Activating Notch with a Notch agonist peptide induces apoptosis in AML patient samples.
Abstract
Although aberrant Notch activation contributes to leukemogenesis in T cells, its role in acute myelogenous leukemia (AML) remains unclear. Here, we report that human AML samples have robust expression of Notch receptors; however, Notch receptor activation and expression of downstream Notch targets are remarkably low, suggesting that Notch is present but not constitutively activated in human AML. The functional role of these Notch receptors in AML is not known. Induced activation through any of the Notch receptors (Notch1–4), or through the Notch target Hairy/Enhancer of Split 1 (HES1), consistently leads to AML growth arrest and caspase-dependent apoptosis, which are associated with B cell lymphoma 2 (BCL2) loss and enhanced p53/p21 expression. These effects were dependent on the HES1 repressor domain and were rescued through reexpression of BCL2. Importantly, activated Notch1, Notch2, and HES1 all led to inhibited AML growth in vivo, and Notch inhibition via dnMAML enhanced proliferation in vivo, thus revealing the physiological inhibition of AML growth in vivo in response to Notch signaling. As a novel therapeutic approach, we used a Notch agonist peptide that led to significant apoptosis in AML patient samples. In conclusion, we report consistent Notch-mediated growth arrest and apoptosis in human AML, and propose the development of Notch agonists as a potential therapeutic approach in AML.
The Notch signaling pathway is highly conserved throughout evolution and has multiple critical roles in neurogenesis, myogenesis, vasculogenesis, and hematopoiesis (Artavanis-Tsakonas et al., 1999). Activation of the Notch pathway has varied effects on proliferation, differentiation, and survival, which are highly cell type specific, though these cell-specific mechanisms have not been elucidated in most systems (Baldi et al., 2004). In cancer, Notch signaling has been shown to play both oncogenic and tumor suppressor roles, depending on the cell type (Koch and Radtke, 2007). Accumulating evidence demonstrates the importance of altered Notch signaling in the growth, differentiation, and apoptosis of human hematopoietic malignancies (Zweidler-McKay and Pear, 2004; Aster et al., 2008; Jundt et al., 2008; Zweidler-McKay, 2008). A central role for Notch signaling in leukemia has been established in T cell acute lymphoblastic leukemia (T-ALL), where Notch pathway–activating mutations are found in 50–70% of children and adults with T-ALL (Weng et al., 2004; Aster et al., 2008). Similarly, Notch receptor mutations have been identified in a range of mature B cell leukemias and lymphomas (Di Ianni et al., 2009; Del Giudice et al., 2011).
In contrast, the roles of Notch signaling on myeloid development and AML remain unclear. In hematopoietic stem cells, Notch signaling can promote self-renewal, induce growth arrest and apoptosis, and induce commitment to the T cell lineage (Carlesso et al., 1999; Ohishi et al., 2002; Maillard et al., 2005; Yu et al., 2006; Chadwick et al., 2007). In conflicting studies, Notch signaling in myeloid precursors has been shown to promote self-renewal, induce/inhibit differentiation to monocytes, or induce apoptosis (Li et al., 1998; Carlesso et al., 1999; Masuya et al., 2002; Schroeder et al., 2003; Sarmento et al., 2005).
Knowledge about the role of Notch in AML is equally poorly understood. Chiaramonte et al. (2005) reported that despite relatively high levels of Notch1 receptor in a panel of primary patient samples, the Notch target gene HES1 was expressed at low levels, suggesting that the Notch pathway was present but not activated. Similarly, Tohda and Nara (2001) demonstrated the presence of Notch1 receptors and only limited evidence of Notch activation. This group has also provided some information on the effects of Notch signaling on AML cells, where exposure of AML patient samples to plate-bound Notch ligand led to a full range of responses, from proliferation to growth arrest, which varied by sample (Tohda et al., 2005). Others have observed that co-culture with Notch ligand-expressing cells does not affect proliferation of an AML cell line, but alters G1–S transition and inhibits mitogen-induced differentiation (Carlesso et al., 1999; Sarmento et al., 2005). In contrast, Chadwick et al. (2008) show that expression of activated Notch1 in TF-1 AML cells leads to growth arrest and apoptosis. Similarly, Yin et al. (2009) demonstrate that expression of activated Notch1 inhibits proliferation and colony formation in K562 chronic myelogenous leukemia (CML) myeloid blast crisis cells. Alternatively, a recent study by Nakahara et al. (2010) demonstrated that expression of HES1 and breakpoint cluster region-Abelson (BCR-ABL) in mouse hematopoietic precursors lead to a CML blast-crisis–like disease.
Given these contrasting findings, we sought to determine whether there was evidence of constitutive Notch activation in human AML and CML myeloid blast crisis cell lines and patient samples; to comprehensively test the consequences of Notch activation and inhibition at multiple levels; to define the downstream mechanisms; and to test the effects of Notch modulation in vivo.
RESULTS
Notch receptors are expressed, but not activated, in human AML samples and cell lines
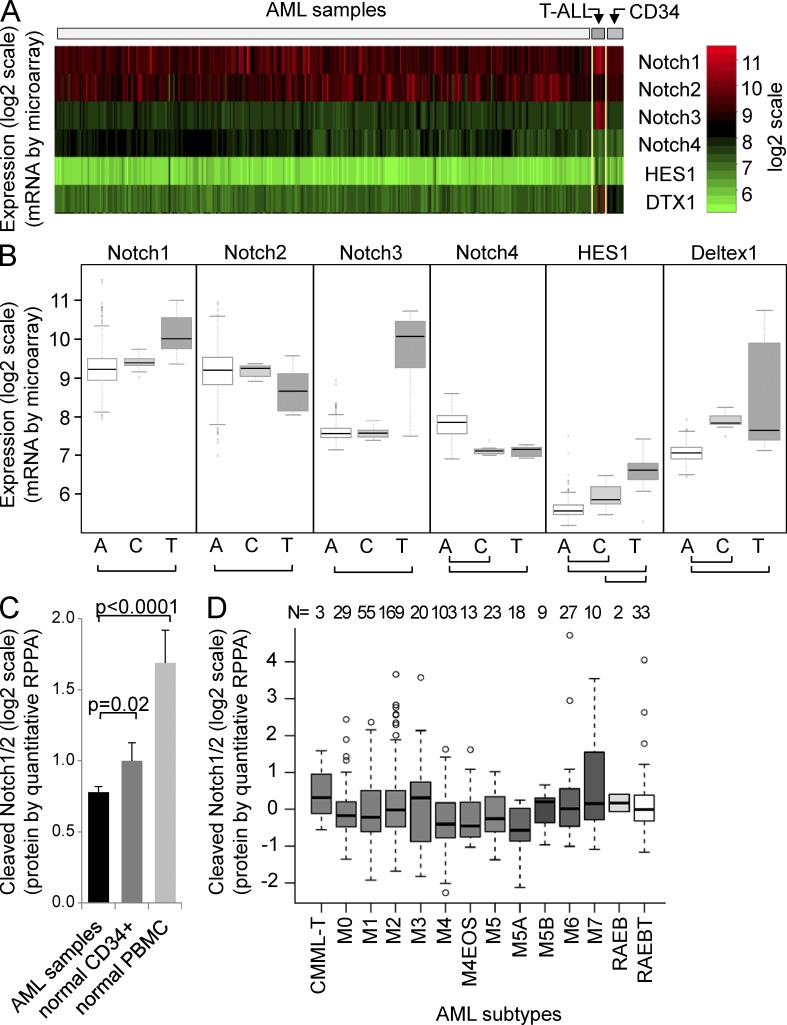
Given the limited studies in the literature as to the expression of Notch pathway members in AML, we first sought to describe the Notch pathway expression patterns in human AML. Using validated cDNA microarray datasets, we compared the mRNA expression of Notch receptors 1–4 and the Notch target genes HES1 and Deltex1 in 344 human AML patient samples, 11 normal CD34-selected bone marrow samples, and 8 human T-ALL patient samples (Figueroa et al., 2009; Verhaak et al., 2009). Overall, Notch1 and Notch2 were the predominant Notch receptors expressed, although Notch3 and 4 were detected at low levels in the majority of AML samples (Fig. 1 A). We observed a higher level of expression of the Notch2 and Notch4 receptors in AML versus T-ALL, and lower expression of the Notch1 and Notch3 receptor (all P < 0.001; Fig. 1 B). Interestingly, AML and normal CD34+ cells expressed similar levels of Notch receptors 1–3, but AML samples expressed significantly higher Notch4, the Notch receptor classically associated with vascular endothelium (Wu et al., 2005) and breast cancer (Harrison et al., 2010).
Figure 1.
Notch pathway expression in human AML patient samples. (A) Heatmap for Notch1–4, HES1, and Deltex1 (DTX1) from Affymetrix HG-U133 Plus 2 microarray data from 344 AML patient samples, 11 normal CD34-selected controls, and 8 T-ALL patient samples, normalized and shown as log2 scale (red, high; green, low). (B) Box plots from data in A. A, AML patient samples; C, CD34-selected controls; T, T-ALL patient samples. Student’s t tests reveal significant differences between AML and T-ALL samples in all four Notch receptors and two target genes (HES1 and Deltex1); bars indicate P < 0.001. A single dataset was analyzed. (C) Column chart showing cleaved Notch1/2 receptor antibody (Val1744; Cell Signaling Technology) data from a validated reverse phase protein lysate array (RPPA) with quantitative dilutions of 560 AML patient samples, 10 normal CD34-selected controls, and 21 normal peripheral blood mononuclear cell controls (mean ± SD). (D) Box plots from RPPA in C showing average and SD for FAB subtypes. Single dataset analyzed.
This difference in Notch receptor expression may be a consequence of myeloid development (Jönsson et al., 2001), and may suggest alternate Notch ligand selectivity (Hicks et al., 2000) or differential consequences of Notch1 and Notch2 signaling, as has been reported to have antagonistic effects in embryonal brain tumors (Fan et al., 2004). Importantly, the Notch target genes HES1 and Deltex1 (Fig. 1 B), as well as HES4 and HES5 (not depicted) were significantly lower in AML when compared with both normal CD34+ cells and T-ALL, suggesting that although Notch receptors are expressed, there is significantly lower constitutive Notch signaling in AML samples.
To determine whether there was protein-level evidence of Notch activation in patient samples, we used an antibody that is selective for the activated (cleaved) forms of either of the Notch1 or 2 receptors. We probed a validated reverse-phase protein lysate array (RPPA) containing duplicate dilutions of total cell lysates from 560 AML patient samples, 10 normal CD34-selected controls, and 21 normal peripheral blood mononuclear cell controls (Tibes et al., 2006; Kornblau and Coombes, 2011). We observed a lower level of detected cleaved (activated) Notch receptors in the AML samples compared with either the CD34+ or peripheral blood mononuclear cell controls (P < 0.02 and <0.0001, respectively; Fig. 1 C), again suggesting that Notch activation is at very low levels in AML cells when compared with normal hematopoietic cells, though the biological relevance of these differences is not yet known. Interestingly, levels of cleaved Notch receptor did not differ significantly between subtypes of AML (Fig. 1 D), although median cleaved Notch1/2 was highest in acute promyelocytic leukemia (FAB M3), consistent with a prior publication that found Notch signaling increased in AML cells expressing promyelocytic leukemia-retinoic acid receptor α (Alcalay et al., 2003).
