to Va11.1/11.2 (RR8-1), Ly51 (BP1), Cy-chrome–conjugated mAbs to CD8a (53–6.7), CD45 (30-F11), PE-Cy7–conjugated CD4 (GK1.5), CD11c (N418), CD45 (30-F11), allophycocyanin (APC)-conjugated mAbs to I-A/I-E (M5/114.15.2), CD326/EpCAM (G8.8), APC-Cy7–conjugated mAbs to CD4 (GK1.5), CD326/EpCAM (G8.8), PE-Cy7–conjugated to streptavidin- (SA) and biotin-conjugated mAbs to CD4 (RM4-5), CD8a (53–6.7), CD80 (16-10A1), and B220 (RA3-6B2) were purchased from BD, BioLegend, or eBioscience. Purified mAb to MHC class II (P7/7) was conjugated to Alexa Fluor 647 using the Alexa Fluor 647 conjugation kit (Molecular Probes). Flow cytometric analyses were performed on a FACSCanto II (BD) using FACSDiva (BD) and FlowJo (Tree Star) software.
In vitro starvation and microscopic analysis.
HEK293T cells were transiently transfected (calcium phosphate method) with pMSCVneo vectors containing the cDNAs encoding for GCL or GCLG120A and seeded onto coverslips immersed in 12-well plates. 48 h after transfection, cells were either transferred to starvation medium (Earles balanced salt solution with 0.5 µM Rapamycin; both from Sigma-Aldrich) or full medium (DMEM with 8% [vol/vol] FCS; Biochrom) and cultured for another 24 h. Subsequently, coverslips were mounted onto glass slides with Prolong Gold antifade reagent containing DAPI (Molecular Probes) and analyzed by fluorescence microscopy.
NFAT-GFP reporter hybridoma cells.
The T1CRP54.4 hybridoma expressing the DEP TCR was infected with the retroviral vector SIN-(NFAT)x-GFP (Hooijberg et al., 2000; provided by H. Spits, Academisch Medisch Centrum, Amsterdam). Subclone 32–14 was used for in vitro stimulation experiments.
Antigen presentation assay.
104 T1CRP-54.4/SIN-(NFAT)x-GFP (clone 32–14) hybridoma cells were co-cultured with 2 × 104 APCs in 150 µl High Glucose Iscove’s modified Dulbecco’s medium (IMDM) w/o Phenol Red (PAA) supplemented with 8% FCS (vol/vol; Biochrom). After 20 h, cells were harvested and GFP expression of hybridoma cells was assessed by flow cytometry.
Preparation of thymic stromal cells.
Thymi were cut into small pieces and digested at 37°C in RPMI (PAA) containing 0.25 mg/ml Collagenase D (Roche), 1 U/ml DispaseI (Roche), 2% FCS (vol/vol), 25 mM Hepes, pH 7.2, and 120 U/ml DNase I (Roche) for 30 min, followed by addition of 5 mM EDTA and incubation for 5 min. Cells were washed and resuspended in Percoll (GE Healthcare; ρ 1.115), followed by a layer of Percoll (ρ 1.050) and PBS as upper phase. Gradients were spun for 30 min at 1,350 g in the cold, and low-density cells were harvested, washed, and stained for FACS analysis or FACS sorting. mTECs were defined as CD45−Ly51−EpCAM+ cells, cTECs as CD45–Ly51+EpCAM+ cells and DCs as CD45+CD11c+ cells. For some experiments, CD45−Ly51–EpCAM+ mTECs were subdivided into mature MHC class IIhiCD80hi cells (mTEChi) and immature MHC class IIloCD80lo cells (mTEClo).
BM chimeras.
BM was depleted of B and T cells using biotinylated mAbs to B220, CD8, and CD4 and streptavidin MACS beads (Miltenyi Biotec). Female recipient mice were irradiated with 2 × 550 rad and reconstituted with 5 × 106 BM cells.
Thymus transplantation.
Embryonic thymi (E14-E16) were transplanted under the kidney capsule of female recipients. Atg5−/− embryos and Atg5+/+ or Atg5+/− controls were obtained from pregnant Atg5+/− females after mating to Atg5+/− males.
Quantitative PCR.
