Expression of the transcription factor Helios identifies thymocyte divergence during positive and negative selection.
Abstract
Acquisition of self-tolerance in the thymus requires T cells to discriminate strong versus weak T cell receptor binding by self-peptide–MHC complexes. We find this discrimination is reported by expression of the transcription factor Helios, which is induced during negative selection but decreases during positive selection. Helios and the proapoptotic protein Bim were coinduced in 55% of nascent CCR7− CD4+ CD69+ thymocytes. These were short-lived cells that up-regulated PD-1 and down-regulated CD4 and CD8 during Bim-dependent apoptosis. Helios and Bim were also coinduced at the subsequent CCR7+ CD4+ CD69+ CD8− stage, and this second wave of Bim-dependent negative selection involved 20% of nascent cells. Unlike CCR7− counterparts, Helios+ CCR7+ CD4+ cells mount a concurrent Card11- and c-Rel–dependent activation response that opposes Bim-mediated apoptosis. This “hollow” activation response consists of many NF-κB target genes but lacks key growth mediators like IL-2 and Myc, and the thymocytes were not induced to proliferate. These findings identify Helios as the first marker known to diverge during positive and negative selection of thymocytes and reveal the extent, stage, and molecular nature of two distinct waves of clonal deletion in the normal thymus.
To understand how self-tolerance is acquired in T cells and how it breaks down as a result of inherited predisposition to autoimmune disease, it is necessary to resolve how thymocytes discriminate weak versus strong TCR engagement by self-peptide–MHC (self-pMHC) antigens. This discrimination is central to determining cell fate, as binding to self-pMHC induces positive selection of thymocytes if it is relatively weak but negative selection if it is stronger (von Boehmer, 1990; Gallegos and Bevan, 2006; Gascoigne and Palmer, 2011; Stritesky et al., 2012). Elucidating this selection process requires identification of molecular markers that qualitatively differentiate the thymocyte response to weak versus strong self-pMHC stimulation.
Qualitative differences in the intracellular location of phosphorylated ERK in response to positive versus negatively selecting pMHC have been revealed in vitro (Daniels et al., 2006), but it is difficult to extend this marker to analyses of the natural thymocyte repertoire in vivo. Differences in the magnitude of induction of specific genes and corresponding proteins, notably Bcl2l11 (Bim), Nr4a1 (Nur77), CD69, and PD-1, have been revealed in thymocytes undergoing positive versus negative selection in vivo (Liston et al., 2004b; Baldwin and Hogquist, 2007; Moran et al., 2011). However, because the magnitude of induction during positive selection grades continuously into negative selection, it is difficult to identify a threshold level of these markers that objectively resolves thymocytes making a positive versus negative selection response.
Understanding the molecular differentiation of positive versus negative selection responses also depends on determining whether these responses occur at the same stage of thymocyte maturation in TCR+ CCR7− cortical thymocytes (CCR7 is a chemokine receptor required for thymocyte migration into the medulla [Ueno et al., 2004; Nitta et al., 2009]) or whether negative selection occurs after positive selection in TCR+ CCR7+ medullary thymocytes. Several in vitro (Swat et al., 1991; Kishimoto and Sprent, 1997) and in vivo (Kappler et al., 1987; Pircher et al., 1989; Surh and Sprent, 1994; Pobezinsky et al., 2012) studies support both possibilities, although the need to perform experiments in tissue culture, in TCR transgenic (Tg) mice, or by tracking T cells reacting to superantigens means that the extent of negative selection in CCR7− or CCR7+ thymocytes in the normal repertoire has yet to be directly measured.
Here we show that the transcription factor Helios qualitatively differentiates CD4+ thymocytes making a positive versus negative selection response in vivo. Helios expression increases during negative selection but decreases during positive selection. Using this marker, we enumerate the fraction of thymocytes undergoing negative selection at different stages of helper T cell development, revealing two main waves of negative selection: one in CCR7− CD24+ CD4+/lo CD8+/lo thymocytes and one in CCR7+ CD4+ CD8− thymocytes. Although Bim is strongly induced and required for negative selection at both stages, it is accompanied by inhibitory versus stimulatory programs at the two stages. Card11 signaling concurrently induces many NF-κB–dependent T cell survival and growth genes during negative selection selectively at the CCR7+ stage, representing a “hollow” activation response comprising many but not all genes known to be induced by strong TCR agonists in mature T cells. This Bim-opposing response does not occur during negative selection at the CCR7− stage, and instead the inhibitory receptor PD-1 is strongly coinduced with Bim.
RESULTS
Helios expression differentiates thymocytes undergoing positive and negative selection
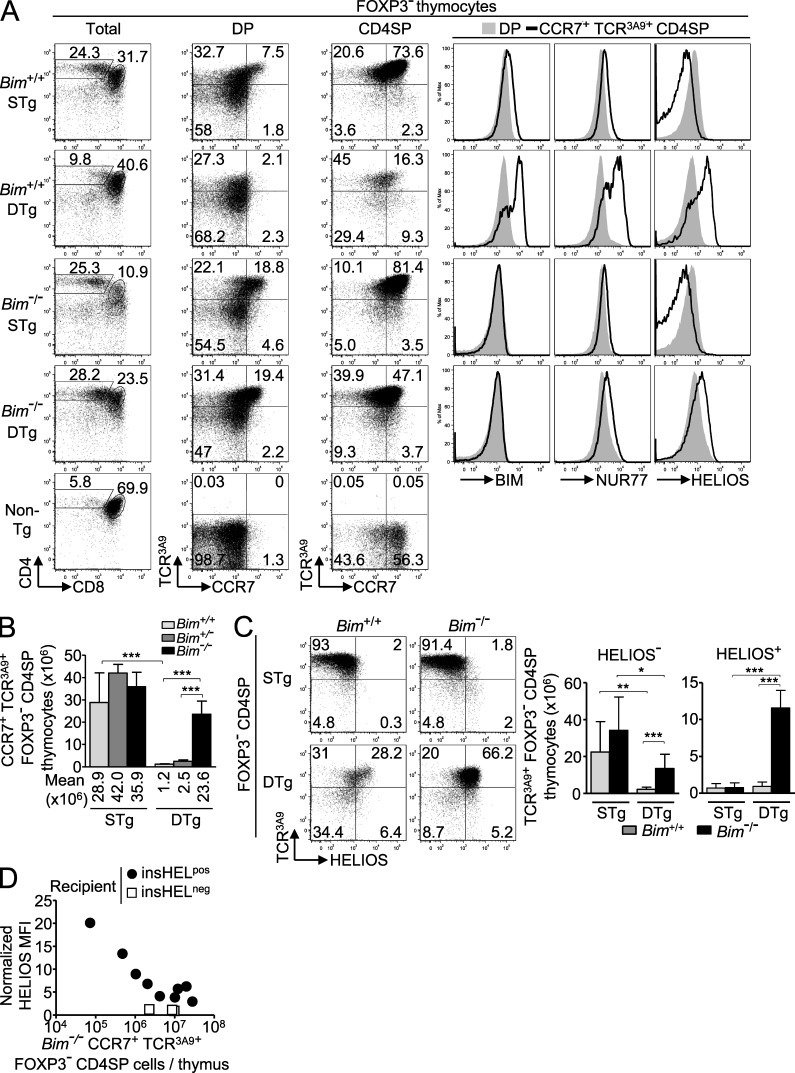
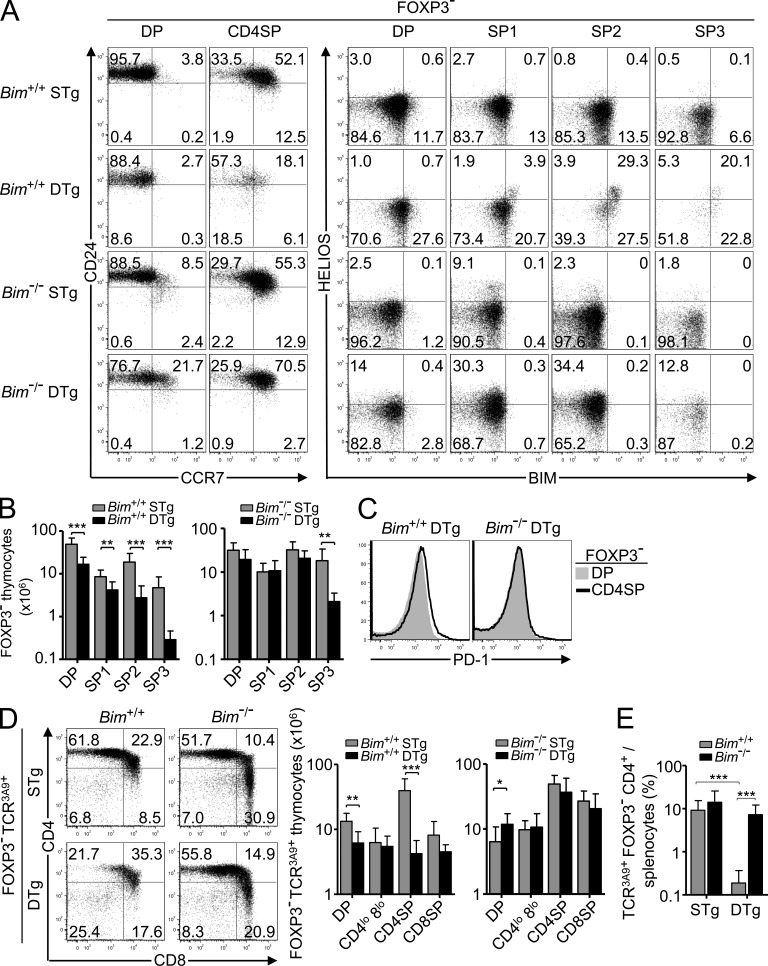
To distinguish CD4+ CD8− (single positive [SP]; CD4SP) thymocytes undergoing negative versus positive selection, we first used flow cytometry to compare protein expression in 3A9 TCR Tg thymocytes. In TCR3A9 single-Tg (STg) mice, weak binding of this TCR to self-peptide–I-Ak complexes promoted positive selection of TCRhi CCR7− CD4+ 8+ (double positive [DP]) cortical thymocytes into CCR7+ CD4SP medullary thymocytes (Fig. 1 A). In contrast, in TCR3A9 double-Tg (DTg) mice that also carry an insulin promoter–driven hen egg lysozyme (HEL [insHEL]) transgene, Aire-dependent expression in medullary thymic epithelial cells generated HEL peptide–I-Ak complexes that were bound strongly by TCR3A9 and triggered negative selection of the CCR7+ subset of SP and DP thymocytes that homed to the medulla or corticomedullary junction, a process of deletion previously shown to require CCR7 ligands (Fig. 1, A and B; Lesage et al., 2002; Liston et al., 2003; Nitta et al., 2009). Elimination of CD4SP thymocytes was severely blunted in Bim−/− DTg mice lacking the proapoptotic protein Bim (Fig. 1, A and B), consistent with previous evidence that Bim is essential for thymocyte apoptosis (Bouillet et al., 2002) and that Bim protein and mRNA are induced to higher levels during negative selection (Liston et al., 2004a, 2007; Baldwin and Hogquist, 2007). Bim protein also increased, albeit to a lesser extent, in CD4SP thymocytes undergoing positive selection in STg mice (Fig. 1 A). A similar graded induction of Nur77 (Fig. 1 A), GITR, CD69, and other proteins was observed (not depicted). However, the magnitudes of these differences were too small and the overlap in the distributions too great to resolve thymocytes undergoing positive and negative selection.
Figure 1.