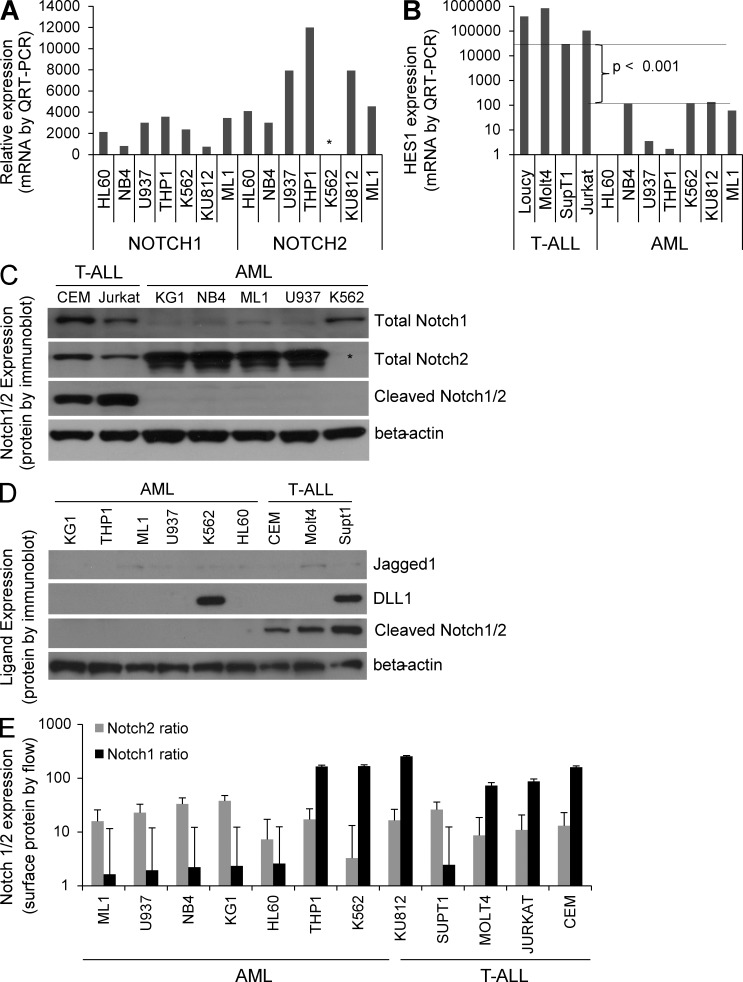
To complement this patient sample data, we assessed the relative mRNA expression levels of Notch1–4 and HES1 in a panel of cell lines representing AML and CML myeloid blast crisis (Table 1) by quantitative RT-PCR. Consistent with the array data, Notch2 and Notch1 were the predominant Notch receptors expressed in most AML cell lines (Fig. 2 A), in that order, and HES1 was expressed at significantly lower levels in AML than in T-ALL (Fig. 2 B).
Table 1.
Human AML cell lines
| Name | FAB | Description | Cytogenetics |
| HL60 | M2 | Progranulocytic AML without features of APML | der(5)t(15;17), but no RARa rearrangement |
| NB4 | M3 | Relapsed APML | t(15;17) PML-RARa |
| ML1 | M4 | Secondary AML with MLL rearrangement | t(6;11) MLL-AF6 |
| THP1 | M5 | Infant AML with MLL rearrangement | t(9;11) MLL |
| U937 | M5 | Monocyte-like histiocytic sarcoma with CD13/C15/CD33 expression | t(10;11), t(1;5) |
| KG1 | M6 | Erythroleukemia | |
| KU812 | CML-B | CML blast crisis, features of basophils | t(9;22) BCR-ABL1 |
| K562 | CML-B | CML blast crisis, undifferentiated granulocytic with erythroid features | t(9;22) BCR-ABL1 |
Figure 2.
Notch pathway expression in human AML lines. (A) mRNA expression of Notch1–4 by QRT-PCR. Asterisk denotes lack of Notch2 mRNA in K562 line. (B) mRNA expression of Notch target gene HES1 in T-ALL and AML lines by QRT-PCR. Log scale was used, and triplicate samples were analyzed. (C) Immunoblot of AML and T-ALL lines probed for Notch1, Notch2, cleaved Notch1/2, and β-actin. Asterisk denotes K562 line with undetectable Notch2 protein expression. Representative of three blots. (D) Immunoblot of AML and T-ALL lines probed for Notch ligands Jagged1 and DLL1, as well as cleaved Notch1/2 and β-actin. Representative of two blots. (E) Flow cytometry data for Notch1 and Notch2 cell surface expression on AML and T-ALL lines, normalized to isotype controls in each cell line (log scale; mean ± SD). Triplicate samples analyzed.
To determine whether the uncleaved (inactive) protein expression levels were consistent with cleaved Notch protein expression levels for cultured human AML cell lines, we performed a characterization of the Notch1/2 receptors in our panel of cell lines. Similar to the aforementioned AML patient sample data, we observed higher levels of Notch2 expression in most AML lines, and generally higher levels of Notch1 in T-ALL lines (Fig. 2 C). Interestingly, the CML myeloid blast crisis line K562 showed undetectable levels of Notch 2 by both quantitative RT-PCR and immunoblot studies. Given the potential role of Notch ligands in trans-Notch activation and/or cis-inhibition of Notch signaling, we measured Jagged1 and Δ-like ligand 1 (DLL1) protein levels in a panel of AML and T-ALL lines. Jagged1 was not easily detected but appeared to be expressed in two T-ALL lines (Fig. 2 D), whereas DLL1 was clearly expressed in one AML (K562) and one T-ALL line, although no correlation with activated (cleaved) Notch1/2 could be made (Fig. 2 D). This differs from the findings of Chiaramonte et al. (2005), which found higher levels of Jagged1 expression in AML, although their data also suggests a lack of Notch activation in AML. Importantly, the cleaved Notch1/2 antibody consistently detected activated Notch in T-ALL lines, but not in any of the AML lines (Fig. 2, B and C). This data confirms a distinct Notch receptor expression pattern in AML, and a lack of constitutive Notch activation in human AML lines.
As Notch receptor protein can be recycled away from the cell surface, thus making them unresponsive to neighboring cell Notch ligand expression, we also sought to determine the relative cell surface expression of Notch1/2 using Notch1 and Notch2 extracellular-specific antibodies in flow cytometry (Fig. 2 E). Generally consistent with the aforementioned protein and mRNA expression data, Notch2 was detected at higher levels on the surface of AML cells than T-ALL cells, with Notch1 having the opposite trend. Interestingly, both CML myeloid blast crisis cell lines expressed higher Notch1 and lower Notch2 cell surface protein, as did an mixed lineage leukemia (MLL)–translocated line, THP1, which was similar to the T-ALL pattern, with K562 having the least Notch1 surface expression, which is consistent with the mRNA and protein results in Fig. 2 (A and C). Notably, the T-ALL line SupT1, which carries the rare Notch1 translocation t(7;9)(q34;q34.3), expresses lower levels of cell surface Notch1 than the other T-ALL lines. This is not unexpected, as the translocation produces excess N-terminally truncated Notch1 transcripts from the involved allele, which contain no extracellular domain of Notch1, thus leaving the extracellular Notch1 expression to the single normal allele (Ellisen et al., 1991).
Although the presence of Notch1 expression in human AML cells has been previously reported (Tohda and Nara, 2001; Chiaramonte et al., 2005; Yin et al., 2009), the comparative expression of all four Notch receptors has not yet been explored. In this study, we report that Notch2 and Notch1 are the predominant Notch receptors in most human AML cells and that Notch3 and Notch4 are present at low levels. This contrasts with the predominant expression of Notch1 over Notch2, and striking Notch3 expression in T cell leukemias (Fig. 1, A and B; Bellavia et al., 2003; Saito et al., 2003). Furthermore, despite protein expression of Notch receptors similar to T-ALL lines, the Notch receptors in AML lines are inactive (uncleaved), and the levels of Notch target genes HES1, HES4, HES5, and Deltex1 are significantly lower than that found in normal CD34+ and T-ALL cells.
Incidentally, Notch1-activating mutations have been found in a very small subset of human AML samples (0.1%, 5 of 553 AML samples; Wouters et al., 2007). These mutations are associated with low/absent CEBPA and are found in T cell/myeloid biphenotypic leukemias. Interestingly, the ML1 AML cell line, which arose from a patient originally diagnosed with T-ALL, was shown to have a Notch1 mutation (Palomero et al., 2006). However, in agreement with Palomero et al. (2006), we found no evidence of constitutive cleavage of Notch receptors (Fig. 2, C and D) and very low levels of the Notch target gene HES1 (Fig. 2 B), suggesting that despite a Notch1 mutation, this line does not demonstrate constitutively active Notch signaling.
Notch signaling inhibits AML cell growth
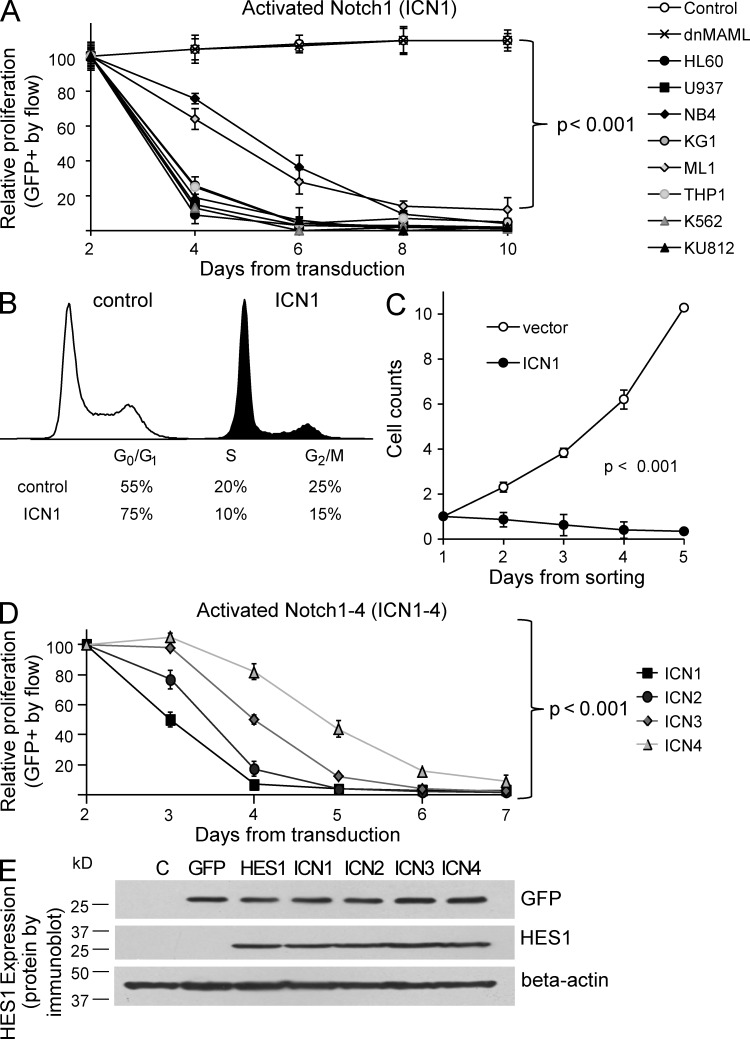
Because Notch signaling appears to be inactive in AML patient samples and lines, we sought to establish the effects of Notch signaling in these AML cells. To test activation of the Notch pathway, we used a construct that encodes the constitutively active, cleaved form of Notch1 (intracellular notch 1 [ICN1]), as well as GFP (Pui et al., 1999). To test inhibition of the Notch pathway, we used a dominant–negative form of the Notch cofactor Mastermind-like1 (dnMAML), which blocks canonical Notch signaling from all four of the Notch receptors (Weng et al., 2003). These constructs, or the GFP-only vector (MigR1), were used to transduce AML cells, as previously described (Zweidler-McKay et al., 2005). In an in vitro competitive proliferation assay, we compared the effect of each construct on the relative growth of AML cells. We found that expression of intracellular Notch1 inhibits the proliferation of all AML lines tested, revealed by a significant >5-fold decrease in the percentage of GFP+ cells over 4–8 d (P < 0.001; Fig. 3 A). In contrast, inhibition of Notch signaling via dnMAML had no effect on proliferation, with no change in GFP percentage over time when compared with control GFP-only vector cells. Representative (GFP) control and dnMAML curves from the HL60 line are shown (Fig. 3 A). These results suggest that constitutive Notch signaling in a panel of AML lines consistently leads to growth inhibition and/or cell loss, whereas pan-Notch inhibition via dnMAML has no effect on proliferation in vitro. In contrast, ICN1 expression had no effect on T-ALL cell line growth, whereas Notch inhibition via expression of dnMAML inhibited the growth of the MOLT4 and SUPT1 T-ALL lines, as expected (Weng et al., 2003, 2004).