RNA was isolated from thymic stromal cells using the miRNAeasy kit including a DNase digest (QIAGEN) and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories). PCR reactions were performed in duplicates on a CFX96 real-time thermal cycler (Bio-Rad Laboratories) using the Ssofast EvaGreen Supermix (Bio-Rad Laboratories). Primers were as follows: Actb forward, 5′-GCCTTCCTTCTTGGGTAT-3′, and reverse, 5′-GGCATAGAGGTCTTTACGG-3′; GCL transgene forward, 5′-TTCTGGGTAGATGGGAAGCC-3′, and reverse, 5′-CCCTTGTATCGCTCTATAATCACT-3′; mPCC and ePCC transgenes forward, 5′-AGCTGAAGGGTTCTCGTA-3′, and reverse, 5′-CTTAGCGGTGGCTTGTTT-3′. Fluorescence was recorded at the annealing step and expression levels were calculated with the comparative Ct method using Actb for normalization.
Statistical analysis.
Statistical significance was assessed using the unpaired two-tailed Student’s t test.
Acknowledgments
We thank C. Federle, S. Höflinger, and S. Kirsch for expert technical assistance and B. Kyewski (Deutsches Krebsforschungszentrum [DFKZ], Heidelberg, Germany) for comments on the manuscript. We also thank N. Mizushima, T. Yoshimori, S. Hedrick, and R. Schwartz for generously providing reagents and/or mouse strains.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; KL 1228/2-1). C. Wu received a PhD fellowship from the China Scholarship Council.
The authors have no competing financial interests.
Author contributions: All experiments were performed jointly by C. Wu and M. Aichinger. J. Nedjic designed the Aire-GCL model and cloned BAC tg constructs. L. Klein, M. Aichinger, C. Wu, and J. Nedjic designed experimental strategies. L. Klein, M. Aichinger, and C.Wu wrote the manuscript.
Footnotes
Abbreviations used:
- Atg5
- autophagy gene 5
- Ciita
- class II transactivator
- CRP
- C-reactive protein
- cTEC
- cortical TEC
- GCL
- GFP-CRP-LC3 fusion protein
- LC3
- light chain 3
- mTEC
- medullary TEC
- PCC
- pigeon cytochrome c
- PTAs
- peripheral tissue antigen
- tg
- transgenic
- TEC
- thymic epithelial cell
References
- Anderson M.S., Venanzi E.S., Klein L., Chen Z., Berzins S.P., Turley S.J., von Boehmer H., Bronson R., Dierich A., Benoist C., Mathis D. 2002. Projection of an immunological self shadow within the thymus by the aire protein. Science. 298:1395–1401 10.1126/science.1075958 [DOI] [PubMed] [Google Scholar]
- Aschenbrenner K., D’Cruz L.M., Vollmann E.H., Hinterberger M., Emmerich J., Swee L.K., Rolink A., Klein L. 2007. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8:351–358 10.1038/ni1444 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazil M.I., Weiss S., Stockinger B. 1997. Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur. J. Immunol. 27:1506–1514 10.1002/eji.1830270629 [DOI] [PubMed] [Google Scholar]
- Chang C.H., Guerder S., Hong S.C., van Ewijk W., Flavell R.A. 1996. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 4:167–178 10.1016/S1074-7613(00)80681-0 [DOI] [PubMed] [Google Scholar]
- Chicz R.M., Urban R.G., Gorga J.C., Vignali D.A., Lane W.S., Strominger J.L. 1993. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J. Exp. Med. 178:27–47 10.1084/jem.178.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D., Gray D., Dierich A., Kaufman J., Lemeur M., Benoist C., Mathis D. 1991. Mice lacking MHC class II molecules. Cell. 66:1051–1066 10.1016/0092-8674(91)90448-8 [DOI] [PubMed] [Google Scholar]
- Dengjel J., Schoor O., Fischer R., Reich M., Kraus M., Müller M., Kreymborg K., Altenberend F., Brandenburg J., Kalbacher H., et al. 2005. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA. 102:7922–7927 10.1073/pnas.0501190102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbinski J., Schulte A., Kyewski B., Klein L. 2001. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat. Immunol. 2:1032–1039 10.1038/ni723 [DOI] [PubMed] [Google Scholar]
- Derbinski J., Gäbler J., Brors B., Tierling S., Jonnakuty S., Hergenhahn M., Peltonen L., Walter J., Kyewski B. 2005. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 202:33–45 10.1084/jem.20050471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongre A.R., Kovats S., deRoos P., McCormack A.L., Nakagawa T., Paharkova-Vatchkova V., Eng J., Caldwell H., Yates J.R., III, Rudensky A.Y. 2001. In vivo MHC class II presentation of cytosolic proteins revealed by rapid automated tandem mass spectrometry and functional analyses. Eur. J. Immunol. 31:1485–1494 [DOI] [PubMed] [Google Scholar]