Helios expression differentiates thymocytes undergoing positive and negative selection. (A) Phenotype of Foxp3− thymocytes from Bim-sufficient (Bim+/+) or Bim-deficient (Bim−/−) 3A9 TCR STg or 3A9 TCR × insHEL DTg mice (denoted far left) with gates defining the CD4+ 8+ (DP) and CD4+ 8− (CD4SP) populations (first column), which were analyzed for expression of TCR3A9 versus CCR7 (second and third columns). Histograms (right) show labeling for Bim, Nur77, or Helios protein on Foxp3− DP or CCR7+ TCR3A9+ CD4SP thymocytes. Data are representative of three experiments. (B) Number of CCR7+ TCR3A9+ Foxp3− CD4SP thymocytes with at least seven mice per group compiled from four experiments. Error bars show SD. (C) Representative plots (left) show gating of Helios+/− subsets of TCR3A9 Foxp3− CD4SP thymocytes, which are enumerated in the graphs (right) with at least six Bim+/+ or Bim−/− STg or DTg mice per group compiled from four experiments. Numbers in plots indicate percentage of cells in gates shown, and columns in graphs show mean and SD. (D) Helios induction in mixed bone marrow chimeras with lower frequencies of TCR3A9+ cells. Irradiated CD45.1+ insHELneg or insHELpos recipient mice were reconstituted with CD45.2+ Bim−/− STg bone marrow cells alone or mixed at a ratio of 1:19 with CD45.1+ non-Tg marrow. Helios mean fluorescence intensity (MFI) on the Bim−/− CD45.2+ CCR7+ TCR3A9+ Foxp3− CD4SP thymocytes is plotted against their absolute number per thymus 4–5 wk after reconstitution. Helios MFIs were normalized to Helios MFI of non-Tg DP thymocytes analyzed in parallel. Data in D were compiled from two separate experiments; each symbol represents an individual chimera. Unpaired Student’s t test p-values: ***, P < 0.001; **, P = 0.001–0.01; *, P = 0.01–0.05.
A survey of a large number of proteins revealed only one, the Ikaros family transcription factor Helios (Ikzf2), that qualitatively differentiated positive and negative selection responses. Helios was highly expressed in nearly all Foxp3+ T regulatory cells (T reg cells) in the thymus, consistent with published results (Thornton et al., 2010), but when Foxp3+ cells were excluded from the flow cytometric analysis, Helios was also selectively induced in Foxp3− TCR3A9+ CD4SP cells undergoing negative selection (Fig. 1 A). In contrast, Helios was expressed at low levels in DP cells and was down-regulated further in positively selected CD4SP cells from STg mice (Fig. 1 A). Helios was induced in TCR3A9+ CD4SP cells from Bim−/− DTg animals, although this induction was slightly blunted when compared with Bim+/+ DTg mice (Fig. 1, A and C). Helios induction increased markedly when Bim−/− HEL-specific thymocytes were present at low frequencies in mixed bone marrow chimeras (Fig. 1 D), suggesting that the slight blunting in Bim−/− DTg mice was caused by increased intraclonal competition for a limited number of HEL-presenting APCs. In Bim-deficient STg mice, positive selection into TCR3A9+ CD4SP cells occurred normally and was still accompanied by Helios down-regulation (Fig. 1 C). Thus, strongly self-reactive Helios+ Foxp3− TCR3A9+ CD4SP thymocytes increased in number 10-fold in Bim−/− compared with Bim+/+ DTg mice, whereas the number of weakly self-reactive Helios− Foxp3− TCR3A9+ CD4SP cells that were positively selected in STg mice was not altered by Bim deficiency (Fig. 1 C).
Helios induction occurs in a large fraction of CCR7− and CCR7+ CD4SP thymocytes
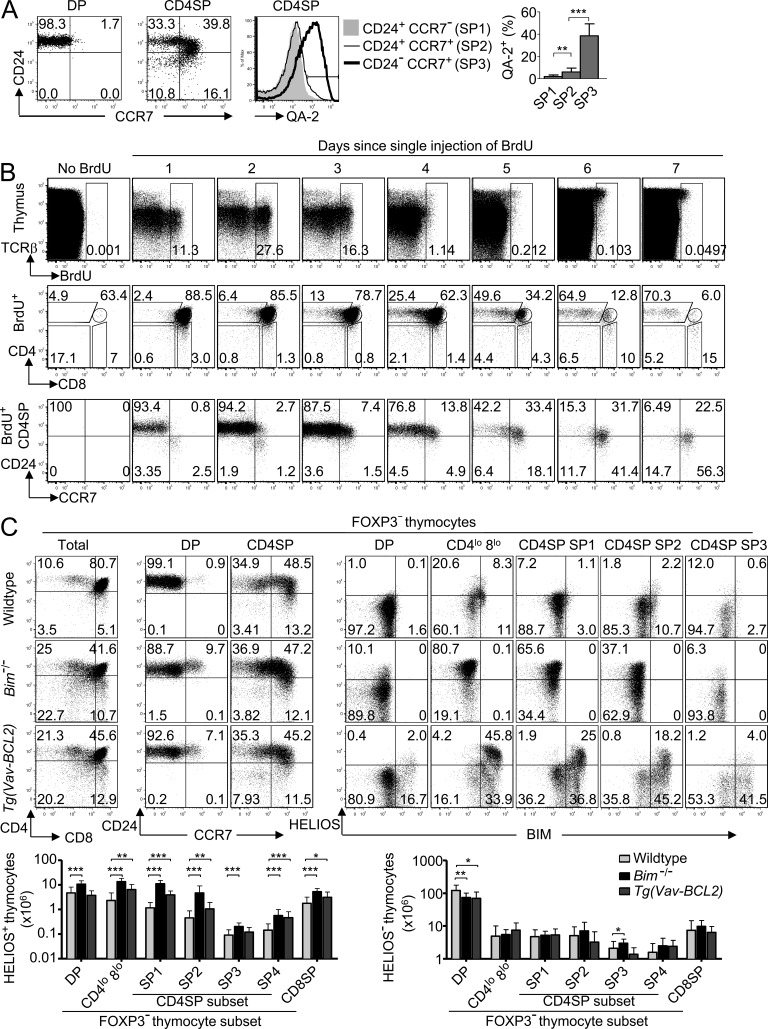
To track Helios expression in thymocytes from mice with natural TCR repertoires, we subdivided CD4SP thymocytes into maturation stages. DP cells have a CD24+ CCR7− phenotype, and these markers divide CD4SP cells into subsets that suggest CCR7 is induced and then CD24 is down-regulated (Fig. 2 A; Davalos-Misslitz et al., 2007). This proposed developmental sequence was corroborated by progressive increase in expression of the thymocyte maturation marker Qa-2 (McCaughtry et al., 2007), allowing resolution of three CD4SP subsets referred to here as SP1–SP3 (Fig. 2 A): CD24+ CCR7− (SP1), CD24+ CCR7+ (SP2), and CD24− CCR7+ (SP3).
Figure 2.
Expanded Helios+ thymocyte populations in apoptosis-deficient mice. (A) CD24/CCR7 phenotype of non-Tg DP and CD4SP thymocytes showing the gates used to define the CD4SP subsets SP1 (top left), SP2 (top right), and SP3 (bottom right), which were examined for Qa-2 expression (middle); summary (right) shows the percentage of Qa-2+ cells within each CD4SP subset compiled from two experiments. Error bars show SD. (B) Phenotypic progression within the CD4SP stage. Non-Tg B6 mice were either left uninjected or injected i.p. with 1 mg BrdU either 1, 2, 3, 4, 5, 6, or 7 d before flow cytometry on thymocytes. Plots show labeling for TCRβ versus BrdU on all thymocytes (top row). BrdU+ thymocytes were analyzed for expression of CD4 versus CD8 (middle row), and BrdU+ CD4SP cells were analyzed for CD24 versus CCR7 expression (bottom row). Data are representative of three experiments. (C) Helios+ thymocyte populations expand in apoptosis-deficient mice with natural TCR repertoires. Representative plots show phenotype of Foxp3− thymocyte subsets in wild-type, Bim−/−, and Vav-BCL2 Tg mice (denoted far left). The first column shows quadrant gates defining four populations on the basis of CD4 and CD8; in the third column, the CD4SP population is further divided into the subsets SP1–SP4. The fourth through eighth columns display the Helios/Bim phenotype of populations denoted above the plots. Graphs (bottom) show number of Helios+ (left) or Helios− (right) Foxp3− thymocytes within each subpopulation as defined using gates represented in the plots above, compiled from eight separate experiments that used a total of 13 Bim−/− and 8 B6.Vav-BCL2 Tg mice. Numbers in plots indicate percentage of cells in gates shown, and columns in graphs show mean and SD. The wild-type group comprises pooled data from 16 B10.BR mice and 10 B6 mice. No statistically significant differences existed between wild-type B10.BR and B6 mice. Paired (in A) or unpaired (in C) Student’s t test p-values: ***, P < 0.001; **, P = 0.001–0.01; *, P = 0.01–0.05.
To confirm the proposed developmental sequence above, we used these markers to extend published experiments (Lucas et al., 1993) tracing thymocyte maturation after in vivo pulse labeling with BrdU, which becomes incorporated into the DNA of proliferating DP thymocytes (Fig. 2 B). Each mouse received a single BrdU injection 1–7 d before sacrifice, so that all thymocyte samples were analyzed on the same day. As expected (Lucas et al., 1993), the BrdU+ thymocytes were predominantly at the DP stage 1–4 d after BrdU injection, whereas by days 6–7, most had a CD4SP phenotype. Subsetting the BrdU+ CD4SP cells showed that most were at the SP1 stage 1–2 d after BrdU injection, and there was a gradual transition to SP2 and SP3 from 3 to 7 d, so that by day 7 most of the BrdU+ CD4SP cells were at the SP3 stage (Fig. 2 B). These results are consistent with a DP–SP1–SP2–SP3 developmental progression during CD4SP maturation.
To extend the evidence above that Helios marks strongly self-reactive thymocytes undergoing negative selection, we analyzed Helios expression in thymocytes in the natural TCR repertoire after gating out Foxp3+ cells. Helios+ Foxp3− thymocytes were detectable in wild-type mice, and their number was dramatically increased when elimination of strongly self-reactive thymocytes was inhibited either in Bim−/− mice or in mice expressing human BCL-2 driven by the mouse Vav promoter (Tg(Vav-BCL2); Fig. 2 C; Bouillet et al., 2002). In contrast, there was little effect on the number of Helios− Foxp3− thymocytes (Fig. 2 C), consistent with the evidence above that Bim deficiency does not alter positive selection. In both Bim−/− and Vav-BCL2 Tg mice, Helios+ cells were strikingly increased among the DP, CD4lo CD8lo, and CD4SP populations, accounting for up to 10% of DP cells, 80% of CD4lo CD8lo cells, 65% of SP1 cells, and 37% of SP2 cells in Bim−/− mice, reflecting 10-fold increases in Helios+ SP1 and SP2 cell numbers (Fig. 2 C). High Helios expression was correlated with high Bim expression at the single cell level, and this was accentuated in thymocytes expressing Tg BCL-2 (Fig. 2 C), which enables Bim to accumulate to high levels in T cells (Jorgensen et al., 2007). Thus, Helios marks thymocytes that have been rescued from Bim-mediated negative selection.