Figure 3.
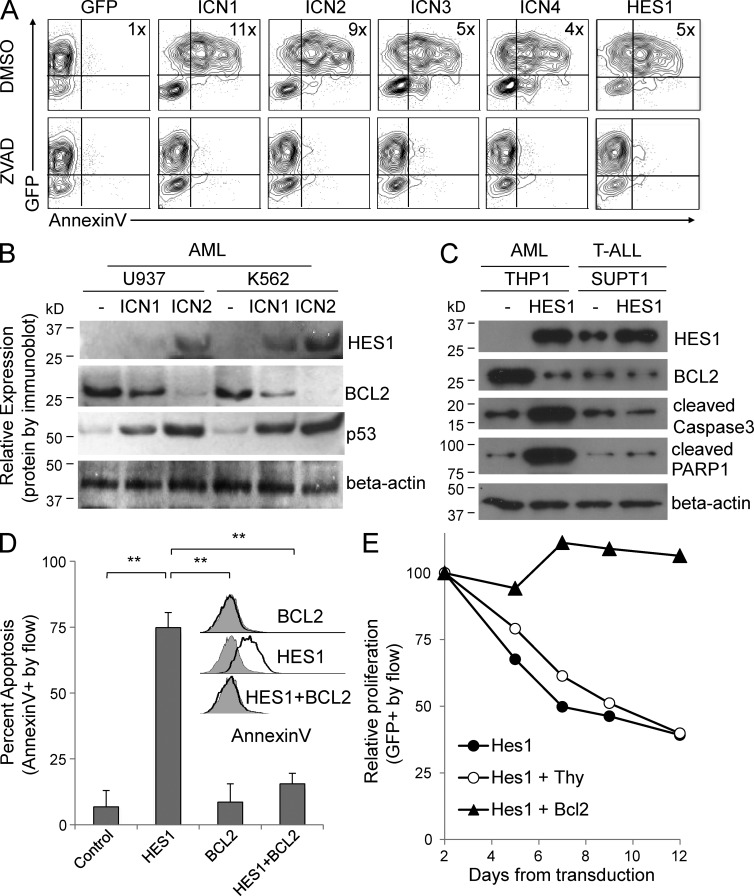
Activated Notch1–4 induce growth arrest and increase HES1 expression in AML cell lines. (A) Flow cytometry data showing changes in GFP when eight AML lines are transduced with retroviral vectors expressing either activated ICN1, the Notch inhibitor dnMAML, or GFP-only control (mean ± SD). Because of differences in transduction efficiency, all experiments were normalized to day 2 levels. Triplicate samples were analyzed. (B) Flow cytometry–based cell cycle analysis via the DNA dye DRAQ5 in U937 AML cells. DNA content of GFP+ (ICN1-transduced) cells are compared to GFP− (untransduced) cells 48 hours after transduction. FlowJo software was used to estimate the percentage of cells in the G0/G1, S, and G2/M phases of the cell cycle. Triplicate samples were analyzed, and similar results were obtained in K562 and HL60 lines. (C) Cell counts measured by hemocytometer. Results from replicate experiments with U937 cells were combined and normalized to day 1. ANOVA reveals significant differences across time points (P < 0.001). Triplicate samples analyzed and similar results were obtained in K562 and HL60 lines. (D) Flow cytometry data showing changes in GFP when U937 cells are transduced with constitutively active forms of Notch1–4, i.e., ICN1–4 (mean ± SD). Triplicate samples analyzed and similar results were obtained in K562 line. (E) Immunoblot of U937 AML cells transduced with GFP, HES1, and ICN1–4, and then probed for GFP, HES1, and β-actin. Representative of two blots.
Next, we evaluated the effect of ICN1 expression on the cell cycle and survival. After transduction, GFP+ (ICN1) cells were smaller than untransduced or vector-transduced controls (not depicted) and had a significant decrease in cells in the S and G2/M phases of the cell cycle and accumulation in the G0/G1 phase (Fig. 3 B), which is consistent with growth arrest. Cell counts of sorted ICN1-transduced and GFP-vector–transduced AML cells demonstrate an ICN1-mediated threefold decrease in cell number, in contrast to a >10-fold increase in cell number in control cells (Fig. 3 C). These results are consistent with the competitive proliferation assays in Fig. 3 A, and demonstrate Notch-induced growth arrest and cell death across a panel of human AML lines.
All four Notch homologues and HES1, a Notch downstream target, is sufficient to inhibit the growth of AML
As we demonstrated that Notch2 was more abundantly expressed in most AML cells, we determined the relative effect of all four activated Notch receptor homologues individually (ICN1–4). We found that each homologue inhibits the growth of AML cells, with differences in potency, i.e., ICN1 > ICN2 > ICN3 > ICN4 (Fig. 3 D). This demonstrates a consistent antiproliferative effect, independent of which activated receptor was expressed, suggesting that a common downstream target may be responsible for these results. The most commonly described Notch downstream target is the basic helix–loop–helix transcription factor Hairy/Enhancer of Split 1 (HES1). And indeed, ICN1–4 all significantly induced protein expression of endogenous HES1 (Fig. 3 E).
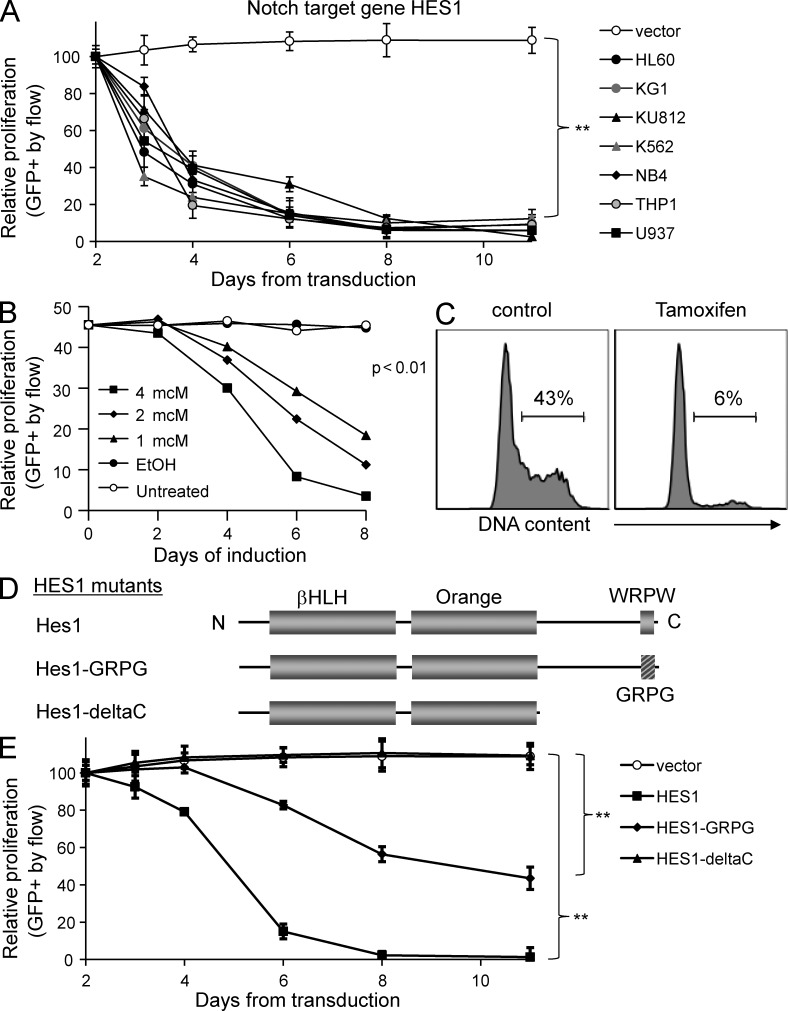
To determine if HES1 was sufficient to reproduce the Notch-mediated effects in AML, we expressed human HES1, compared with another well-described Notch target Deltex1. Similar to ICN1–4, HES1 was able to inhibit the growth of AML cells with a fivefold decrease in transduced cells, demonstrating that HES1 is sufficient to induce the growth-inhibiting effects of Notch activation (P < 0.001; Fig. 4 A). In contrast, neither Deltex1 (not depicted) nor the GFP control (MigR1) had any effect on the growth or survival of any of the AML lines. To confirm these results, we used a tamoxifen-inducible form of HES1 (ER-HES1; Zweidler-McKay et al., 2005) and found that induction of HES1 led to significant inhibition of proliferation in a dose-dependent manner (Fig. 4 B) associated with G0/G1 growth arrest (Fig. 4, B and C). These results demonstrate that the Notch target HES1 is a likely mechanism of Notch-mediated effects in AML.
Figure 4.
The Notch target gene HES1 induces growth arrest via its repressor domain. (A) Flow cytometry–based competitive proliferation assay showing changes in GFP after HES1 transduction in seven AML lines (mean ± SD). Triplicate samples were analyzed. (B) Flow cytometry–based assay showing changes in GFP in cells stably transduced with a tamoxifen-inducible ER-HES1 and exposed to 4-OH tamoxifen at the doses and the time durations indicated. Representative of two experiments. (C) Flow cytometry–based cell cycle histogram showing DNA content measured by propidium iodide (PI) in ER-HES1 cells exposed to 4 µM 4-OH tamoxifen for 48 h. Representative of three experiments. (D) HES1 mutants lacking either the conserved WRPW motif (HES1-GRPG) or the C-terminal repressor domain (HES1-deltaC) were constructed. (E) HES1 mutants were transduced into AML cells. A competitive proliferation assay reveals growth effects by wild-type HES1, the GRPG mutant, and the HES1-deltaC mutant (mean ± SD). **, P < 0.001. Triplicate samples analyzed and similar results were obtained in K562 and HL60 lines.