- Dörfel D., Appel S., Grünebach F., Weck M.M., Müller M.R., Heine A., Brossart P. 2005. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood. 105:3199–3205 10.1182/blood-2004-09-3556 [DOI] [PubMed] [Google Scholar]
- Gallegos A.M., Bevan M.J. 2004. Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. J. Exp. Med. 200:1039–1049 10.1084/jem.20041457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerder S., Viret C., Luche H., Ardouin L., Malissen B. 2012. Differential processing of self-antigens by subsets of thymic stromal cells. Curr. Opin. Immunol. 24:99–104 10.1016/j.coi.2012.01.008 [DOI] [PubMed] [Google Scholar]
- Hinterberger M., Aichinger M., Prazeres da Costa O., Voehringer D., Hoffmann R., Klein L. 2010. Autonomous role of medullary thymic epithelial cells in central CD4(+) T cell tolerance. Nat. Immunol. 11:512–519 10.1038/ni.1874 [DOI] [PubMed] [Google Scholar]
- Hooijberg E., Bakker A.Q., Ruizendaal J.J., Spits H. 2000. NFAT-controlled expression of GFP permits visualization and isolation of antigen-stimulated primary human T cells. Blood. 96:459–466 [PubMed] [Google Scholar]
- Hubert F.X., Kinkel S.A., Davey G.M., Phipson B., Mueller S.N., Liston A., Proietto A.I., Cannon P.Z., Forehan S., Smyth G.K., et al. 2011. Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood. 118:2462–2472 10.1182/blood-2010-06-286393 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai M., Tanida I., Ueno T., Kominami E., Seki S., Ikeda T., Mizuochi T. 2009. Autophagic compartments gain access to the MHC class II compartments in thymic epithelium. J. Immunol. 183:7278–7285 10.4049/jimmunol.0804087 [DOI] [PubMed] [Google Scholar]
- Kaushik S., Cuervo A.M. 2012. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 22:407–417 10.1016/j.tcb.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J., Hsu M.L., Sauron M.E., Jameson S.C., Gascoigne N.R., Hedrick S.M. 1989. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 341:746–749 10.1038/341746a0 [DOI] [PubMed] [Google Scholar]
- Kim I., Rodriguez-Enriquez S., Lemasters J.J. 2007. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462:245–253 10.1016/j.abb.2007.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L., Klein T., Rüther U., Kyewski B. 1998. CD4 T cell tolerance to human C-reactive protein, an inducible serum protein, is mediated by medullary thymic epithelium. J. Exp. Med. 188:5–16 10.1084/jem.188.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L., Roettinger B., Kyewski B. 2001. Sampling of complementing self-antigen pools by thymic stromal cells maximizes the scope of central T cell tolerance. Eur. J. Immunol. 31:2476–2486 [DOI] [PubMed] [Google Scholar]
- Klein L., Hinterberger M., Wirnsberger G., Kyewski B. 2009. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9:833–844 10.1038/nri2669 [DOI] [PubMed] [Google Scholar]
- Klein L., Hinterberger M., von Rohrscheidt J., Aichinger M. 2011. Autonomous versus dendritic cell-dependent contributions of medullary thymic epithelial cells to central tolerance. Trends Immunol. 32:188–193 10.1016/j.it.2011.03.002 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Ohsumi Y. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:1–32 10.1146/annurev.cellbio.15.1.1 [DOI] [PubMed] [Google Scholar]
- Koble C., Kyewski B. 2009. The thymic medulla: a unique microenvironment for intercellular self-antigen transfer. J. Exp. Med. 206:1505–1513 10.1084/jem.20082449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. 2004. The role of autophagy during the early neonatal starvation period. Nature. 432:1032–1036 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- Kyewski B., Klein L. 2006. A central role for central tolerance. Annu. Rev. Immunol. 24:571–606 10.1146/annurev.immunol.23.021704.115601 [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2011. Autophagy in protein and organelle turnover. Cold Spring Harb. Symp. Quant. Biol. 76:397–402 10.1101/sqb.2011.76.011023 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Klionsky D.J. 2007. Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 27:19–40 10.1146/annurev.nutr.27.061406.093749 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 15:1101–1111 10.1091/mbc.E03-09-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münz C. 2009. Enhancing immunity through autophagy. Annu. Rev. Immunol. 27:423–449 10.1146/annurev.immunol.021908.132537 [DOI] [PubMed] [Google Scholar]