Helios, PD-1, and coreceptor dulling mark T cells that bind strongly to self-antigens during the CCR7-negative stage
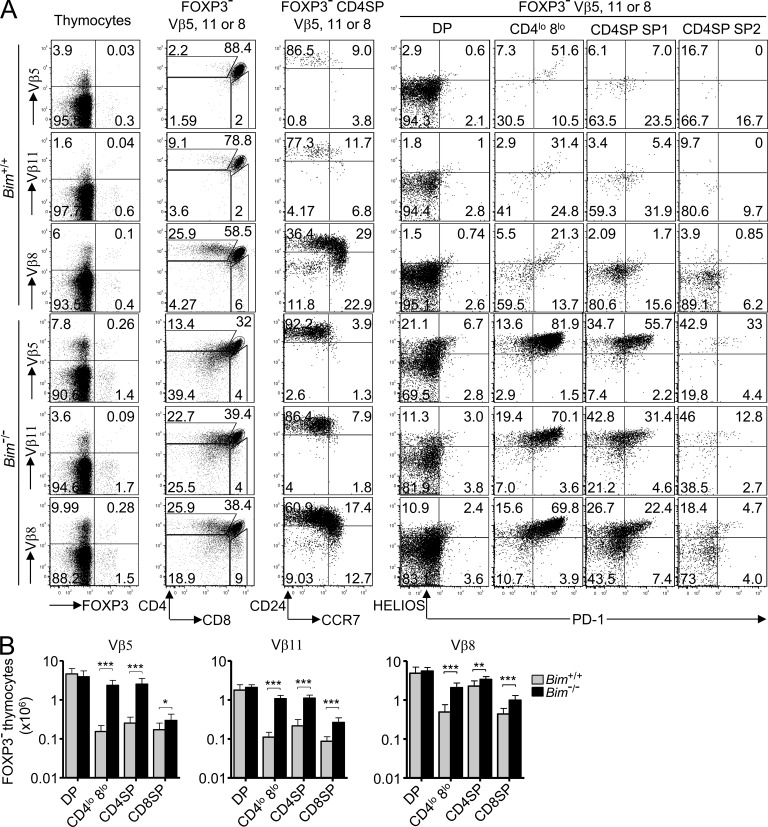
To study cells known to be strongly self-reactive in B10.BR mice, we focused on thymocytes using the Vβ5 or Vβ11 TCR V-regions that bind self-superantigens and I-Ek (Dyson et al., 1991; Woodland et al., 1991). In Bim+/+ thymi, there was a 10–20-fold decrease in the number of CD4SP cells expressing Vβ5 or Vβ11 relative to the corresponding numbers at the DP stage, so that only 2% of Vβ5+ and 9% of Vβ11+ cells were CD4SP and these were mostly immature CCR7− SP1 cells (Fig. 3, A [second and third columns] and B). In contrast, in both the TCRβ+ CD4SP population as a whole (equivalent to Fig. 2 C and not depicted) and in the subset expressing Vβ8 that does not bind self-superantigen, CD4SP thymocytes were distributed normally across the SP1–SP3 stages.
Figure 3.
Superantigen recognition by CCR7− thymocytes induces Helios, PD-1, and acquisition of a CD4lo 8lo phenotype. (A) Each row displays thymocytes expressing TCR Vβ5, Vβ11, or Vβ8 from Bim+/+ or Bim−/− B10.BR mice (denoted far left). The gated subpopulations are denoted above the plots, and the markers displayed are below. Numbers in plots indicate percentage of cells in gates shown. Data are representative of five experiments. (B) Enumeration of Foxp3− thymocytes expressing TCR Vβ5, Vβ11, or Vβ8 with a DP, CD4lo 8lo, CD4SP, or CD8SP phenotype as gated in A compiled from five experiments with a total of 9 Bim−/− and 11 Bim+/+ mice. Columns in graphs show mean and SD. Unpaired Student’s t test p-values: ***, P < 0.001; **, P = 0.001–0.01; *, P = 0.01–0.05.
The dramatic drop in Vβ5+ and Vβ11+ cell numbers at the CD4SP stage was almost completely abolished in Bim−/− mice, whereas there was no effect of Bim deficiency on numbers at the DP stage (Fig. 3 B). The rescued Vβ5+ and Vβ11+ CD4SP cells nevertheless showed little evidence of maturing past the CCR7− SP1 stage (Fig. 3 A, third column), and many fell outside the CD4SP or DP gates exhibiting a CD4lo CD8lo “dulled” phenotype (Fig. 3 A, second column). This dulled phenotype matches that of DP thymocytes undergoing negative selection in vitro (Swat et al., 1991; Vasquez et al., 1992; Page et al., 1993) or in vivo (McCaughtry et al., 2008; Pobezinsky et al., 2012). 75% or more of the Vβ5+ or Vβ11+ CD4lo CD8lo and SP1 thymocytes in Bim−/− mice expressed high levels of Helios, and many of these also expressed the inhibitory receptor PD-1 (Fig. 3 A, fifth and sixth columns), which is induced during negative selection of MHC I–restricted DP thymocytes (Baldwin and Hogquist, 2007; McCaughtry et al., 2008). Many non-superantigen–reactive Vβ8+ thymocytes in Bim−/− mice were also Helios+, accounting for 13% of DP cells, 85% of CD4lo CD8lo, and 50% of SP1 thymocytes (Fig. 3 A, bottom row). These frequencies and the fraction of CD4lo CD8lo (DP dull) cells among Vβ8+ Bim−/− thymocytes were equivalent to those in polyclonal TCR-expressing cells shown in Fig. 2 C, which was similar to when all TCRβ+ thymocytes were examined (not depicted). Despite the prominent increase in superantigen-reactive thymocytes (Fig. 3 B), Bim deficiency resulted in only a fourfold increase in Vβ11+ Foxp3− CD4+ splenocytes and no significant increase in Vβ5+ Foxp3− CD4+ splenocytes (not depicted). These findings demonstrate that, as well as Bim-mediated apoptosis, Helios and PD-1 induction and coreceptor dulling characterize the response of strongly self-reactive CCR7− thymocytes to self-antigen.
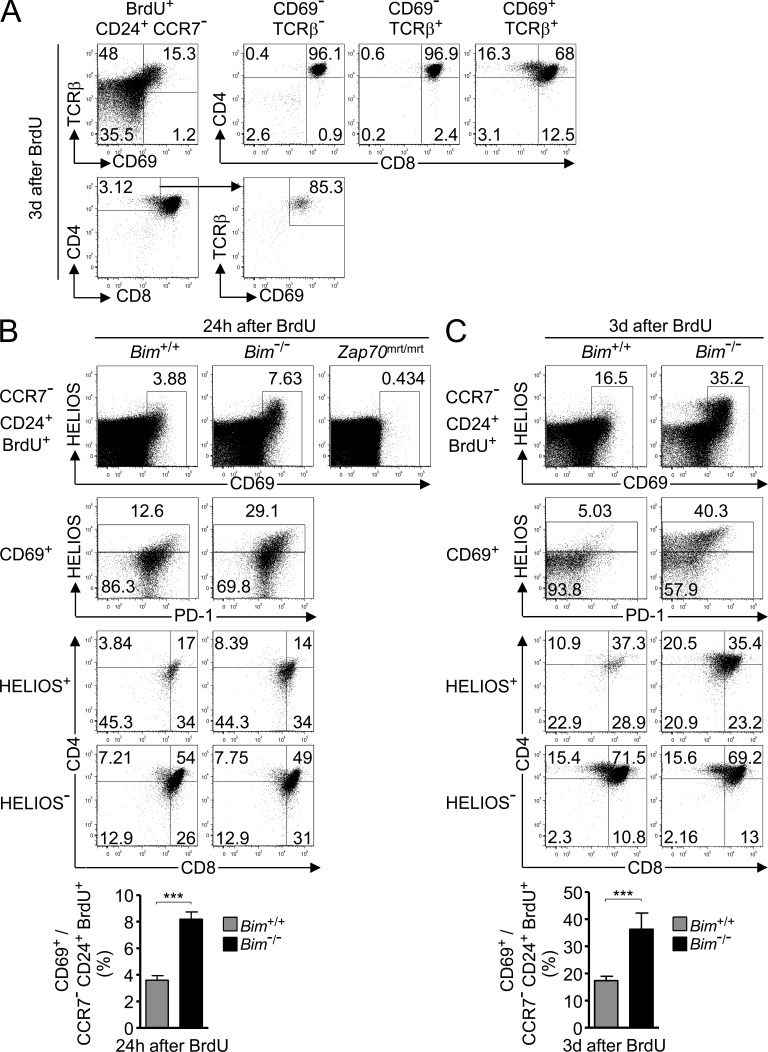
The results above implied that more than half of the nascent T cells in the natural repertoire are strongly self-reactive and normally deleted by Bim, but steady-state analysis alone may not allow for the most accurate estimate if strongly and weakly self-reactive cells dwell for different periods of time in the thymus of Bim−/− mice. To address this, we first used in vivo BrdU labeling and flow cytometry to identify TCR-signaled cells within the pool of recently proliferating (BrdU+) nascent (CD24+ CCR7−) thymocytes. 3 d after BrdU injection, 96–97% of CD69− TCRβ− and CD69− TCRβ+ cells were DP cells (Fig. 4 A), whereas 32% of the CD69+ TCRβ+ population fell outside the DP gate, including 16% in the CD4SP gate (Fig. 4 A, top). By comparison, CD69 was not induced in BrdU+ thymocytes from Zap70mrt/mrt mice, which express a catalytically mutant form of Zap-70 at 25% of the wild-type quantity and fail to be positively selected past the DP stage (Fig. 4 B, top row; Siggs et al., 2007), confirming that CD69 is a specific marker of thymocytes that have received a pMHC-TCR signal (Hare et al., 1999). Of the newly formed CD4SP thymocytes, 85% were CD69+ TCRβ+ (Fig. 4 A, bottom), indicating that CD69 up-regulation is also a sensitive marker of thymocytes that had progressed to the CD4SP stage. Based on these data, we used CD69 up-regulation as a marker of TCR-signaled thymocytes.
Figure 4.
Approximately 55% of TCR-signaled thymocytes die by Bim-dependent negative selection at the CCR7− stage. (A) Analysis of nascent BrdU+ CCR7− CD24+ thymocytes from wild-type mice 3 d after BrdU injection. Quadrants in top left panel define three major CD69 versus TCRβ subsets that are each analyzed for CD4 and CD8 expression on the right. Bottom panels reverse the gating order to show percentage of CD4SP in the nascent pool and their expression of CD69 and TCRβ. Plots were concatenated from four samples, representative of two experiments each analyzing at least four mice individually. (B and C) Analysis of nascent BrdU+ CCR7− CD24+ thymocytes concatenated from four Bim+/+ or four Bim−/− mice on B10.BR background 24 h (B) or 3 d (C) after BrdU treatment. Two Zap70mrt/mrt mice were also analyzed to test TCR signaling. Top row: percentage of CD69+ cells. Second row: Helios and PD-1 expression on CD69+ subset. Bottom rows: CD4 and CD8 expression on Helios+ and Helios− subsets of CD69+ cells. Percentage of gated cells is shown in the plots. Graphs show mean and SD percentage of CD69+ cells among BrdU+ CD24+ CCR7− thymocytes from at least five mice per group compiled from two experiments in B and at least four mice per group from two experiments in C. Unpaired Student’s t test p-value: ***, P < 0.001.
In Bim−/− mice, CD69+ cells were more than twice as frequent among nascent BrdU+ cells 24 h after BrdU injection (mean ± SEM: 8.2 ± 0.3%, n = 4 Bim−/− mice; 3.6 ± 0.2%, n = 5 Bim+/+; Fig. 4 B), suggesting that in wild-type animals, 56% of the TCR-signaled nascent thymocytes had been deleted. The newly selected CD69+ population included a Helios+ subset that was markedly expanded in Bim−/− mice (Fig. 4, B and C), indicating many of the cells rescued from death by Bim deficiency were Helios+. Nearly all of these Helios+ cells fell within the CD69+ gate 24 h after BrdU, consistent with evidence that CD69 is rapidly induced during negative selection as well as during positive selection (McCaughtry et al., 2008). When the analysis was performed 3 d after BrdU injection, the difference in nascent CD69+ cell frequencies between Bim−/− mice (36.3 ± 2.7%, n = 5) and Bim+/+ mice (17.3 ± 0.6%, n = 7; Fig. 4 C) suggested that 52% of CD69+ TCR-signaled thymocytes had been deleted in the Bim+/+ animals. Many, though not all, of the Helios+ CD69+ cells were also PD-1+, and they expressed less CD4 and CD8 than their Helios− CD69+ counterparts (Fig. 4, B and C). In contrast with this response, ∼15% of Helios− CD69+ cells had differentiated into CD4SP cells by 3 d after BrdU injection, and this was unaffected by Bim deficiency (Fig. 4 C). These findings extend the evidence above that Helios discriminates thymocytes destined for negative versus positive selection, including in the polyclonal TCR repertoire, and suggest that the fraction of TCR-signaled thymocytes that die by Bim-dependent negative selection at the CCR7− stage is ∼55%. This is close to the 65.6% Helios+ cells observed among steady-state SP1 Foxp3− CD4SP cells in Bim−/− mice (Fig. 2 C).