HES1 mutants suggest a role for co-repressors
HES1 is a transcriptional repressor whose function is enhanced by the recruitment of co-repressors, e.g. transducin-like enhancer of split (TLE). To test whether the repressor domain of HES1 is required for the Notch/HES1 effects on AML cells, we generated two C-terminal mutants of HES1 (Fig. 4 D). First, HES1-GRPG has two point mutations in the WRPW motif, known to be the binding site for TLE co-repressors and conserved in all HES family proteins. Second, HES1-ΔC truncates the HES1 protein after the conserved Orange domain, removing the entire repressor domain. We observed that the HES1-GRPG mutant had a moderate decrease in potency as compared with wild-type HES1 with a 40% reduction in GFP+ cells versus >90% reduction, respectively, 10 d after transduction (Fig. 4 E). More strikingly, the HES1-ΔC lost all ability to inhibit AML growth. These results suggest that the repressor function of HES1 and the association of TLE co-repressors are critical for HES1-mediated effects in AML.
Notch/HES1 induces apoptosis via down-regulation of B cell lymphoma 2 (BCL2)
To determine the ability of Notch signaling to induce apoptosis in AML lines, we transduced AML cells with ICN1–4 or HES1 and stained with Annexin V (Fig. 5 A). Consistent with Fig. 3 D, ICN1 had the highest percentage of Annexin V–positive cells among the Notch receptors, followed by ICN2 > ICN3 > ICN4, whereas HES1 had the highest percentage of Annexin V–positive staining in this assay. Importantly, incubation with the pan-caspase inhibitor ZVAD mitigated the Annexin V binding, suggesting that Notch/HES1 induce caspase-mediated apoptosis in AML (Fig. 5 A). Confirmatory experiments demonstrate enhanced caspase activity in ICN1-expressing AML cells (not depicted). To determine which growth and survival pathways were effected by Notch signaling in AML, we purified GFP-only and ICN1- and ICN2-transfected AML cells and found significant changes in two pathways, namely a decrease in BCL2 protein levels and an increase in p53 levels (Fig. 5 B). Next, we expressed HES1 in an AML line and a T-ALL line and found that BCL2 was again down-regulated coincident with up-regulation of the cleaved forms of caspase 3 and poly ADP-ribose polymerase 1 (PARP1), only in the AML line, indicating Notch/HES1-induced, caspase-mediated apoptosis in AML, but not T-ALL (Fig. 5 C). Interestingly HES1-mediated caspase activity could be prevented with co-incubation with a caspase 8 or 9 inhibitor (not depicted), suggesting proapoptotic crosstalk between the extrinsic and intrinsic pathways. To test whether BCL2 expression could rescue Notch-mediated apoptosis in AML cells, we expressed human BCL2 (coexpressing the marker Thy1, detected by anti–Thy1-APC), HES1-GFP, or the combination and measured Annexin V-PE binding in AML cells. Importantly, overexpression of BCL2 in AML lines was able to rescue the proapoptotic effects of HES1 expression (Fig. 5 D). Furthermore, expression of BCL2 in AML cells coexpressing HES1 rescued the growth-inhibiting effects of HES1 in a competitive proliferation assay (Fig. 5 E). Collectively, these data suggest that AML-specific down-regulation of BCL2 plays a critical role in Notch-mediated apoptosis in AML, although whether this is a direct or indirect HES1 effect is not yet clear.
Figure 5.
Notch activation induces caspase-mediated apoptosis with BCL2 down-regulation, and HES1-mediated growth inhibition in AML cells is rescued by BCL2 re-expression. (A) Flow cytometry–based contour plots of AML cells transduced with ICN1–4 or HES1 or GFP-only vector and treated with the pan-caspase inhibitor ZVAD, or DMSO vehicle control. Fold increase in Annexin V MFI is noted in each graph (4–11×). Representative of two experiments, and similar results were obtained in the K562 line. (B) Immunoblot of AML cells transduced with GFP vector, ICN1, or ICN2 probed for HES1, BCL2, p53, and β-actin. Representative of two blots. (C) Immunoblot of AML and T-ALL cells transduced with GFP vector or HES1 probed for HES1, BCL2, cleaved caspase 3, cleaved PARP1, and β-actin. Representative of two blots. (D) Flow cytometry–based graph of Annexin V binding percentage in AML cells expressing GFP vector, HES1, BCL2, or HES1 + BCL2 (mean ± SD). Inset shows flow cytometry–based histograms of Annexin V binding from same experiment. Triplicate samples were analyzed and similar results were obtained in an additional cell line. (E) Competitive proliferation assay showing changes in GFP percentage after transduction with HES1, HES1 + Thy vector control, and HES1 + BCL2. Representative of two experiments.
Inhibiting Notch signaling via dnMAML enhances AML proliferation in vivo, which appears to be linked to p53
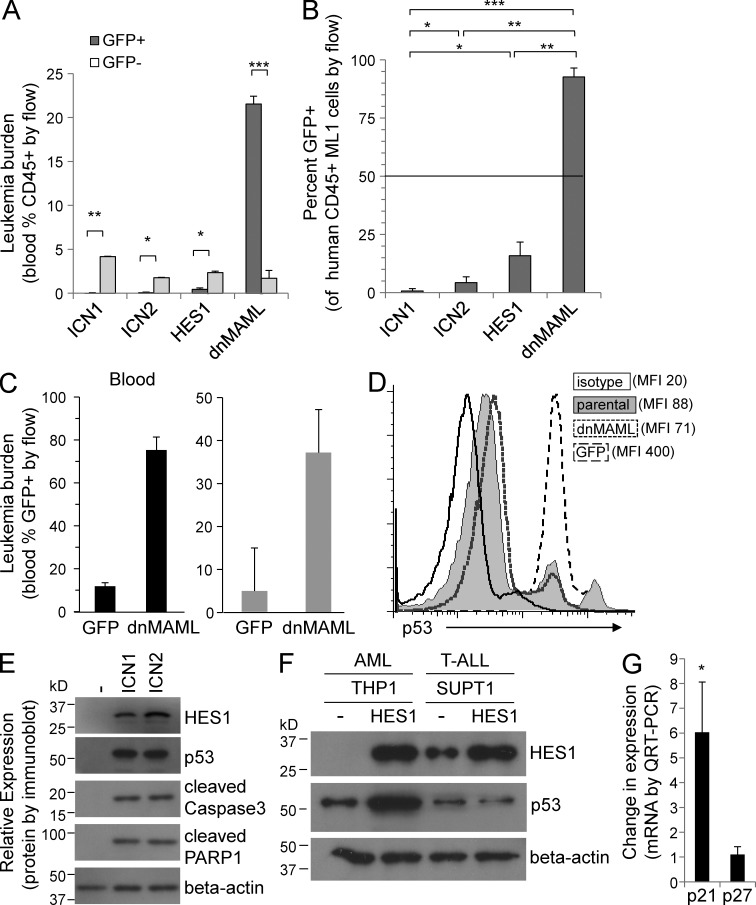
To determine whether Notch signaling would have growth-inhibiting effects in vivo, we transduced ML1 AML cells with ICN1, ICN2, HES1, or the pan-Notch inhibitor dnMAML and performed in vivo competitive proliferation assays. After transduction, each vector type was sorted to 50% GFP+ (containing the gene of interest) and GFP− (untransduced control cells). Groups of NOD-SCID-IL2Rg− (NSG) mice were injected with these 50:50 mixtures of ML1 cells, and peripheral blood levels of GFP− and GFP+ cells were measured with flow cytometry for anti–human CD45 and GFP. By 5 weeks, control peripheral blood engraftment of human CD45+ GFP− ML1 cells was similar in all mice (2–5%; Fig. 6 A, light gray bars). In contrast, as seen in in vitro assays (Fig. 3, A and D; and Fig. 4 A), ICN1, ICN2, and HES1 all led to decreased numbers of the percentage of GFP+ cells, with <1% peripheral blood engraftment in all three (Fig. 6 A, dark gray bars). Indeed, the percentage of GFP+ ICN1, ICN2, and HES1 cells decreased from the original sorted 50:50 to 50:1 (1%), 50:2 (4%), and 50:8 (16%), respectively (Fig. 6 B), indicating a significant growth inhibition in vivo, as seen in vitro. Importantly, dnMAML, which had little effect on proliferation in vitro (Fig. 3 A), led to dramatic increases in leukemia burden, with 23% peripheral blood engraftment (Fig. 6 A) compared with <5% in controls, and <1% for ICN1/2 and HES1. The ratio and percentage of GFP+ dnMAML cells increased from 50:50 to 4:50 (93%; Fig. 5 F), demonstrating a strong selective advantage for dnMAML-expressing ML1 in vivo. This suggests a previously unreported concept, namely that host-based, Notch ligand–mediated signaling inhibits AML growth in vivo. To confirm these findings, we transduced another AML line, THP1, with GFP only or dnMAML; sorted stably transduced cells to >99% purity; and injected equal numbers of GFP+ and dnMAML-GFP+ cells into separate cohorts of NSG mice. At 3 weeks, we detected ∼12% GFP+ THP1 cells in the GFP-only control mice (Fig. 6 C), which is consistent with prior engraftment experience. In contrast, mice injected with dnMAML-GFP+ THP1 cells had 75% peripheral blood engraftment (Fig. 6 C). The mice were sacrificed, and bone marrow leukemia burden revealed similar advantage for the dnMAML-transduced THP1 cells over the controls, 37 versus 5% (Fig. 6 C). These two engraftment models demonstrate that the pan-Notch inhibitor dnMAML enhances the growth of AML lines in vivo, suggesting the novel hypothesis that microenvironment-based Notch activation inhibits AML growth. As we found that p53 was up-regulated by Notch signaling in vitro (Fig. 5 B), we measured p53 levels by intracellular flow cytometry on peripheral blood in these THP1-engrafted mice. Indeed, p53 staining was significantly increased in control GFP-only THP1 cells in vivo, compared with parental THP1 cells growing in vitro, mean fluorescence intensity (MFI; MFI, 400 versus 88; Fig. 6 D), suggesting that an endogenous factor induced p53 in AML cells when grown in vivo. Strikingly, dnMAML-expressing THP1 cells had >5-fold lower p53 expression than GFP-only THP1 cells in vivo (MFI 71 versus 400), reaching the low levels seen in control in vitro parental THP1 cells (MFI 71 versus 88; Fig. 6 D). These data suggest that in vivo activation of Notch signaling may inhibit AML growth, in part through induction of p53-mediated growth arrest.
Figure 6.
Notch inhibitor dnMAML enhances in vivo engraftment and increases p53/p21 levels. (A) In vivo competitive proliferation/engraftment assay: all transduced ML1 cells were sorted to 50:50 GFP+:GFP− prior to injection. Graph shows percent peripheral blood engraftment with GFP− (control) and GFP+ (transduced) cells. Mean ± SD. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. (B) Graph shows percentage of human CD45+ ML1 which express GFP (mean ± SD). Dashed 50% line shows starting 50:50 mixture for all groups revealing disadvantage for ICN1, ICN2, and HES1 and significant advantage for dnMAML. Mean ± SD. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. Five mice per cohort. (C) In vivo proliferation/engraftment assay: all transduced THP1 cells were sorted to >99% GFP+ prior to injection. Graph shows percent peripheral blood engraftment with GFP+ THP1 cells in peripheral blood and bone marrow (mean ± SD). (*, P < 0.01; **, P < 0.001). Five mice per cohort. (D) Flow cytometry–based histogram shows levels of intracellular p53 staining in peripheral blood GFP+ cells for GFP-vector control and dnMAML mice. MFI indicated by each layered histogram, revealing high p53 levels in control GFP cells, and lower p53 levels in dnMAML-expressing cells. Representative of three mice. (E) Immunoblot of AML cells transduced with GFP, ICN1, or ICN2 and probed for HES1, p53, cleaved caspase 3, cleaved PARP1, and β-actin. Representative of two blots. (F) Immunoblot of AML and T-ALL cells transduced with GFP or HES1 and probed for HES1, p53, and β-actin. Representative of two blots. (G) Graph showing change in mRNA levels of p21 and p27 by QRT-PCR comparing GFP and HES1-transduced AML cells. *, P < 0.01. Triplicate samples analyzed.