- Nedjic J., Aichinger M., Emmerich J., Mizushima N., Klein L. 2008. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 455:396–400 10.1038/nature07208 [DOI] [PubMed] [Google Scholar]
- Nedjic J., Aichinger M., Mizushima N., Klein L. 2009. Macroautophagy, endogenous MHC II loading and T cell selection: the benefits of breaking the rules. Curr. Opin. Immunol. 21:92–97 10.1016/j.coi.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F., Milosevic S., Behrends U., Jaffee E.M., Pardoll D.M., Bornkamm G.W., Mautner J. 2003. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur. J. Immunol. 33:1250–1259 10.1002/eji.200323730 [DOI] [PubMed] [Google Scholar]
- Oehen S., Feng L., Xia Y., Surh C.D., Hedrick S.M. 1996. Antigen compartmentation and T helper cell tolerance induction. J. Exp. Med. 183:2617–2626 10.1084/jem.183.6.2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C., Pullner A., King S.B., Drexler I., Meier S., Brocker T., Voehringer D. 2009. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J. Exp. Med. 206:549–559 10.1084/jem.20082394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oukka M., Colucci-Guyon E., Tran P.L., Cohen-Tannoudji M., Babinet C., Lotteau V., Kosmatopoulos K. 1996. CD4 T cell tolerance to nuclear proteins induced by medullary thymic epithelium. Immunity. 4:545–553 10.1016/S1074-7613(00)80481-1 [DOI] [PubMed] [Google Scholar]
- Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Münz C. 2005. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 307:593–596 10.1126/science.1104904 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Petrie H.T., Zúñiga-Pflücker J.C. 2007. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu. Rev. Immunol. 25:649–679 10.1146/annurev.immunol.23.021704.115715 [DOI] [PubMed] [Google Scholar]
- Riedel A., Nimmerjahn F., Burdach S., Behrends U., Bornkamm G.W., Mautner J. 2008. Endogenous presentation of a nuclear antigen on MHC class II by autophagy in the absence of CRM1-mediated nuclear export. Eur. J. Immunol. 38:2090–2095 10.1002/eji.200737900 [DOI] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., Münz C. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 26:79–92 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Birkeland H.C., Gillooly D.J., Mizushima N., Kuma A., Yoshimori T., Slagsvold T., Brech A., Stenmark H. 2004. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 117:4239–4251 10.1242/jcs.01287 [DOI] [PubMed] [Google Scholar]
- Smith K.M., Olson D.C., Hirose R., Hanahan D. 1997. Pancreatic gene expression in rare cells of thymic medulla: evidence for functional contribution to T cell tolerance. Int. Immunol. 9:1355–1365 10.1093/intimm/9.9.1355 [DOI] [PubMed] [Google Scholar]
- Starr T.K., Jameson S.C., Hogquist K.A. 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21:139–176 10.1146/annurev.immunol.21.120601.141107 [DOI] [PubMed] [Google Scholar]
- Strawbridge A.B., Blum J.S. 2007. Autophagy in MHC class II antigen processing. Curr. Opin. Immunol. 19:87–92 10.1016/j.coi.2006.11.009 [DOI] [PubMed] [Google Scholar]
- Wirnsberger G., Mair F., Klein L. 2009. Regulatory T cell differentiation of thymocytes does not require a dedicated antigen-presenting cell but is under T cell-intrinsic developmental control. Proc. Natl. Acad. Sci. USA. 106:10278–10283 10.1073/pnas.0901877106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Li P., Lin Y., Lott J.M., Hislop A.D., Canaday D.H., Brutkiewicz R.R., Blum J.S. 2005. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 22:571–581 10.1016/j.immuni.2005.03.009 [DOI] [PubMed] [Google Scholar]