A distinct hollow activation response accompanies Helios and Bim induction in strongly self-reactive thymocytes at the CCR7+ stage
As the results above showed that CCR7− thymocytes that were induced to express Helios also induced PD-1 and failed to progress to the CCR7+ stage, we asked whether a comparable response occurred within the Helios+ CCR7+ SP2 thymocytes that bound strongly to self-pMHC in the thymus medulla. We first analyzed SP1 and SP2 subsets in TCR3A9 × insHEL DTg mice, where most CD4SP cells bind strongly to self-pMHC produced selectively in Aire+ medullary thymic epithelial cells (Liston et al., 2003, 2004a). 10-fold fewer SP2 cells were present in Bim+/+ DTg mice compared with STg mice (Fig. 5 B), and those present appeared transitional between the SP1 and SP2 stages as they had yet to fully up-regulate CCR7 (Fig. 5, A and B). Induction of Helios and Bim occurred predominantly in these SP2 cells in Bim+/+ DTg mice (Fig. 5 A). In Bim−/− DTg mice, CCR7 up-regulation was restored (Fig. 5 A) so that Bim−/− STg and Bim−/− DTg mice had a similar number of SP2 cells, ∼10-fold greater than Bim+/+ DTg mice (Fig. 5 B). Unlike the response of CCR7− SP1 thymocytes, the CCR7+ self-reactive cells did not express PD-1 (Fig. 5 C) and were not developmentally arrested as CD4lo 8lo cells (Fig. 5 D) when rescued from deletion in Bim−/− DTg mice. Instead, many reached the periphery, resulting in a 38-fold increase in TCR3A9+ Foxp3− CD4+ splenocytes in Bim−/− DTg mice compared with Bim+/+ DTg mice (Fig. 5 E).
Figure 5.
Organ-specific antigen in thymic medulla induces Helios and Bim in CCR7+ thymocytes, and Bim is required to prevent organ-specific CD4+ T cell peripheralization. (A) The first and second columns show the CD24/CCR7 phenotype of Foxp3− DP and CD4SP thymocytes in Bim+/+ or Bim−/− STg or DTg mice (denoted far left); the third through sixth columns show the Helios/Bim phenotype on the Foxp3− thymocyte subsets DP–SP3. Plots are representative of three separate experiments. (B) Number of Foxp3− thymocytes at each maturation stage from DP to SP3. (C) PD-1 is not induced in DTg thymocytes. Histograms show PD-1 expression on Foxp3− DP and Foxp3− CD4SP thymocytes from Bim+/+ or Bim−/− DTg mice, representative of three experiments. (D) CD4lo 8lo thymocyte number is not increased in Bim−/− DTg mice. CD4/CD8 phenotype (left) and number (right) of Foxp3− TCR3A9+ thymocyte subsets in Bim+/+ or Bim−/− STg or DTg mice are shown. (E) Frequency of TCR3A9+ Foxp3− CD4+ cells among splenic lymphocytes in Bim+/+ or Bim−/− STg or DTg mice. All column graphs show mean and SD for at least six STg mice and nine DTg mice per Bim genotype compiled from at least three experiments. Unpaired Student’s t test p-values: ***, P < 0.001; **, P = 0.001–0.01; *, P = 0.01–0.05.
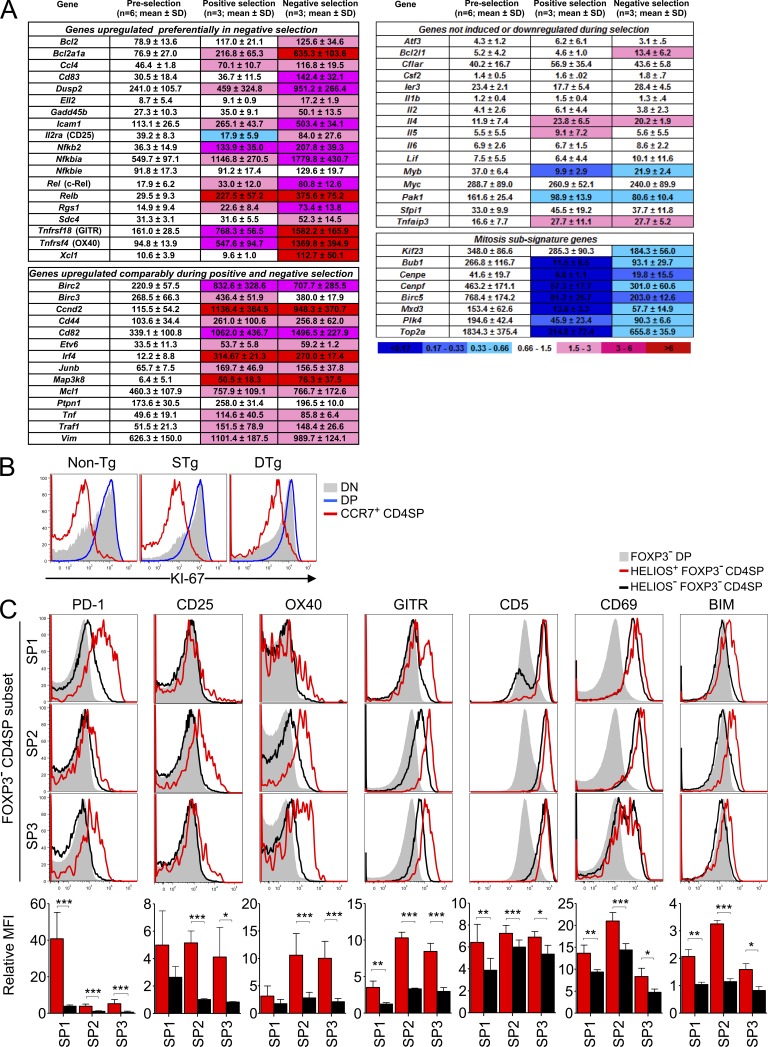
Although the inhibitory receptor PD-1 was not induced in strongly self-reactive CD4SP thymocytes at the CCR7+ SP2 stage of maturation, many other mature T cell activation genes were strongly induced. We analyzed DTg versus STg thymocyte microarray data from sorted CD69+ CD4SP cells representing a mixture of SP1 and SP2 cells (Liston et al., 2007) for the expression of 49 mRNAs known to be induced in primary mature T cells activated for 3 h (Fig. 6 A; Shaffer et al., 2001). Of these 49 mRNAs, 19 were preferentially induced in CD4+ CD8lo CD69+ TCR3A9+ thymocytes from DTg mice compared with their counterparts that bound self-pMHC weakly in STg mice. These included the mRNAs encoding CD25, OX40, GITR, and c-Rel but also other mediators of the mature T cell growth-activating response: the integrin ligand ICAM-1, the antiapoptosis proteins Bcl-2 and A1/Bfl1 (Bcl2a1a), the transcription factor partners NF-κB2 and Rel-B, the chemokines MIP-1β (Ccl4) and lymphotactin (Xcl1), and the signaling molecule Gadd45β. Another 14 mature T cell activation genes were induced to similar levels in CD4+ CD8lo CD69+ TCR3A9+ thymocytes that were strongly or weakly self-reactive from DTg and STg mice, respectively. Only 16 of the 49 mature T cell activation genes examined were not induced in CD4+ CD8lo CD69+ TCR3A9+ thymocytes from DTg mice, although it was striking that those missing included two critical factors for T cell proliferation, Il2 and Myc (Fig. 6 A). The absence of Il2 and Myc (Trumpp et al., 2001; Dose et al., 2009) implied that the T cell activation response induced by strong binding to self-pMHC in CCR7+ CD4SP thymocytes was hollow in that it lacked the capacity for autocrine cell growth and proliferation. Consistent with this, the proliferation-associated protein Ki-67 was highly expressed in double-negative and DP cells but down-regulated in CCR7+ CD4SP cells in DTg mice (Fig. 6 B), and the mRNA levels of eight genes comprising a mitosis subsignature (Shaffer et al., 2001) were significantly down-regulated relative to preselection thymocytes (Fig. 6 A).
Figure 6.
A distinct hollow activation response accompanies Helios and Bim induction in strongly self-reactive thymocytes at the CCR7+ stage. (A) A microarray dataset (Liston et al., 2007) was mined for 53 mRNAs previously shown to be induced in primary human T cells activated for 3 h and 12 mRNAs associated with mitosis (Shaffer et al., 2001). 47 and 9 of these mRNAs, respectively, were represented on the array. For comparison, mRNAs encoding OX40 and GITR were added to this set. Sorted thymocytes analyzed by microarray were preselection CD4+ CD8+ CD69− TCR3A9− thymocytes from STg and DTg mice, CD4+ CD8lo CD69+ TCR3A9+ thymocytes from STg mice (undergoing positive selection), and CD4+ CD8lo CD69+ TCR3A9+ thymocytes from DTg mice (undergoing negative selection). T cell activation genes were categorized into three groups using the p-values of one-way ANOVA followed by Tukey’s multiple comparison tests (see Materials and methods). For each of the nine mitosis genes analyzed, the p-value of ANOVAs testing preselection with either positive or negative selection thymocytes was <0.05. Shading indicates fold change of the mean relative to preselection thymocytes. (B) Flow cytometry for proliferation antigen Ki-67 in CD4− CD8− (DN), CD4+ CD8+ (DP), and CCR7+ CD4SP thymocytes from non-Tg, STg, and DTg mice, representative of two separate experiments. (C) Histogram overlays and column graphs compare expression of the indicated proteins on Helios+ (red) and Helios− (black) Foxp3− CD4SP populations at SP1–SP3 maturation stages with corresponding DP cells (shaded histograms) from non-Tg mice. Column graphs below show the mean fluorescence intensity (MFI) for each marker on the indicated SP subsets relative to the MFI of Foxp3− DP thymocytes from the same sample. Histograms in B and C are representative of at least three separate experiments; column graphs in C show mean ± SD for at least four mice compiled from at least two experiments. Paired Student’s t tests p-values: ***, P < 0.001; **, P = 0.001–0.01; *, P = 0.01–0.05.
To confirm that strong binding to self-pMHC induced a distinct response in CCR7− SP1 versus CCR7+ SP2 thymocytes, we compared expression of a subset of these T cell activation proteins on Helios− and Helios+ SP1 and SP2 cells in the normal repertoire by flow cytometry (Fig. 6 C). PD-1 was strongly induced on Helios+ SP1 cells but was hardly induced on Helios+ SP2 cells. In contrast, CD25, OX40, and GITR were strongly induced on Helios+ SP2 cells, consistent with the DTg microarray data, but only weakly or not at all on Helios+ SP1 cells. Other TCR-induced proteins such as CD5, CD69, and Bim were induced on Helios+ cells at both stages, although to slightly higher levels on SP2 cells. These data reveal that, in the natural repertoire, the phenotypes of Helios+ Foxp3− CD4SP cells vary with maturation stage, with PD-1 induction a feature at the SP1 stage, whereas the induction of GITR, OX40, and CD25 accompanies Helios and Bim induction at the SP2 stage.