To further examine the ability of Notch signaling to induce p53 expression, we transduced and sorted AML cells in vitro and found significantly increased levels of p53 protein, along with increased cleaved caspase3 and PARP1 (Fig. 6 E). Furthermore, when the immunoblot from Fig. 5 C was probed for p53, a significant increase in p53 was seen in the AML but not in the T-ALL line in response to HES1 expression (Fig. 6 F). Finally, HES1 induced a sixfold increase in p21 mRNA expression (Fig. 6 G), suggesting that Notch/HES1 induce AML-specific, p53/p21-mediated growth inhibition, though the mechanism remains to be determined.
Notch ligand co-culture confirms critical roles for HES1, BCL2, and p53 in Notch-mediated apoptosis
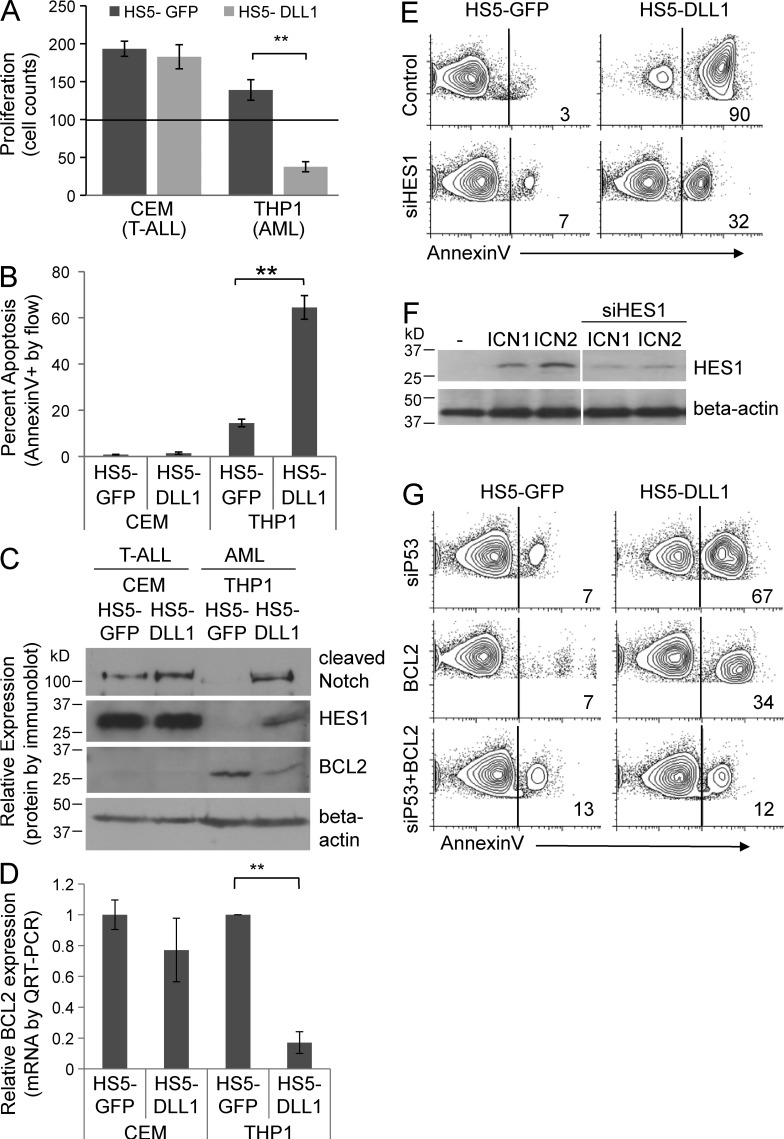
As our overexpression approaches do not depend on the endogenous Notch receptors present on AML cells and may not produce physiological levels of Notch signaling, we established a Notch ligand co-culture assay using the human stromal line HS5 (CRL-11882; American Type Culture Collection) expressing human Notch ligand DLL1. Co-culture of AML cells on control HS5-GFP cells led to a 40% increase in leukemia cell counts over 72 h (Fig. 7 A). However, co-culture on HS5-DLL1 cells led to a decrease of >60% in cell number in the same time period, demonstrating that exposure to the Notch ligand DLL1 inhibits the growth and survival of AML cells. When co-cultured AML cells were stained with Annexin V, HS5-DLL1 cells induced >60% Annexin V positivity (Fig. 7 B), suggesting a strong induction of apoptosis in response to DLL1. On immunoblot, co-culture with HS5-DLL1 cells induced cleaved Notch and HES1 (Fig. 7 C), demonstrating the activation of Notch signaling in this co-culture. Furthermore, BCL2 levels were decreased in AML cells co-cultured with HS5-DLL1 cells (Fig. 7 C), confirming BCL2 down-regulation as a potential mechanism for the induction of apoptosis. Through quantitative RT-PCR, we found BCL2 mRNA transcripts were decreased after exposure to HS5-DLL1 cells in AML cells (Fig. 7 D), but not T-ALL cells, further supporting an AML-specific, Notch-mediated proapoptotic effect.
Figure 7.
Notch ligand DLL1 co-culture inhibits AML growth, induces apoptosis, down-regulates BCL2, and reveals the critical roles of HES1, BCL2, and p53 in Notch-mediated apoptosis in AML. (A) Cell counts after 72 h of co-culture on human stromal line HS5 transduced with GFP-only or Notch ligand DLL1 GFP. 100,000 AML or T-ALL cells were placed over confluent HS5-GFP or HS5-DLL1 cells. (**, P < 0.001). Triplicate samples analyzed. (B) Flow cytometry–based apoptosis assay in AML and T-ALL cells stained with Annexin V after co-culture with HS5-GFP or HS5-DLL1 cells for 24 h. **, P < 0.001. Triplicate samples were analyzed. (C) Immunoblot of AML and T-ALL cells after co-culture with HS5-GFP or HS5-DLL1 cells for 48 h. Representative of two blots. (D) mRNA expression of BCL2 in AML and T-ALL co-cultured with HS5-GFP or HS5-DLL1 cells for 24 h. **, P < 0.001. Triplicate samples analyzed. (E) Flow cytometry–based apoptosis assay of AML cells transfected with siRNA to HES1 or control, and then co-cultured on HS5-GFP or HS5-DLL1 for 48 h. Representative of two experiments. (F) Immunoblot showing effect of transfection with siRNA to HES1 on level of HES1 protein induced by transduction with ICN1. Representative of two blots. (G) Flow cytometry–based apoptosis assay of AML cells transfected with BCL2, siP53, or BCL2 + siP53 was then co-cultured on HS5-GFP or HS5-DLL1 for 48 h. Representative of two experiments.
To determine whether HES1 was necessary for the Notch-mediated apoptosis in AML, we transfected AML cells with HES1 siRNA before co-culture with either HS5-GFP or HS5-DLL1 cells, and then measured Annexin V by flow cytometry. Consistent with Fig. 7 B, co-culture with HS5-DLL1 significantly increased Annexin V binding (Fig. 7 E). Transfection with siRNA to HES1 lowered the percentage of Annexin V–positive cells from 90 to 32% (Fig. 7 E). Importantly, siRNA transfection of these cells did not completely inhibit ICN1-mediated induction of HES1, but led to a ∼50% decrease in expression (Fig. 7 F). Thus, the significant decrease in Annexin V binding after partial knockdown of HES1 via siHES1, suggests that HES1 plays an important role in DLL1/Notch-mediated apoptosis. To determine whether BCL2 expression could rescue DLL1/Notch-induced apoptosis in this co-culture approach, we expressed human BCL2 as in Fig. 5 (D and E). Here, BCL2 expression decreased Annexin V binding from 90 to 34%, a similar magnitude to the siHES1, suggesting an important role for BCL2 in this proapoptotic mechanism. Finally, we transfected AML cells with siRNA to p53, and found a modest effect, with Annexin V positivity decreasing from 90 to 67% (Fig. 7 G), which was not unexpected as p53 likely has more profound effects on cell cycle than on apoptosis. Interestingly, the combination of BCL2 expression and siP53 prevented the majority of apoptosis with Annexin V binding, decreasing from 90 to 12% (Fig. 7 G). The data suggest that ligand-based Notch activation effectively down-regulates BCL2 mRNA/protein in AML cells, and HES1, BCL2, and p53 all contribute to Notch-mediated apoptosis.