Card11 and c-Rel mutations perturb Helios+ Foxp3− CD4SP thymocytes selectively at the SP2 and SP3 stages
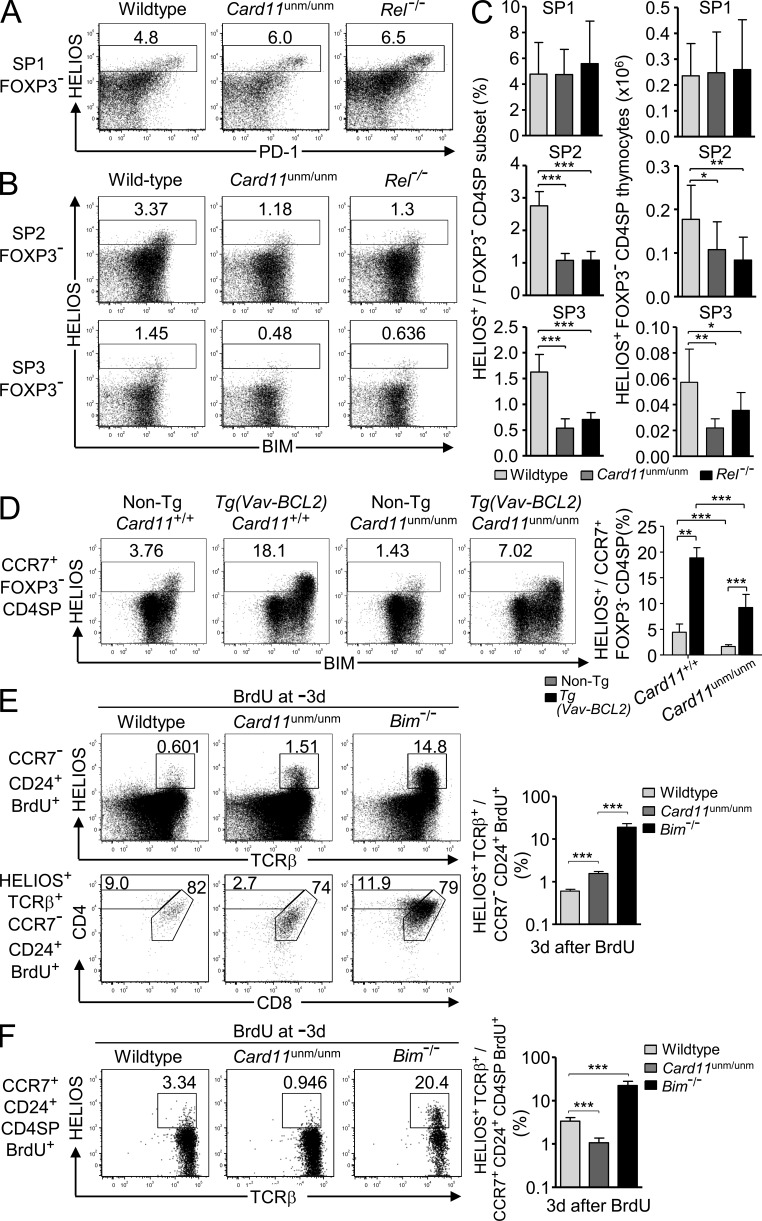
Many of the T cell activation genes induced in strongly self-reactive CCR7+ (SP2 and SP3) thymocytes are transcriptionally induced by NF-κB in mature T cells (Pahl, 1999; Shaffer et al., 2001), but these genes were not induced in Helios+ cells at the CCR7− SP1 stage. This implied that NF-κB might have a distinct function in the activation response of strongly self-reactive CCR7+ CD4SP thymocytes. To test this, we used mice bearing a partial loss of function mutation in TCR signaling to NF-κB, Card11unmodulated (Card11unm), which reduces but does not abolish CD25 induction by anti-CD3/CD28 stimulation in mature CD4+ T cells (Jun et al., 2003). In parallel, we analyzed mice lacking one member of the NF-κB transcription factor family, c-Rel (Köntgen et al., 1995). The formation of SP1 Helios+ Foxp3− CD4SP thymocytes that strongly express PD-1 was intact in Card11unm/unm and Rel−/− mice (Fig. 7, A and C). In contrast, although Helios was still induced in Card11unm/unm and Rel−/− CCR7+ thymocytes, the frequency and number of Helios+ SP2 and SP3 Foxp3− thymocytes were significantly decreased by these mutations (Fig. 7, B and C). The number of Helios− CD4SP thymocytes at each maturation stage SP1-SP3 was not significantly affected by homozygous mutations in Card11 or Rel (not depicted). These data demonstrate that Card11 and c-Rel are dispensable for the formation of CCR7− SP1 Helios+ Foxp3− CD4SP thymocytes, but both are required for normal accumulation of Helios+ strongly self-reactive cells at the CCR7+ SP2 and SP3 stages in the normal TCR repertoire.
Figure 7.
Card11 and c-Rel mutations perturb Helios+ Foxp3− CD4SP thymocytes selectively at the SP2 and SP3 stages. (A) Efficient SP1 Helios+ Foxp3− CD4SP formation. Helios versus PD-1 expression on SP1 (CD24+ CCR7−) Foxp3− CD4SP thymocytes from wild-type, Card11unm/unm, and Rel−/− mice. (B) Reduced SP2 and SP3 Helios+ Foxp3− CD4SP formation in Card11unm/unm and Rel−/− mice. Helios versus Bim expression on SP2 (CD24+ CCR7+) and SP3 (CD24+ CCR7−) Foxp3− CD4SP thymocytes in the same set of samples shown in A. (C) Graphs show the percentage and number of Helios+ cells in the SP1, SP2, and SP3 Foxp3− CD4SP thymocyte populations for wild-type, Card11unm/unm, and Rel−/− mice on the B6 background with at least eight mice per group compiled from five separate experiments, two of which included mice of all three genotypes. (D) Plots (left) show Helios versus Bim expression on Foxp3− CCR7+ CD4SP (SP2 and SP3) thymocytes from non-Tg Card11unm/unm mice (n = 5), age-matched wild-type B6 controls (n = 5), and Vav-BCL2 Tg Card11+/+ (n = 4) or Card11unm/unm (n = 5) littermates on the B6 background with a summary showing the mean and SD frequencies of Helios+ cells as gated in the plots, compiled from two separate experiments. (E) Card11unm/unm or Bim−/− mice and their wild-type littermates on the B10.BR background were injected i.p. with 1 mg BrdU 3 d before flow cytometry on thymocytes. Top panel shows CCR7− CD24+ BrdU+ cells (concatenated from four to five mice per genotype) with the gate defining the Helios+ TCRβ+ population, which was analyzed for CD4/CD8 expression in the bottom panel. Graph (right) shows frequency of Helios+ TCRβ+ cells among CCR7− CD24+ BrdU+ thymocytes. (F) Helios versus TCRβ expression (left) on nascent SP2 (CD24+ CCR7+ CD4SP BrdU+) thymocytes in the same set of samples shown in E; graph (right) shows frequency of Helios+ TCRβ+ cells as gated on plots. Data in E and F represent four to five mice per group in a single experiment. Numbers in plots indicate percentage of cells in gates shown, and columns show mean and SD. Unpaired Student’s t test p-values: ***, P < 0.001; **, P = 0.001–0.01; *, P = 0.01–0.05.
To clarify the requirement for Card11 in accumulation of Helios+ CCR7+ Foxp3− CD4SP cells, we tested the consequences of inhibiting clonal deletion in Card11unm/unm Tg(Vav-BCL2) mice. These had much larger numbers of Helios+ CCR7+ Foxp3− CD4SP cells than Card11unm/unm non-Tg controls, confirming that Helios was still strongly induced in self-reactive cells with defective NF-κB signaling (Fig. 7 D). However, their frequency was half that observed in Card11+/+ Tg(Vav-BCL2) littermates. These data support the conclusion that Card11 signaling to NF-κB is required for Helios+ medullary thymocytes to survive strong TCR signaling.
To extend the genetic evidence of a distinct response induced by strong binding to self-pMHC in CCR7− versus CCR7+ CD4+ thymocytes, mice were analyzed 3 d after BrdU, when the numbers of BrdU+ SP1 and SP2 cells peak (Fig. 2 B) and before many have emigrated (McCaughtry et al., 2007). 3 d after BrdU injection, Card11unm/unm mice had a small but significant increase in the frequency of Helios+ TCRβ+ cells among nascent CCR7− thymocytes, demonstrating that Card11 is required for efficient apoptosis of these cells, although their frequency was still 10 times lower in Card11unm/unm mice than in Bim−/− mice (Fig. 7 E). The characteristic dulling of CD4 and CD8 expression by CCR7− Helios+ TCRβ+ thymocytes was intact in Card11unm/unm mice (Fig. 7 E). Although mutations in Card11 and Bim both increased the frequency of nascent Helios+ thymocytes at the CCR7− stage, these mutations had opposite effects on the frequency of Helios+ cells among nascent CCR7+ CD24+ CD4SP thymocytes, being decreased threefold in Card11unm/unm mice and increased sixfold in Bim−/− mice (Fig. 7 F). In Bim−/− animals, Helios+ cells accounted for 20% of BrdU-labeled SP2 cells (Fig. 7 F).
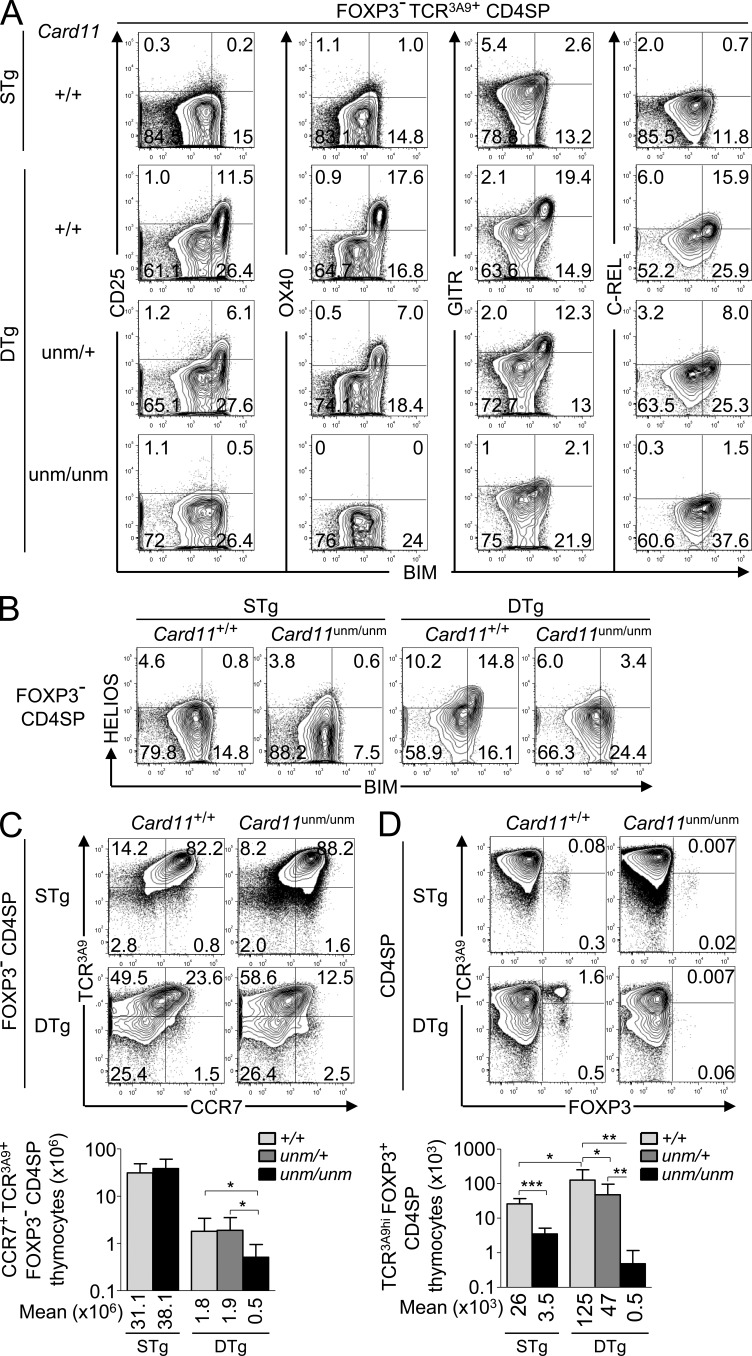
A potential explanation for the reduced Helios+ cell frequencies at the SP2 and SP3 stages in Card11unm/unm and Rel−/− mice is that Card11 activation of NF-κB delivers a survival signal that opposes the clonal deletion of CCR7+ CD4SP thymocytes that receive a strong TCR signal. For example, signaling to NF-κB induces T cell survival genes such as Bcl2, Bcla1a (A1), and Bcl2l1 (Bcl-X), and these were preferentially induced in strongly self-reactive SP2 cells in DTg mice (Fig. 6 A). To determine whether there was concurrent induction of Bim and NF-κB response proteins in strongly self-reactive SP2 cells, we analyzed Foxp3− CCR7+ CD4SP thymocytes undergoing clonal deletion at the SP2 stage in Card11unm/unm DTg mice. Confirming the microarray data, we found that high levels of CD25, OX40, GITR, and c-Rel were induced selectively in the Bimhi subset of Foxp3− TCR3A9+ CD4SP thymocytes from DTg but not STg mice (Fig. 8 A). The induction of each of these proteins was abolished in Card11unm homozygotes and markedly decreased in heterozygotes (Fig. 8 A), establishing that these are exquisitely dependent on Card11 signaling. Homozygosity for Card11unm diminished the accumulation of Helios+ CCR7+ SP2 cells (Fig. 8, B and C), so that these were 3.6-fold lower in Card11unm/unm DTg mice than in Card11+/+ DTg mice (Fig. 8 C). In contrast, Card11 was not required for accumulation of CCR7+ cells in STg mice where they bound self-pMHC weakly. Collectively, these results indicate that strongly self-reactive CD4 cells are deleted earlier in the SP1–SP2 transition when they are unable to make a concurrent NF-κB survival response.
Figure 8.
A Card11-dependent response induced concurrently with Bim in self-reactive Foxp3− CCR7+ CD4SP thymocytes curbs clonal deletion and is rate limiting for thymic T reg cell differentiation. (A) Expression of the T cell activation markers CD25, OX40, GITR, and c-Rel versus Bim in Foxp3− TCR3A9+ CD4SP thymocytes from STg or DTg mice of the indicated Card11 genotypes. Data are representative of four experiments with a total of at least nine mice of each Card11 genotype. (B and C) Reduced formation of mature phenotype Foxp3− CD4SP thymocytes in Card11unm/unm DTg mice. Phenotype of Foxp3− CD4SP thymocytes from Card11+/+ or Card11unm/unm STg or DTg mice displaying Helios versus Bim (representative of two experiments; B) or TCR3A9+ versus CCR7 (C). Summary (bottom) accompanies the data in C, showing the total number of cells in the top right quadrant (TCR3A9+ CCR7+) per thymus, compiled from five experiments with at least 8 STg and 11 DTg mice per Card11 genotype. (D) Representative plots (top) show phenotype, and summary (bottom) shows number of TCR3A9hi Foxp3+ CD4SP thymocytes compiled from six experiments with at least 6 STg and 14 DTg mice per Card11 genotype. Numbers in plots indicate percentage of cells in gates shown, and summaries show mean and SD. Student’s t tests p-values: ***, P < 0.001; **, P = 0.001–0.01; *, P = 0.01–0.05.
Thymic selection of CD4+ T cells also encompasses differentiation of Foxp3+ T reg cells, which are crucial for self-tolerance (Rudensky, 2011; Sakaguchi, 2011). Although T reg cells have heightened affinity for self–pMHC II ligands (Jordan et al., 2001; Hsieh et al., 2004), it has been debated whether this reflects active induction of T reg cell differentiation by strong recognition of self-pMHC (Cozzo Picca et al., 2011) or heightened resistance of T reg cell precursors to clonal deletion (van Santen et al., 2004). On a Card11+/+ background, DTg mice had almost fivefold more TCR3A9hi Foxp3+ CD4SP thymocytes than STg mice (Fig. 8 D), arguing that strong binding to self-pMHC actively induced the formation of organ-specific T reg cells in the thymus of DTg mice. The number of TCR3A9hi Foxp3+ CD4SP thymocytes was decreased by 2.6-fold in Card11unm/+ heterozygous DTg mice and by 250-fold in Card11unm/unm DTg mice (Fig. 8 D). These data show that many Foxp3− CCR7+ CD4SP cells that are Bimhi and destined to die nevertheless mount a Card11-dependent program that has the dual function of curbing clonal deletion and enabling T reg cell differentiation.
DISCUSSION
The findings above address several key questions about thymic negative selection. The results identify the Ikaros family member Helios as a unique molecular marker that qualitatively differentiates the thymocyte response to weak or strong self-pMHC stimulation in vivo. Flow cytometric measurement of Helios provides a way to enumerate and analyze the fraction of nascent thymocytes that are strongly self-reactive, both in normal mouse repertoires and in animals that are genetically predisposed to autoimmune disease. We show that Helios and Bim are coinduced in strongly self-reactive thymocytes at two major waves of Bim-mediated deletion of CD4+ cells: one in CCR7-negative thymocytes that is induced in 55% of newly formed cells and a second in 20% of cells that reach the CCR7+ CD4SP stage. This in turn enabled analysis of the other proteins and mRNAs that are induced in strongly self-reactive thymocytes at each of these stages, addressing the question of how the thymocyte response to strong TCR stimulation varies with maturation stage.
Helios was shown to be induced selectively in strongly self-reactive thymocytes and concurrently with Bim in several settings: in the subset of cells in the normal repertoire that are normally eliminated by Bim, in Vβ5+ and Vβ11+ thymocytes that bind strongly to self-superantigens, and in TCR Tg thymocytes that bind strongly to self-pMHC expressed in AIRE+ medullary thymic epithelium. In contrast, Helios was down-regulated during positive selection of CD4+ thymocytes that bind self-pMHC weakly. This splitting of the response distinguishes Helios from other TCR-induced proteins like Bim, Nur77, CD69, and GITR, which are induced during both positive and negative selection and differentiate the weak and strong response only by their magnitude of induction. T cell development was reported to be normal in the absence of Helios (Cai et al., 2009; Thornton et al., 2010), although it is possible that when the Helios protein is absent this allows other Ikaros family members such as Ikaros itself to fill the niche and substitute, as was found to be the case from comparison of mice with null alleles and non-DNA–binding alleles of Ikaros (Papathanasiou et al., 2003). Acute retroviral-mediated expression of Helios decreased the proliferation of activated splenic CD4+ T cells (Zhang et al., 2007), raising the possibility that Helios has an antiproliferative function. Consistent with this possibility, non-DNA–binding dominant-negative short isoforms of Helios have been associated with T cell leukemia in humans (Sun et al., 2002; Fujii et al., 2003) and induce T cell leukemia in mice (Zhang et al., 2007). Continuous activating receptor stimulation has recently been shown to induce Helios in NK cells and dampen their responsiveness (Narni-Mancinelli et al., 2012).
Helios expression provides a direct way to measure the percentage of thymocytes that bear strongly self-reactive TCRs and are susceptible to deletion. Our analysis of CCR7− thymocytes, including CD69+ TCRβ+ cells selected from BrdU-labeled precursors in the previous 24 or 72 h, suggests that 55% of all TCR-signaled thymocytes are deleted by Bim at the CCR7− stage. At the subsequent CCR7+ stage, 20% of nascent BrdU+ SP2 cells were Helios+ in Bim−/− mice versus only 3% in wild-type controls, indicating that additional CD4SP thymocytes are deleted at this stage. Although a few Helios+ SP2 cells may derive from Helios+ SP1 precursors, it seems unlikely that many Helios+ SP1 cells up-regulate CCR7 to become SP2 cells, given that the superantigen-reactive Vβ5+ and Vβ11+ populations in Bim−/− mice had lower Helios+ frequencies at the SP2 stage than at the SP1 stage (Fig. 3). After accounting for the 55% of TCR-signaled CCR7− cells that arrest their maturation and fail to up-regulate CCR7 in Bim−/− mice, we calculate that the 20% of nascent SP2 cells that were Helios+ in Bim−/− mice (Fig. 7 F) represents 9% of the pool that was initially selected. It is conceivable that these cells bound weakly to self-pMHC in the cortex but then bound strongly to a self-pMHC ligand confined to the medulla, as occurs in the DTg mice. It was recently suggested that the absence of both Bim and Puma (gene name Bbc3) results in more autoreactive thymocytes escaping negative selection than when only Bim is absent (Gray et al., 2012). As the percentage and number of CD69+ thymocytes at the DP stage, a population shown here to include cells TCR-signaled for positive or negative selection, were similar in Bim-deficient and Bim/Puma double-deficient mice, Puma would not seem to delete additional autoreactive cells in the early wave of negative selection. Bim/Puma double-deficient mice had a striking accumulation of CD24lo CD4SP cells (Gray et al., 2012), which indicates that Puma mediates cell death of Bim-deficient CD4+ cells that are more mature than the SP2 stage. Our estimate that a total of ∼64% of TCR-signaled CD4+ thymocytes undergo Bim-dependent negative selection either at the CCR7− (55%) or CCR7+ (9%) stage agrees with earlier estimates based on the frequency of mature CD4SP thymocytes (Tourne et al., 1997; van Meerwijk et al., 1997) or the proportion of CD4+ T cells with self-reactive TCRs (Ignatowicz et al., 1996) in mice in which deletion was defective.
The results here provide molecular insight into the acquisition of self-tolerance by analyzing the thymocyte response to strong TCR stimulation and how it differs from the mature T cell response, independent of the normally rapid Bim-mediated deletion. At the CCR7− stage, strongly self-reactive T cells make a unique response, even compared with CCR7+ thymocytes, which is dominated by developmental arrest at the CCR7− stage, PD-1 induction, CD4 and CD8 down-regulation, and lack of any induction of CD25 or OX40. The induction of PD-1 and dulling of CD4 and CD8 have been observed in HYcd4 TCR Tg thymocytes encountering MHC I–presented antigen on cortical epithelium (McCaughtry et al., 2008), in thymocytes lacking CD28 co-stimulation (Pobezinsky et al., 2012), and in TCR-activated DP thymocytes undergoing apoptosis in vitro (Swat et al., 1991; Vasquez et al., 1992; Page et al., 1993). The fact that Helios+ CCR7− cells remain developmentally arrested at the CCR7− stage in the thymus when Bim-mediated deletion is abolished and that they are not found in the periphery as shown for Vβ5+ and Vβ11+ CD4 cells is consistent with a comparable developmental arrest shown for MHC I–restricted T cells binding strongly to self-pMHC in the thymus cortex (Hu et al., 2009; Kovalovsky et al., 2010; Suen and Baldwin, 2012). This unique response thus appears to be a general, stage-specific response program that may either be intrinsic to very immature CCR7− thymocytes or reflect the nature of the APCs they encounter at that stage. Thymocyte negative selection at this stage has interesting parallels with negative selection of immature B cells that are strongly self-reactive in the bone marrow, which also become developmentally arrested at a stage where they cannot home to lymph nodes when their apoptotic response is blocked (Hartley et al., 1993).