Notch ligand peptide induces Notch signaling in AML
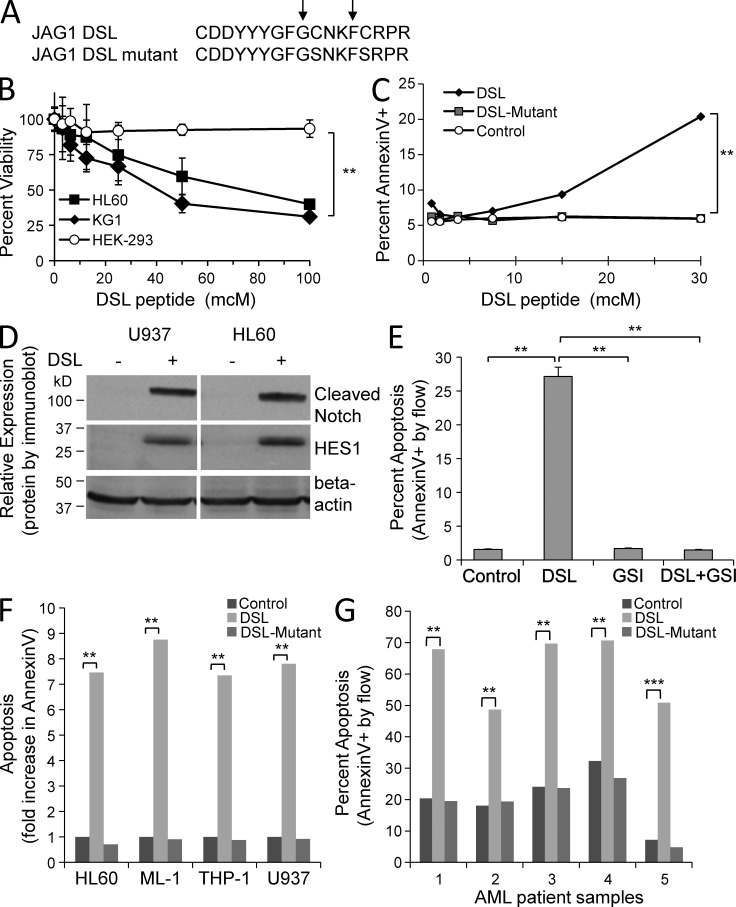
Given our findings of a potent Notch-induced growth arrest and apoptosis in a panel of human AML lines, we wondered whether forced activation of Notch signaling could be used as a therapeutic approach for AML. To induce Notch signaling directly, via a clinically feasible method, we chose to use a 17-aa peptide with Notch agonist activity, based on the highly conserved Jagged1 delta/Serrate/Lag-2 (DSL) domain (Fig. 8 A; Li et al., 1998). Exposure to varying concentrations led to a dose-dependent decrease in viability in two AML lines (HL60, KG1), but had no effect on the nonhematopoietic line HEK-293 (Fig. 8 B). Interestingly, this peptide contains two internal cysteines that are predicted to be important for proper folding and binding to the epidermal growth factor repeats of the Notch receptors (Cordle et al., 2008). Therefore, we generated a DSL mutant peptide in which both cysteines were mutated to serine, as these two amino acids are structurally nearly identical, with a substitution of a hydroxyl for a sulfhydryl group (Fig. 8 A). Treatment of U937 cells with DSL peptide resulted in a dose-dependent increase in apoptosis (Annexin V+), whereas untreated and DSL mutant–treated cells had no change in apoptosis, suggesting specificity for the intact DSL peptide (Fig. 8 C). To confirm DSL-mediated Notch signaling, we evaluated the effect of DSL exposure on Notch receptor cleavage and HES1 expression. We observed a dramatic increase in cleaved Notch and HES1 expression after 6 h (Fig. 8 D), as well as a significant 18-fold increase in mRNA by quantitative RT-PCR (not depicted). To determine whether this DSL effect was dependent on Notch receptor activation, we tested the ability of a γ-secretase inhibitor (GSI) to block DSL-induced apoptosis (Fig. 8 E). Indeed, pretreatment with the GSI abolished the proapoptotic effect of the DSL peptide, demonstrating that this DSL peptide induces apoptosis through a Notch agonist effect (P < 0.001). In a panel of AML lines, we found consistent increases in Annexin V positivity with exposure to the DSL peptide, but not the DSL mutant peptide (Fig. 8 F). We next tested the DSL peptide on a panel of human primary AML patient samples (Table 2). When these AML patient samples were cultured with DSL peptide for 24 h, the percentage of Annexin V+ cells reached 49–71%, compared with control and DSL mutant-treated cells at 8–32% (P < 0.001; Fig. 8 G). Similar results were seen with a DSL peptide derived from the corresponding region of another Notch ligand, i.e., DLL1 (not depicted). These data demonstrate that Notch ligand mimetic peptides may be used to induce Notch signaling and apoptosis in AML and warrant further development.
Figure 8.
Notch agonist peptide induces apoptosis in AML cell lines and patient samples. (A) A 17-aa peptide derived from the human Jagged1 DSL domain is shown (DSL). A mutant with two cysteines replaced by serines (DSL mutant) is also shown. (B) Dose−response curves after 48-h exposure to DSL peptide for AML lines (HL60, KG1) and nonhematopoietic line HEK-293. alamarBlue staining was used to measure percent viability of cells after exposure to peptide. Mean ± SD, ANOVA; **, P < 0.001. Triplicate samples were analyzed and are representative of two experiments. (C) Dose response curves after 48 h exposure to DSL and DSL mutant peptides for U937 cells, with percentage of apoptosis measured by Annexin V binding. Mean ± SD, ANOVA **, P < 0.001. Triplicate samples were analyzed and are representative of two experiments. (D) Immunoblot showing induction of cleaved Notch receptors and HES1 expression in AML lines after 24 h of exposure to the DSL peptide. Representative of two blots. (E) The effect of GSI on DSL-mediated apoptosis (**, P < 0.001). Triplicate samples analyzed. (F) Human AML lines treated for 48 h with DSL peptide, DSL mutant peptide, or vehicle control. (**, P < 0.001). Triplicate samples were analyzed. (G) Primary human AML patient samples (Table 2) treated for 48 h with DSL peptide, DSL mutant peptide, or vehicle control. Mean ± SD; **, P < 0.001; ***, P < 0.0001. Triplicate samples were analyzed.
Table 2.
AML patient samples
| Patient | FAB | Description | Specimen Type | % blasts | Collection date |
| 1 | M3 | 15-yr-old APML with t(15;17) | BMA | 73% | 11-24-05 |
| 2 | M5A | 6-yr-old relapsed monoblastic AML without differentiation | BMA | 76% | 5-10-05 |
| 3 | M4 | 14-yr-old myelomonocytic AML with t(9;11) | CD34+ BMA | 97% | 12-7-07 |
| 4 | M1 | 12-yr-old relapsed undifferentiated AML | leukapheresis | 94% | 7-15-04 |
| 5 | M4 | 16-yr-old myelomonocytic AML | BMA | 68% | 8-2-05 |
AML, acute myelogenous leukemia; APML, acute promyelocytic leukemia; BMA, bone marrow aspirate.
DISCUSSION
To address the contrasting findings on the role of Notch in AML, we undertook a comprehensive characterization of the expression of Notch pathway genes, the effects of Notch receptors, target genes, in vivo engraftment, ligand co-culture, and ligand mimetics on AML growth and survival, and downstream consequences of Notch activation in AML. We found a generalized lack of constitutive Notch signaling in AML patient samples and cell lines, yet AML cells express at least one Notch receptor homologue and consistently demonstrate growth inhibition and apoptosis after Notch activation at multiple levels. This mechanism appears to be dependent on the HES1 repressor domain and co-repressors, which directly or indirectly lead to BCL2 down-regulation and increased expression of p53/p21. Consistent with this finding, Notch-mediated growth arrest was reported in murine hematopoietic progenitor cell lines and was found to be p53/p21-dependent (Henning et al., 2008). Furthermore, p53 has been shown to repress BCL2 at the transcriptional level though binding the proximal BCL2 promoter region in hematopoietic cells (Wu et al., 2001). Although we do not yet know if this specific mechanism is occurring in our system, the findings are consistent with our observations.
Perhaps the most intriguing of our results was the selective advantage given to AML cells that expressed the pan-Notch inhibitor dnMAML in two AML xenografts in vivo, but not in vitro. This result suggests that host-based endogenous Notch ligands are capable of inhibiting AML growth via induction of Notch signaling, providing evidence for a microenvironment-based, leukemia-inhibiting effect for Notch in AML. Although hematopoietic studies using dnMAML have not reported myeloid abnormalities (Maillard et al., 2008), inhibition of Notch ligands through defective fucosylation (Zhou et al., 2008) or impaired endocytosis (Kim et al., 2008), or inhibition of Notch receptor processing in hematopoietic progenitors (Yoda et al., 2011) led to myeloproliferative disease in murine models. These studies imply that Notch signaling can inhibit normal myeloid proliferation in vivo, which together with our data support our conclusion that Notch signaling inhibits AML proliferation.
A leukemia-inhibiting role for Notch in AML would predict that Notch pathway alterations might be found, at least in a subset of AML cases; indeed, Notch pathway deletions, mutations, and a translocation have been found. A recent study found that deletion of MAML1 at 5q35.3 to be frequently lost in interstitial deletions of 5q (Jerez et al., 2012). Furthermore, this group found point mutations of MAML1 in three patients with aggressive 5q-AML (Jerez et al., 2012). Mutations have also been found in NOTCH2, MAML1, Nicastrin, and APH1A in patients with chronic myelomonoblastic leukemia (CMML), a myeloproliferative leukemia (Klinakis et al., 2011). Furthermore, Nemoto et al. (2007) reported a Notch-inhibiting MLL–MAML2 translocation in a case of secondary AML. These Notch-inhibiting alterations support our conclusion that Notch signaling inhibits AML growth and survival and that a subset of AML cells delete, mutate, or acquire Notch-inhibiting fusions, which may contribute to myelodysplastic syndrome, AML, and/or MPD/CMML.
Controversy will remain, as we do not understand the complexity of the Notch pathway and tools to specifically modulate the Notch pathway are still limited. Further studies assessing the levels of Notch activation and inhibition in situ still need to be carried out. However, we believe our data represents the first comprehensive evaluation of the Notch pathway in human AML, which reveal a novel leukemia-inhibiting role for Notch/HES1 signaling, the concept of host-based Notch ligand–mediated inhibition of AML, and offers the rationale and method for a Notch-agonist approach for AML therapy.
MATERIALS AND METHODS
Gene expression array.
Affymetrix HG-U133 Plus 2 microarray data from 344 primary human AML samples, 11 normal CD34-selected samples, and 8 T-ALL patient samples, as described previously (deposited as GEO datasets GSE6891 and GSE14479; Figueroa et al., 2009; Verhaak et al., 2009). CEL files were downloaded from the GEO repository. Robust multiarray normalization was performed across all samples using the Affy package from Bioconductor (Gentleman et al., 2004). The Student’s t test was used to compare groups of sample data points.
Cell culture.
Human AML cell lines HL-60, NB4, ML1, THP1, and KG1; the myeloid histiocytic sarcoma line U937; and the CML myeloid blast crisis lines KU812 and K562, were maintained in C10 media, as described previously (J. Chandra, University of Texas M.D. Anderson Cancer Center, Houston, TX; Zweidler-McKay et al., 2005). Similarly T-ALL lines Molt4, Jurkat, Loucy, and CEM were maintained in C10 media (RPMI-1640 [Invitrogen], plus 10% heat-inactivated FCS (HyClone), 1 mM Hepes, 1 mM glutamine, 1 mM Na pyruvate, and 1× nonessential amino acids [Invitrogen]). HL-60, U937, KG1, KU812, K562, Loucy, CEM, and HS-5 lines were obtained directly from the American Type Culture Collection. The remainder of the lines were confirmed to be human myeloid or T phenotype with CD34/CD33/CD14 or CD3/CD4/CD8 flow cytometry. AML patient samples were acquired from the MDACC Leukemia Bank, with approval from the M.D. Anderson Cancer Center Internal Review Board. AML samples were thawed, grown in StemSpan SFEM (serum-free expansion media), and supplemented with human recombinant cytokines (Flt3L, SCF, IL-3, and IL-6; 1× CC100; STEMCELL Technologies).
Plasmids and retroviral transduction.
Human cDNAs for genes encoding the intracellular domains of human Notch1–4 (ICN1–4), full-length HES1, dnMAML, and full-length Deltex1 were cloned into a MigR1 (MSCV-based) retroviral vector (W. Pear, University of Pennsylvania, Philadelphia, PA; J. Aster, Brigham and Women’s Hospital, Boston, MA; Pui et al., 1999; Zweidler-McKay et al., 2005). This bicistronic vector co-expresses GFP after an internal ribosome entry site (IRES), providing a surrogate marker for gene-of-interest expression. The production of retroviral supernatant and transduction of each cell line was performed following previously established methods (Pear et al., 1993). For transduction, 0.1–2 × 106 cells are plated in 24-well tissue culture plate with 250–500 μl of retroviral supernatant and 4–8 μg/ml of polybrene (Sigma-Aldrich). After centrifugation at 1,000 g for 90 min, the cells are incubated at 37°C in 5% CO2 for 3–6 h before addition of fresh C10 media. At 2 d and thereafter, GFP expression was measured by flow cytometry (FL1 on a FACSCalibur; BD). Constructs that express mutated or truncated forms of human HES1 were made as follows: HES1-GRPG is identical to wild-type HES1 except for site-directed mutagenesis of the WRPW to GRPG, destroying the conserved TLE co-repressor–binding motif. HES1-deltaC retains the N terminus, basic helix–loop–helix and the ORANGE domain, but truncates the remaining C terminus of the protein, removing the putative CtBP co-repressor binding region and the WRPW motif. The human BCL2-expressing vector (MSCV-hBCL2 IRES Thy1) expressing a truncated Thy1 antigen via an IRES, which was detected with an anti–Thy1-APC antibody (H. Leighton Grimes, Cincinnati Children’s Hospital, Cincinnati, OH).
In vivo competitive proliferation assays.
ML1 and THP1 AML cells were used for these in vivo experiments because of their reproducible engraftment and progression in the NOD-SCID-IL2Rγ (NSG; stock 005557; Jackson ImmunoResearch Laboratories) mouse model and their permissiveness to retroviral transduction. Initially, ML1 cells were transduced with ICN1, ICN2, HES1, and dnMAML virus as above. After 48 h, each transduction experiment was sorted to 50:50 mixture of GFP+ (expressing the gene of interest) and GFP− (control untransduced) cells. 10,000 cells of these 50:50 mixtures were injected by tail vein into unirradiated NSG mice. Weekly tail vein bleeds and dual-color flow cytometry for human CD45 and GFP revealed >1% engraftment in all mice by 5 wk after injection. The GFP+ percentages of human CD45+ ML1 cells were used to determine relative in vivo engraftment/proliferation. Similarly, relative amounts of CD45+/GFP− control cells and GFP+/CD45+ transduced cells were compared with murine peripheral blood cells. Similarly, THP1 cells were transduced with MigR1 (GFP-only) and dnMAML virus and sorted to >99% purity at 48 h after transduction. NSG mice were injected with 50,000 cells by tail vein and peripheral blood was monitored for engraftment via flow cytometry for GFP. All animal experiments were performed with prior approval from the M.D. Anderson Cancer Center Institutional Animal Care and Use Committee in compliance with the US Department of Health and Human Services Guide for the Care and Use of Laboratory Animals.
Cell cycle and apoptosis.
Transduced cells were permeabilized and stained in 0.1% Triton X-100 and 50 µg/ml propidium iodide in PBS for >30 min. Alternatively, DRAQ5 (Enzo Life Sciences) was added to cells to a final concentration of 5 µM for >5 min. Cell cycle data were collected by flow cytometer (FL2 on a FACSCalibur; BD) measurement of DNA content, cells with <2N DNA content (subdiploid) were considered undergoing cell death. Annexin V binding was measured by flow cytometry after incubation with Annexin V conjugated to PE or APC (BD). Dependence of Notch/HES1-mediated cell death on caspase activity was tested via incubation with the pan-caspase inhibitor ZVAD. Pan-caspase activity was measured after incubation with SR-FLICA reagent (Immunochemistry Technologies). Caspase-3 activity was measured using a fluorogenic caspase-3 substrate (DEVD-AMC), as described previously (Miller et al., 2007). In brief, after induction of HES1, cells were lysed and incubated with 50 µM DEVD-AMC. AMC fluorescence was measured by spectrofluorometer (excitation 355 nm, emission 460 nm). Fluorescence generated by the cleavage of this fluorogenic peptide is a measure of caspase-3 activity.
RNA extraction and RT-PCR.
Total RNA was prepared using an RNeasy kit (QIAGEN) and primed with random hexamers to synthesize complementary DNA using AMV reverse transcription (GE Healthcare) according to the manufacturer’s instructions. TaqMan PCR primers and probes specific for human Notch pathway genes and controls β-actin/GAPDH were purchased commercially and used per the manufacturer’s instructions (Applied Biosystems). The quantitative PCR reactions were performed on an iCycler (Bio-Rad Laboratories). All values were normalized to a control gene using 2Δ-Δ Ct method.
Western SDS-PAGE and RPPA.
Whole-cell lysates were prepared according to established methods using Triton X-100 or RIPA lysis buffer. Bradford assay was used to determine protein concentration. Antibodies directed against Beta-Actin (Sigma-Aldrich), Notch1 (sc-6014; Santa Cruz Biotechnology, Inc.; bTAN20; Hybridoma Bank at the University of Iowa), Notch2 (Hybridoma Bank at the University of Iowa, C651.6DBHN), cleaved Notch1/2 (Val1744; Cell Signaling Technology), HES1 (gift from T. Sudo, Toray Industries, Tokyo, Japan) and HES1 (ab71559; Abcam), Jagged1 (sc11376; Santa Cruz Biotechnology, Inc.), DLL1 (sc73899; Santa Cruz Biotechnology, Inc.), p53 (9282; Cell Signaling Technology), BCL2 (2876; Cell Signaling Technology), cleaved caspase 3 (9661L; Cell Signaling Technology), cleaved PARP1 (9544S; Cell Signaling Technology), GFP (ab290; Abcam).
Co-culture.
HS5 cells were transduced with MigR1 (GFP) control vector or DLL1, sorted to purity, and plated in 6-well tissue culture plates. AML and CEM (T-ALL) cells were counted and 100,000 cells were plated onto GFP or DLL1-expressing HS5 cells. One day after AML cells were plated, cells were collected and stained with Annexin5 using standard protocol techniques. RNA was also isolated and was reverse transcribed using standard techniques. QRT-PCR was performed using Hes1, p53, Bcl2, and GAPDH TaqMan probes from Applied Biosystems. 2 d after co-incubation, cells were collected; whole-cell lysates were made and run on SDS-PAGE as indicated above. 3 d after cells were co-cultured with GFP or DLL1 HS5 cells were counted by trypan blue staining.
Statistical analysis.
Student’s t test or multiple measures ANOVA was used to assess statistical significance. All experiments were performed at least in triplicate unless otherwise noted.
Acknowledgments
The authors would like to thank Drs. Warren Pear (University of Pennsylvania), Jon Aster (Brigham and Women’s Hospital), Leighton Grimes (Cincinnati Children’s Hospital), and T. Sudo (Toray Industries) for their kind gifts of constructs and HES1 antibody, respectively. We also thank Dr. Joya Chandr for her kind gifts of cells lines. We would also like to acknowledge the support of the M.D. Anderson Pediatric Hematology/Oncology Fellowship Program and the Division of Pediatrics.
This research was partially supported by the M.D. Anderson Physician-Scientist Program.
The authors have no financial conflicts.
Author contributions: S. Kannan performed research and analyzed data; R.M. Sutphin designed research, performed research, and contributed to writing the manuscript; M. Hall performed research, analyzed data, and contributed to editing of the manuscript; L. Golfman performed research; Wendy Fang, performed research; R. Nolo performed research; L.J. Akers performed research; R. Hammitt and J. McMurray contributed vital new reagents; A. Melnick and M. Figueroa contributed data and did analyses; P.A. Zweidler-McKay designed research, performed research, analyzed data, and wrote the manuscript.
Footnotes
Abbreviations used:
- AML
- acute myelogenous leukemia
- BCL2
- B cell lymphoma 2
- BCR-ABL
- breakpoint cluster region-Abelson
- CML
- chronic myelogenous leukemia
- DLL
- delta-like ligand
- dnMAML
- dominant–negative MasterMind-like
- DSL
- delta/Serrate/Lag-2
- GSI
- γ secretase inhibitor
- HES
- Hairy/Enhancer of Split
- ICN
- intracellular notch
- MFI
- mean fluorescence intensity
- MLL
- mixed lineage leukemia
- PARP1
- poly ADP-ribose polymerase 1
- SCID
- severe combined immunodeficiency
- T-ALL
- T cell acute lymphoblastic leukemia
- TLE
- transducin-like enhancer of split
References
- Alcalay M., Meani N., Gelmetti V., Fantozzi A., Fagioli M., Orleth A., Riganelli D., Sebastiani C., Cappelli E., Casciari C., et al. 2003. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Invest. 112:1751–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776 10.1126/science.284.5415.770 [DOI] [PubMed] [Google Scholar]
- Aster J.C., Pear W.S., Blacklow S.C. 2008. Notch signaling in leukemia. Annu. Rev. Pathol. 3:587–613 10.1146/annurev.pathmechdis.3.121806.154300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi A., De Falco M., De Luca L., Cottone G., Paggi M.G., Nickoloff B.J., Miele L., De Luca A. 2004. Characterization of tissue specific expression of Notch-1 in human tissues. Biol. Cell. 96:303–311 [DOI] [PubMed] [Google Scholar]
- Bellavia D., Campese A.F., Vacca A., Gulino A., Screpanti I. 2003. Notch3, another Notch in T cell development. Semin. Immunol. 15:107–112 10.1016/S1044-5323(03)00007-1 [DOI] [PubMed] [Google Scholar]
- Carlesso N., Aster J.C., Sklar J., Scadden D.T. 1999. Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood. 93:838–848 [PubMed] [Google Scholar]
- Chadwick N., Nostro M.C., Baron M., Mottram R., Brady G., Buckle A.M. 2007. Notch signaling induces apoptosis in primary human CD34+ hematopoietic progenitor cells. Stem Cells. 25:203–210 10.1634/stemcells.2005-0303 [DOI] [PubMed] [Google Scholar]
- Chadwick N., Fennessy C., Nostro M.C., Baron M., Brady G., Buckle A.M. 2008. Notch induces cell cycle arrest and apoptosis in human erythroleukaemic TF-1 cells. Blood Cells Mol. Dis. 41:270–277 10.1016/j.bcmd.2008.06.003 [DOI] [PubMed] [Google Scholar]
- Chiaramonte R., Basile A., Tassi E., Calzavara E., Cecchinato V., Rossi V., Biondi A., Comi P. 2005. A wide role for NOTCH1 signaling in acute leukemia. Cancer Lett. 219:113–120 10.1016/j.canlet.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Cordle J., Redfieldz C., Stacey M., van der Merwe P.A., Willis A.C., Champion B.R., Hambleton S., Handford P.A. 2008. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J. Biol. Chem. 283:11785–11793 10.1074/jbc.M708424200 [DOI] [PubMed] [Google Scholar]
- Del Giudice I., Rossi D., Chiaretti S., Marinelli M., Tavolaro S., Gabrielli S., Laurenti L., Marasca R., Rasi S., Fangazio M., Guarini A., Gaidano G., Foa R. 2011. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica.97:437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ianni M., Baldoni S., Rosati E., Ciurnelli R., Cavalli L., Martelli M.F., Marconi P., Screpanti I., Falzetti F. 2009. A new genetic lesion in B-CLL: a NOTCH1 PEST domain mutation. Br. J. Haematol. 146:689–691 10.1111/j.1365-2141.2009.07816.x [DOI] [PubMed] [Google Scholar]
- Ellisen L.W., Bird J., West D.C., Soreng A.L., Reynolds T.C., Smith S.D., Sklar J. 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 66:649–661 10.1016/0092-8674(91)90111-B [DOI] [PubMed] [Google Scholar]
- Fan X., Mikolaenko I., Elhassan I., Ni X., Wang Y., Ball D., Brat D.J., Perry A., Eberhart C.G. 2004. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 64:7787–7793 10.1158/0008-5472.CAN-04-1446 [DOI] [PubMed] [Google Scholar]