At the CCR7+ stage, however, the thymocyte response to strong pMHC stimulation was found to be surprisingly similar to mature T cell activation, with induction of a broad program of T cell survival and growth genes through Card11 signaling to NF-κB. This hollow activation response opposes Bim-mediated deletion, and by activating c-Rel and CD25, it sets the stage for Foxp3 expression and diversion of a minority of cells to the Foxp3+ T reg cell lineage. The observation that this Card11-mediated hollow activation response is required to oppose Bim-mediated deletion explains why a constitutively active STAT5 transgene failed to rescue thymic T reg cell formation in Card11-deficient mice (Molinero et al., 2009). It is significant that the CCR7+ thymocyte response to strong self-antigen lacks key growth mediators like IL-2 and Myc that are central to mature T cell activation and that there was no evidence that the thymocytes were induced into cell cycle. Because IL-2 and Myc are both direct NF-κB targets in mature T cells, their failure to be activated in CCR7+ SP2 thymocytes may reflect stage-specific programming, for example at the level of chromatin accessibility, or the effect of Helios itself as Ikaros suppresses Il2 transcription in anergic T cells (Bandyopadhyay et al., 2007; Thomas et al., 2007). Collectively, these results reveal that the acquisition of self-tolerance in CD4SP cells in the thymic medulla walks a fine line between T cell activation and Bim-mediated deletion and would appear to provide much scope for inherited variability in susceptibility to autoimmune disease.
MATERIALS AND METHODS
Mice.
Mice were bred and housed in the Australian Phenomics Facility. Non-Tg mice were either B10.BR (H-2k) or C57BL/6 (B6; H-2b), and all mice were 6–14 wk old when used. 3A9 TCR Tg (Ho et al., 1994), insHEL Tg (originally called ILK-3; Akkaraju et al., 1997), Bim-deficient (Bcl2l11−/−, here referred to as Bim−/−; Bouillet et al., 1999), Vav-BCL2 Tg (Ogilvy et al., 1999), Zap70mrt (Siggs et al., 2007), Card11unm (Jun et al., 2003), and Rel−/− (Köntgen et al., 1995) mice have been described previously. 3A9 TCR, insHEL, and Vav-BCL2 Tg mice were hemizygous for the relevant transgenes and genotyped by PCR. Mice bearing the Bim− allele were genotyped by PCR. A mutagenically separated PCR assay (Rust et al., 1993) was used to genotype mice for the Card11unm allele. Except for Card11unm/unm mice on the B6 background in Fig. 7, all Bim−/− and Card11unm/unm mice were offspring of at least the third generation backcross to B10.BR, and at least half of the Card11+/+ or Bim+/+ animals represented in each figure panel were their littermates, whereas the remainder were wild-type controls from our facility. Mice from a homozygous Rel−/− strain were used. To make bone marrow chimeras, CD45.1+ insHELneg/pos mice were irradiated with x rays (two doses of 4.5 Gy given 4 h apart) and then injected intravenously with 3 × 106 bone marrow cells from a Bim−/− STg donor alone or mixed at a ratio of 1:19 with CD45.1+ non-Tg marrow. They were allowed to reconstitute for 4–5 wk before analysis. The Animal Experimentation Ethics Committee of the Australian National University approved all procedures.
Flow cytometry.
Single cell thymocyte suspensions were incubated for 60 min at 37°C in prewarmed FACS buffer (PBS containing 2% vol/vol heat-inactivated bovine serum and 0.01% m/v sodium azide) or culture supernatant from the 1G12 hybridoma (specific for TCR3A9; American Type Culture Collection) containing anti-CCR7. Cells were then pelleted by centrifugation and incubated for 30 min at 4°C in FACS buffer containing assortments of fluorochrome- or biotin-conjugated monoclonal antibodies against cell surface proteins or mouse IgG1 (to detect 1G12). After washing in FACS buffer, cells were fixed, permeabilized, and stained for intracellular proteins using the eBioscience Foxp3 staining buffers or the FITC BrdU Flow kit (BD) with anti-Helios (clone 22F6; BioLegend; Thornton et al., 2010) included in the same incubation step as anti–BrdU-FITC. Finally, cells were washed in FACS buffer and then incubated for 15 min at 4°C in FACS buffer containing Qdot 605–streptavidin conjugate (Invitrogen) to detect biotinylated antibodies. Except for anti-Bim (clone 3C5; a gift from A. Strasser, Walter and Eliza Hall Institute of Medical Research, Melbourne, Victoria, Australia), which was conjugated to Alexa Fluor 647 using a Protein Labeling kit (Molecular Probes), antibodies were purchased from BD, eBioscience, BioLegend, or Santa Cruz Biotechnology, Inc. (anti–c-Rel–A488). Data were acquired with an LSR II flow cytometer (BD) and analyzed using FlowJo software (Tree Star).
Microarray analysis.
A microarray dataset (Liston et al., 2007) accessible through GEO Series accession no. GSE3997 was mined for T cell activation genes (Shaffer et al., 2001). Thymocyte populations analyzed by microarray were preselection (PreS; CD4+ CD8+ CD69− TCR3A9− from STg and DTg mice), positive selection (S+; CD4+ CD8lo CD69+ TCR3A9+ from STg mice), and negative selection (S−; CD4+ CD8lo CD69+ TCR3A9+ from DTg mice). Microarray analyses were performed before the establishment of the SP1–SP3 categorization system; analysis indicates the percentages of SP1 and SP2 cells were ∼40% and 55%, respectively, in the S+ population, and 75% and 22%, respectively, in the S− population. The genes were categorized using p-values of one-way ANOVA followed by Tukey’s multiple comparison tests. The three categories were (1) genes up-regulated preferentially in negative selection (PreS vs. S− <0.05 and either S− vs. S+ <0.05 or PreS vs. S+ >0.05), (2) genes up-regulated comparably during positive and negative selection (PreS vs. S+ <0.05 and S+ vs. S− >0.05; Irf4 was included in category 2 even though S+ vs. S− was <0.05 on the grounds that S+ exceeded S− by only 1.16-fold), and (3) genes not induced (PreS vs. S+ and PreS vs. S− >0.05) or down-regulated during selection (PreS significantly exceeded both S+ and S−).
Statistical analysis.
Prism version 5.00 for Windows (GraphPad Software) was used to conduct unpaired or paired Student’s t tests as described in the figure legends and for one-way ANOVA followed by Tukey’s multiple comparison tests on microarray data.
Acknowledgments
We thank Debbie Howard for technical assistance, Andreas Strasser for the gift of the anti-Bim antibody, and Steve Gerondakis for making Rel−/− mice available.
This work was supported by a National Health and Medical Research Ccouncil Program Grant and Australia Fellowship and by the Juvenile Diabetes Research Foundation and involved mutant strains funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health and the Wellcome Trust.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- B6
- C57BL/6
- DP
- double positive
- DTg
- double Tg
- HEL
- hen egg lysozyme
- insHEL
- insulin promoter–driven HEL
- pMHC
- peptide–MHC
- SP
- single positive
- STg
- single Tg
- Tg
- transgenic
References
- Akkaraju S., Ho W.Y., Leong D., Canaan K., Davis M.M., Goodnow C.C. 1997. A range of CD4 T cell tolerance: partial inactivation to organ-specific antigen allows nondestructive thyroiditis or insulitis. Immunity. 7:255–271 10.1016/S1074-7613(00)80528-2 [DOI] [PubMed] [Google Scholar]
- Baldwin T.A., Hogquist K.A. 2007. Transcriptional analysis of clonal deletion in vivo. J. Immunol. 179:837–844 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Duré M., Paroder M., Soto-Nieves N., Puga I., Macián F. 2007. Interleukin 2 gene transcription is regulated by Ikaros-induced changes in histone acetylation in anergic T cells. Blood. 109:2878–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P., Metcalf D., Huang D.C., Tarlinton D.M., Kay T.W., Köntgen F., Adams J.M., Strasser A. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 286:1735–1738 10.1126/science.286.5445.1735 [DOI] [PubMed] [Google Scholar]
- Bouillet P., Purton J.F., Godfrey D.I., Zhang L.C., Coultas L., Puthalakath H., Pellegrini M., Cory S., Adams J.M., Strasser A. 2002. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 415:922–926 10.1038/415922a [DOI] [PubMed] [Google Scholar]
- Cai Q., Dierich A., Oulad-Abdelghani M., Chan S., Kastner P. 2009. Helios deficiency has minimal impact on T cell development and function. J. Immunol. 183:2303–2311 10.4049/jimmunol.0901407 [DOI] [PubMed] [Google Scholar]
- Cozzo Picca C., Simons D.M., Oh S., Aitken M., Perng O.A., Mergenthaler C., Kropf E., Erikson J., Caton A.J. 2011. CD4+CD25+Foxp3+ regulatory T cell formation requires more specific recognition of a self-peptide than thymocyte deletion. Proc. Natl. Acad. Sci. USA. 108:14890–14895 10.1073/pnas.1103810108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels M.A., Teixeiro E., Gill J., Hausmann B., Roubaty D., Holmberg K., Werlen G., Holländer G.A., Gascoigne N.R., Palmer E. 2006. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 444:724–729 10.1038/nature05269 [DOI] [PubMed] [Google Scholar]
- Davalos-Misslitz A.C., Worbs T., Willenzon S., Bernhardt G., Förster R. 2007. Impaired responsiveness to T-cell receptor stimulation and defective negative selection of thymocytes in CCR7-deficient mice. Blood. 110:4351–4359 10.1182/blood-2007-01-070284 [DOI] [PubMed] [Google Scholar]
- Dose M., Sleckman B.P., Han J., Bredemeyer A.L., Bendelac A., Gounari F. 2009. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. USA. 106:8641–8646 10.1073/pnas.0812255106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson P.J., Knight A.M., Fairchild S., Simpson E., Tomonari K. 1991. Genes encoding ligands for deletion of V beta 11 T cells cosegregate with mammary tumour virus genomes. Nature. 349:531–532 10.1038/349531a0 [DOI] [PubMed] [Google Scholar]
- Fujii K., Ishimaru F., Nakase K., Tabayashi T., Kozuka T., Naoki K., Miyahara M., Toki H., Kitajima K., Harada M., Tanimoto M. 2003. Over-expression of short isoforms of Helios in patients with adult T-cell leukaemia/lymphoma. Br. J. Haematol. 120:986–989 10.1046/j.1365-2141.2003.04216.x [DOI] [PubMed] [Google Scholar]
- Gallegos A.M., Bevan M.J. 2006. Central tolerance: good but imperfect. Immunol. Rev. 209:290–296 10.1111/j.0105-2896.2006.00348.x [DOI] [PubMed] [Google Scholar]
- Gascoigne N.R., Palmer E. 2011. Signaling in thymic selection. Curr. Opin. Immunol. 23:207–212 10.1016/j.coi.2010.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D.H., Kupresanin F., Berzins S.P., Herold M.J., O’Reilly L.A., Bouillet P., Strasser A. 2012. The BH3-only proteins Bim and Puma cooperate to impose deletional tolerance of organ-specific antigens. Immunity. 37:451–462 10.1016/j.immuni.2012.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare K.J., Jenkinson E.J., Anderson G. 1999. CD69 expression discriminates MHC-dependent and -independent stages of thymocyte positive selection. J. Immunol. 162:3978–3983 [PubMed] [Google Scholar]
- Hartley S.B., Cooke M.P., Fulcher D.A., Harris A.W., Cory S., Basten A., Goodnow C.C. 1993. Elimination of self-reactive B lymphocytes proceeds in two stages: arrested development and cell death. Cell. 72:325–335 10.1016/0092-8674(93)90111-3 [DOI] [PubMed] [Google Scholar]