- Figueroa M.E., Wouters B.J., Skrabanek L., Glass J., Li Y., Erpelinck-Verschueren C.A., Langerak A.W., Löwenberg B., Fazzari M., Greally J.M., et al. 2009. Genome-wide epigenetic analysis delineates a biologically distinct immature acute leukemia with myeloid/T-lymphoid features. Blood. 113:2795–2804 10.1182/blood-2008-08-172387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison H., Farnie G., Howell S.J., Rock R.E., Stylianou S., Brennan K.R., Bundred N.J., Clarke R.B. 2010. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 70:709–718 10.1158/0008-5472.CAN-09-1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning K., Heering J., Schwanbeck R., Schroeder T., Helmbold H., Schäfer H., Deppert W., Kim E., Just U. 2008. Notch1 activation reduces proliferation in the multipotent hematopoietic progenitor cell line FDCP-mix through a p53-dependent pathway but Notch1 effects on myeloid and erythroid differentiation are independent of p53. Cell Death Differ. 15:398–407 10.1038/sj.cdd.4402277 [DOI] [PubMed] [Google Scholar]
- Hicks C., Johnston S.H., diSibio G., Collazo A., Vogt T.F., Weinmaster G. 2000. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat. Cell Biol. 2:515–520 10.1038/35019553 [DOI] [PubMed] [Google Scholar]
- Jerez A., Gondek L.P., Jankowska A.M., Makishima H., Przychodzen B., Tiu R.V., O’Keefe C.L., Mohamedali A.M., Batista D., Sekeres M.A., et al. 2012. Topography, clinical, and genomic correlates of 5q myeloid malignancies revisited. J. Clin. Oncol. 30:1343–1349 10.1200/JCO.2011.36.1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson J.I., Xiang Z., Pettersson M., Lardelli M., Nilsson G. 2001. Distinct and regulated expression of Notch receptors in hematopoietic lineages and during myeloid differentiation. Eur. J. Immunol. 31:3240–3247 10.1002/1521-4141(200111)31:113.0.CO;2-E [DOI] [PubMed] [Google Scholar]
- Jundt F., Schwarzer R., Dörken B. 2008. Notch signaling in leukemias and lymphomas. Curr. Mol. Med. 8:51–59 10.2174/156652408783565540 [DOI] [PubMed] [Google Scholar]
- Kim Y.W., Koo B.K., Jeong H.W., Yoon M.J., Song R., Shin J., Jeong D.C., Kim S.H., Kong Y.Y. 2008. Defective Notch activation in microenvironment leads to myeloproliferative disease. Blood. 112:4628–4638 10.1182/blood-2008-03-148999 [DOI] [PubMed] [Google Scholar]
- Klinakis A., Lobry C., Abdel-Wahab O., Oh P., Haeno H., Buonamici S., van De Walle I., Cathelin S., Trimarchi T., Araldi E., et al. 2011. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 473:230–233 10.1038/nature09999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U., Radtke F. 2007. Notch and cancer: a double-edged sword. Cell. Mol. Life Sci. 64:2746–2762 10.1007/s00018-007-7164-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblau S.M., Coombes K.R. 2011. Use of reverse phase protein microarrays to study protein expression in leukemia: technical and methodological lessons learned. Methods Mol. Biol. 785:141–155 10.1007/978-1-61779-286-1_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Milner L.A., Deng Y., Iwata M., Banta A., Graf L., Marcovina S., Friedman C., Trask B.J., Hood L., Torok-Storb B. 1998. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 8:43–55 10.1016/S1074-7613(00)80457-4 [DOI] [PubMed] [Google Scholar]
- Maillard I., Fang T., Pear W.S. 2005. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23:945–974 10.1146/annurev.immunol.23.021704.115747 [DOI] [PubMed] [Google Scholar]
- Maillard I., Koch U., Dumortier A., Shestova O., Xu L., Sai H., Pross S.E., Aster J.C., Bhandoola A., Radtke F., Pear W.S. 2008. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2:356–366 10.1016/j.stem.2008.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuya M., Katayama N., Hoshino N., Nishikawa H., Sakano S., Araki H., Mitani H., Suzuki H., Miyashita H., Kobayashi K., et al. 2002. The soluble Notch ligand, Jagged-1, inhibits proliferation of CD34+ macrophage progenitors. Int. J. Hematol. 75:269–276 10.1007/BF02982040 [DOI] [PubMed] [Google Scholar]
- Miller C.P., Ban K., Dujka M.E., McConkey D.J., Munsell M., Palladino M., Chandra J. 2007. NPI-0052, a novel proteasome inhibitor, induces caspase-8 and ROS-dependent apoptosis alone and in combination with HDAC inhibitors in leukemia cells. Blood. 110:267–277 10.1182/blood-2006-03-013128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara F., Sakata-Yanagimoto M., Komeno Y., Kato N., Uchida T., Haraguchi K., Kumano K., Harada Y., Harada H., Kitaura J., et al. 2010. Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood. 115:2872–2881 10.1182/blood-2009-05-222836 [DOI] [PubMed] [Google Scholar]
- Nemoto N., Suzukawa K., Shimizu S., Shinagawa A., Takei N., Taki T., Hayashi Y., Kojima H., Kawakami Y., Nagasawa T. 2007. Identification of a novel fusion gene MLL-MAML2 in secondary acute myelogenous leukemia and myelodysplastic syndrome with inv(11)(q21q23). Genes Chromosomes Cancer. 46:813–819 10.1002/gcc.20467 [DOI] [PubMed] [Google Scholar]
- Ohishi K., Varnum-Finney B., Bernstein I.D. 2002. Delta-1 enhances marrow and thymus repopulating ability of human CD34(+)CD38(-) cord blood cells. J. Clin. Invest. 110:1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T., McKenna K., O-Neil J., Galinsky I., Stone R., Suzukawa K., Stiakaki E., Kalmanti M., Fox E.A., Caligiuri M.A., et al. 2006. Activating mutations in NOTCH1 in acute myeloid leukemia and lineage switch leukemias. Leukemia. 20:1963–1966 10.1038/sj.leu.2404409 [DOI] [PubMed] [Google Scholar]
- Pear W.S., Nolan G.P., Scott M.L., Baltimore D. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 90:8392–8396 10.1073/pnas.90.18.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui J.C., Allman D., Xu L., DeRocco S., Karnell F.G., Bakkour S., Lee J.Y., Kadesch T., Hardy R.R., Aster J.C., Pear W.S. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 11:299–308 10.1016/S1074-7613(00)80105-3 [DOI] [PubMed] [Google Scholar]
- Saito T., Chiba S., Ichikawa M., Kunisato A., Asai T., Shimizu K., Yamaguchi T., Yamamoto G., Seo S., Kumano K., et al. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 18:675–685 10.1016/S1074-7613(03)00111-0 [DOI] [PubMed] [Google Scholar]
- Sarmento L.M., Huang H., Limon A., Gordon W., Fernandes J., Tavares M.J., Miele L., Cardoso A.A., Classon M., Carlesso N. 2005. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J. Exp. Med. 202:157–168 10.1084/jem.20050559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T., Kohlhof H., Rieber N., Just U. 2003. Notch signaling induces multilineage myeloid differentiation and up-regulates PU.1 expression. J. Immunol. 170:5538–5548 [DOI] [PubMed] [Google Scholar]
- Tibes R., Qiu Y., Lu Y., Hennessy B., Andreeff M., Mills G.B., Kornblau S.M. 2006. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 5:2512–2521 10.1158/1535-7163.MCT-06-0334 [DOI] [PubMed] [Google Scholar]
- Tohda S., Nara N. 2001. Expression of Notch1 and Jagged1 proteins in acute myeloid leukemia cells. Leuk. Lymphoma. 42:467–472 10.3109/10428190109064603 [DOI] [PubMed] [Google Scholar]
- Tohda S., Kogoshi H., Murakami N., Sakano S., Nara N. 2005. Diverse effects of the Notch ligands Jagged1 and Delta1 on the growth and differentiation of primary acute myeloblastic leukemia cells. Exp. Hematol. 33:558–563 10.1016/j.exphem.2005.01.010 [DOI] [PubMed] [Google Scholar]
- Verhaak R.G., Wouters B.J., Erpelinck C.A., Abbas S., Beverloo H.B., Lugthart S., Löwenberg B., Delwel R., Valk P.J. 2009. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 94:131–134 10.3324/haematol.13299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A.P., Nam Y., Wolfe M.S., Pear W.S., Griffin J.D., Blacklow S.C., Aster J.C. 2003. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol. Cell. Biol. 23:655–664 10.1128/MCB.23.2.655-664.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Wouters B.J., Jordà M.A., Keeshan K., Louwers I., Erpelinck-Verschueren C.A., Tielemans D., Langerak A.W., He Y., Yashiro-Ohtani Y., Zhang P., et al. 2007. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 110:3706–3714 10.1182/blood-2007-02-073486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Mehew J.W., Heckman C.A., Arcinas M., Boxer L.M. 2001. Negative regulation of bcl-2 expression by p53 in hematopoietic cells. Oncogene. 20:240–251 10.1038/sj.onc.1204067 [DOI] [PubMed] [Google Scholar]
- Wu J., Iwata F., Grass J.A., Osborne C.S., Elnitski L., Fraser P., Ohneda O., Yamamoto M., Bresnick E.H. 2005. Molecular determinants of NOTCH4 transcription in vascular endothelium. Mol. Cell. Biol. 25:1458–1474 10.1128/MCB.25.4.1458-1474.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D.D., Fan F.Y., Hu X.B., Hou L.H., Zhang X.P., Liu L., Liang Y.M., Han H. 2009. Notch signaling inhibits the growth of the human chronic myeloid leukemia cell line K562. Leuk. Res. 33:109–114 10.1016/j.leukres.2008.06.023 [DOI] [PubMed] [Google Scholar]
- Yoda M., Kimura T., Tohmonda T., Uchikawa S., Koba T., Takito J., Morioka H., Matsumoto M., Link D.C., Chiba K., et al. 2011. Dual functions of cell-autonomous and non-cell-autonomous ADAM10 activity in granulopoiesis. Blood. 118:6939–6942 10.1182/blood-2011-06-357210 [DOI] [PubMed] [Google Scholar]
- Yu X., Alder J.K., Chun J.H., Friedman A.D., Heimfeld S., Cheng L., Civin C.I. 2006. HES1 inhibits cycling of hematopoietic progenitor cells via DNA binding. Stem Cells. 24:876–888 10.1634/stemcells.2005-0598 [DOI] [PubMed] [Google Scholar]
- Zhou L., Li L.W., Yan Q., Petryniak B., Man Y., Su C., Shim J., Chervin S., Lowe J.B. 2008. Notch-dependent control of myelopoiesis is regulated by fucosylation. Blood. 112:308–319 10.1182/blood-2007-11-115204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-McKay P.A. 2008. Notch signaling in pediatric malignancies. Curr. Oncol. Rep. 10:459–468 10.1007/s11912-008-0071-2 [DOI] [PubMed] [Google Scholar]
- Zweidler-McKay P.A., Pear W.S. 2004. Notch and T cell malignancy. Semin. Cancer Biol. 14:329–340 10.1016/j.semcancer.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Zweidler-McKay P.A., He Y., Xu L., Rodriguez C.G., Karnell F.G., Carpenter A.C., Aster J.C., Allman D., Pear W.S. 2005. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 106:3898–3906 10.1182/blood-2005-01-0355 [DOI] [PMC free article] [PubMed] [Google Scholar]