- Ho W.Y., Cooke M.P., Goodnow C.C., Davis M.M. 1994. Resting and anergic B cells are defective in CD28-dependent costimulation of naive CD4+ T cells. J. Exp. Med. 179:1539–1549 10.1084/jem.179.5.1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.S., Liang Y., Tyznik A.J., Self S.G., Liggitt D., Rudensky A.Y. 2004. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 21:267–277 10.1016/j.immuni.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Hu Q., Sader A., Parkman J.C., Baldwin T.A. 2009. Bim-mediated apoptosis is not necessary for thymic negative selection to ubiquitous self-antigens. J. Immunol. 183:7761–7767 10.4049/jimmunol.0902181 [DOI] [PubMed] [Google Scholar]
- Ignatowicz L., Kappler J., Marrack P. 1996. The repertoire of T cells shaped by a single MHC/peptide ligand. Cell. 84:521–529 10.1016/S0092-8674(00)81028-4 [DOI] [PubMed] [Google Scholar]
- Jordan M.S., Boesteanu A., Reed A.J., Petrone A.L., Holenbeck A.E., Lerman M.A., Naji A., Caton A.J. 2001. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat. Immunol. 2:301–306 10.1038/86302 [DOI] [PubMed] [Google Scholar]
- Jorgensen T.N., McKee A., Wang M., Kushnir E., White J., Refaeli Y., Kappler J.W., Marrack P. 2007. Bim and Bcl-2 mutually affect the expression of the other in T cells. J. Immunol. 179:3417–3424 [DOI] [PubMed] [Google Scholar]
- Jun J.E., Wilson L.E., Vinuesa C.G., Lesage S., Blery M., Miosge L.A., Cook M.C., Kucharska E.M., Hara H., Penninger J.M., et al. 2003. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 18:751–762 10.1016/S1074-7613(03)00141-9 [DOI] [PubMed] [Google Scholar]
- Kappler J.W., Roehm N., Marrack P. 1987. T cell tolerance by clonal elimination in the thymus. Cell. 49:273–280 10.1016/0092-8674(87)90568-X [DOI] [PubMed] [Google Scholar]
- Kishimoto H., Sprent J. 1997. Negative selection in the thymus includes semimature T cells. J. Exp. Med. 185:263–271 10.1084/jem.185.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köntgen F., Grumont R.J., Strasser A., Metcalf D., Li R., Tarlinton D., Gerondakis S. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9:1965–1977 10.1101/gad.9.16.1965 [DOI] [PubMed] [Google Scholar]
- Kovalovsky D., Pezzano M., Ortiz B.D., Sant’Angelo D.B. 2010. A novel TCR transgenic model reveals that negative selection involves an immediate, Bim-dependent pathway and a delayed, Bim-independent pathway. PLoS ONE. 5:e8675 10.1371/journal.pone.0008675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S., Hartley S.B., Akkaraju S., Wilson J., Townsend M., Goodnow C.C. 2002. Failure to censor forbidden clones of CD4 T cells in autoimmune diabetes. J. Exp. Med. 196:1175–1188 10.1084/jem.20020735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Lesage S., Wilson J., Peltonen L., Goodnow C.C. 2003. Aire regulates negative selection of organ-specific T cells. Nat. Immunol. 4:350–354 10.1038/ni906 [DOI] [PubMed] [Google Scholar]
- Liston A., Gray D.H., Lesage S., Fletcher A.L., Wilson J., Webster K.E., Scott H.S., Boyd R.L., Peltonen L., Goodnow C.C. 2004a. Gene dosage—limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J. Exp. Med. 200:1015–1026 10.1084/jem.20040581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Lesage S., Gray D.H., O’Reilly L.A., Strasser A., Fahrer A.M., Boyd R.L., Wilson J., Baxter A.G., Gallo E.M., et al. 2004b. Generalized resistance to thymic deletion in the NOD mouse; a polygenic trait characterized by defective induction of Bim. Immunity. 21:817–830 [DOI] [PubMed] [Google Scholar]
- Liston A., Hardy K., Pittelkow Y., Wilson S.R., Makaroff L.E., Fahrer A.M., Goodnow C.C. 2007. Impairment of organ-specific T cell negative selection by diabetes susceptibility genes: genomic analysis by mRNA profiling. Genome Biol. 8:R12 10.1186/gb-2007-8-1-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas B., Vasseur F., Penit C. 1993. Normal sequence of phenotypic transitions in one cohort of 5-bromo-2′-deoxyuridine-pulse-labeled thymocytes. Correlation with T cell receptor expression. J. Immunol. 151:4574–4582 [PubMed] [Google Scholar]
- McCaughtry T.M., Wilken M.S., Hogquist K.A. 2007. Thymic emigration revisited. J. Exp. Med. 204:2513–2520 10.1084/jem.20070601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughtry T.M., Baldwin T.A., Wilken M.S., Hogquist K.A. 2008. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J. Exp. Med. 205:2575–2584 10.1084/jem.20080866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero L.L., Yang J., Gajewski T., Abraham C., Farrar M.A., Alegre M.L. 2009. CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J. Immunol. 182:6736–6743 10.4049/jimmunol.0900498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., Hogquist K.A. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E., Jaeger B.N., Bernat C., Fenis A., Kung S., De Gassart A., Mahmood S., Gut M., Heath S.C., Estellé J., et al. 2012. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science. 335:344–348 10.1126/science.1215621 [DOI] [PubMed] [Google Scholar]
- Nitta T., Nitta S., Lei Y., Lipp M., Takahama Y. 2009. CCR7-mediated migration of developing thymocytes to the medulla is essential for negative selection to tissue-restricted antigens. Proc. Natl. Acad. Sci. USA. 106:17129–17133 10.1073/pnas.0906956106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvy S., Metcalf D., Print C.G., Bath M.L., Harris A.W., Adams J.M. 1999. Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc. Natl. Acad. Sci. USA. 96:14943–14948 10.1073/pnas.96.26.14943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page D.M., Kane L.P., Allison J.P., Hedrick S.M. 1993. Two signals are required for negative selection of CD4+CD8+ thymocytes. J. Immunol. 151:1868–1880 [PubMed] [Google Scholar]
- Pahl H.L. 1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 18:6853–6866 10.1038/sj.onc.1203239 [DOI] [PubMed] [Google Scholar]
- Papathanasiou P., Perkins A.C., Cobb B.S., Ferrini R., Sridharan R., Hoyne G.F., Nelms K.A., Smale S.T., Goodnow C.C. 2003. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity. 19:131–144 10.1016/S1074-7613(03)00168-7 [DOI] [PubMed] [Google Scholar]
- Pircher H., Bürki K., Lang R., Hengartner H., Zinkernagel R.M. 1989. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 342:559–561 10.1038/342559a0 [DOI] [PubMed] [Google Scholar]
- Pobezinsky L.A., Angelov G.S., Tai X., Jeurling S., Van Laethem F., Feigenbaum L., Park J.H., Singer A. 2012. Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nat. Immunol. 13:569–578 10.1038/ni.2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudensky A.Y. 2011. Regulatory T cells and Foxp3. Immunol. Rev. 241:260–268 10.1111/j.1600-065X.2011.01018.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust S., Funke H., Assmann G. 1993. Mutagenically separated PCR (MS-PCR): a highly specific one step procedure for easy mutation detection. Nucleic Acids Res. 21:3623–3629 10.1093/nar/21.16.3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S. 2011. Regulatory T cells: history and perspective. Methods Mol. Biol. 707:3–17 10.1007/978-1-61737-979-6_1 [DOI] [PubMed] [Google Scholar]
- Shaffer A.L., Rosenwald A., Hurt E.M., Giltnane J.M., Lam L.T., Pickeral O.K., Staudt L.M. 2001. Signatures of the immune response. Immunity. 15:375–385 10.1016/S1074-7613(01)00194-7 [DOI] [PubMed] [Google Scholar]
- Siggs O.M., Miosge L.A., Yates A.L., Kucharska E.M., Sheahan D., Brdicka T., Weiss A., Liston A., Goodnow C.C. 2007. Opposing functions of the T cell receptor kinase ZAP-70 in immunity and tolerance differentially titrate in response to nucleotide substitutions. Immunity. 27:912–926 10.1016/j.immuni.2007.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritesky G.L., Jameson S.C., Hogquist K.A. 2012. Selection of self-reactive T cells in the thymus. Annu. Rev. Immunol. 30:95–114 10.1146/annurev-immunol-020711-075035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen A.Y., Baldwin T.A. 2012. Proapoptotic protein Bim is differentially required during thymic clonal deletion to ubiquitous versus tissue-restricted antigens. Proc. Natl. Acad. Sci. USA. 109:893–898 10.1073/pnas.1114834109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Kerawalla H., Wu X., Lehnert M.S., Uckun F.M. 2002. Expression of a unique helios isoform in human leukemia cells. Leuk. Lymphoma. 43:841–849 10.1080/10428190290016980 [DOI] [PubMed] [Google Scholar]
- Surh C.D., Sprent J. 1994. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 372:100–103 10.1038/372100a0 [DOI] [PubMed] [Google Scholar]
- Swat W., Ignatowicz L., von Boehmer H., Kisielow P. 1991. Clonal deletion of immature CD4+8+ thymocytes in suspension culture by extrathymic antigen-presenting cells. Nature. 351:150–153 10.1038/351150a0 [DOI] [PubMed] [Google Scholar]
- Thomas R.M., Chunder N., Chen C., Umetsu S.E., Winandy S., Wells A.D. 2007. Ikaros enforces the costimulatory requirement for IL2 gene expression and is required for anergy induction in CD4+ T lymphocytes. J. Immunol. 179:7305–7315 [DOI] [PubMed] [Google Scholar]
- Thornton A.M., Korty P.E., Tran D.Q., Wohlfert E.A., Murray P.E., Belkaid Y., Shevach E.M. 2010. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184:3433–3441 10.4049/jimmunol.0904028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourne S., Miyazaki T., Oxenius A., Klein L., Fehr T., Kyewski B., Benoist C., Mathis D. 1997. Selection of a broad repertoire of CD4+ T cells in H-2Ma0/0 mice. Immunity. 7:187–195 10.1016/S1074-7613(00)80522-1 [DOI] [PubMed] [Google Scholar]
- Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G.R., Bishop J.M. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 414:768–773 10.1038/414768a [DOI] [PubMed] [Google Scholar]
- Ueno T., Saito F., Gray D.H., Kuse S., Hieshima K., Nakano H., Kakiuchi T., Lipp M., Boyd R.L., Takahama Y. 2004. CCR7 signals are essential for cortex–medulla migration of developing thymocytes. J. Exp. Med. 200:493–505 10.1084/jem.20040643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meerwijk J.P., Marguerat S., Lees R.K., Germain R.N., Fowlkes B.J., MacDonald H.R. 1997. Quantitative impact of thymic clonal deletion on the T cell repertoire. J. Exp. Med. 185:377–383 10.1084/jem.185.3.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen H.M., Benoist C., Mathis D. 2004. Number of T reg cells that differentiate does not increase upon encounter of agonist ligand on thymic epithelial cells. J. Exp. Med. 200:1221–1230 10.1084/jem.20041022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez N.J., Kaye J., Hedrick S.M. 1992. In vivo and in vitro clonal deletion of double-positive thymocytes. J. Exp. Med. 175:1307–1316 10.1084/jem.175.5.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Boehmer H. 1990. Developmental biology of T cells in T cell-receptor transgenic mice. Annu. Rev. Immunol. 8:531–556 10.1146/annurev.iy.08.040190.002531 [DOI] [PubMed] [Google Scholar]
- Woodland D.L., Happ M.P., Gollob K.J., Palmer E. 1991. An endogenous retrovirus mediating deletion of alpha beta T cells? Nature. 349:529–530 10.1038/349529a0 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Swindle C.S., Bates J.T., Ko R., Cotta C.V., Klug C.A. 2007. Expression of a non-DNA-binding isoform of Helios induces T-cell lymphoma in mice. Blood. 109:2190–2197 10.1182/blood-2005-01-031930 [DOI] [PMC free article] [PubMed] [Google Scholar]