Abstract
The respiratory chain or oxidative phosphorylation system (OxPhos) generates most of the chemical energy (ATP) used by our cells. The cytochrome c oxidase (CcO) is one of three protein complexes of OxPhos building up a proton gradient across the inner mitochondrial membrane, which is ultimately used by the ATP synthase to produce ATP. We present molecular dynamic simulations of CcO in a mimic of the mitochondrial membrane, and identify precise binding sites of cardiolipin (CL, signature phospholipid of mitochondria) on the protein surface. Two of these CL binding sites reveal pathways linking CLs to the entrance of the D and H proton channels across CcO. CLs being able to carry protons our results strongly support an involvement of CLs in the proton delivery machinery to CcO. The ubiquitous nature of CL interactions with the components of the OxPhos suggests that this delivery mechanism might extend to the other respiratory complexes.
Mitochondria are the intra-cellular organelles that produce most of the chemical energy consumed by a cell and are therefore often refered to as the “power plant” of the cell. The energy is produced through the oxidative phosphorylation (OxPhos) system. The OxPhos system is embedded in the mitochondrial inner-membrane and consists of four large membrane protein assemblies, the so-called “respiratory chain complexes” (i.e.: NADH quinone oxidoreductase, complex I; cytochrome bc1, complex III; cytochrome c oxidase, complex IV; ATP synthase, complex V) and by some small electron carriers (quinones and cytochrome c). Complexes I, III and IV operate a series of electron transfers whose tortuous paths through the respiratory chain system is coupled with the translocation of protons from the inside (matrix) to the mitochondrial intermembrane space (IMS), leading to an electro-chemical gradient ultimately utilized by the ATP synthase to transform ADP into ATP.
Cardiolipin (CL), the signature phospholipid of mitochondrial membranes, has crucial implications in mitochondrial processes, and on the OxPhos in particular1,2. CLs are formed by two phosphatidyl-glycerol molecules linked by an additional glycerol moiety and possess an anionic charge (−2 e). They are found in several parts of the mitochondria and constitute about 10% of the phospholipids of, for instance, the bovine heart mitochondrial inner membrane3,4. CL deficiency correlates with numerous diseases5 including the Barth syndrome6 and heart failure7. Notably, the formation, stability, and function of individual respiratory chain complexes and of respiratory chain supercomplexes strongly relies on the presence of CLs in the membrane8,9,10,11,12. CLs bound to individual complexes have been reported by several structural studies on various organisms13,14,15, leading to hypotheses on their possible involvement in a proton uptake pathway16 and/or in assuring the structural integrity of individual complexes16,17,18,19 and supercomplexes15.
The focus of the present work is on bovine cytochrome c oxydase (CIV). It is a transmembrane protein complex that consists of 13 subunits labeled A to M as depicted in Fig. 1A (The subunit's nomenclature following a roman number code as defined by Kadenbach20 is given in Supplementary Information). Primarily based on purification and delipidation by chromatography and binding affinity assays13,17,18,21,22 two classes of CL binding sites have been defined: two sites with high-affinity and one to two additional sites with a lower affinity. The latter have been associated with the structural integrity of CIV and of its dimeric form because these CLs stabilize the subunits G and H (VIa and VIb according to Kadenbach nomenclature20), which are mandatory for the formation of the dimer17,18. The two CLs binding CIV with high affinity have been associated with the regulation of the electron activity of the enzyme21,22.
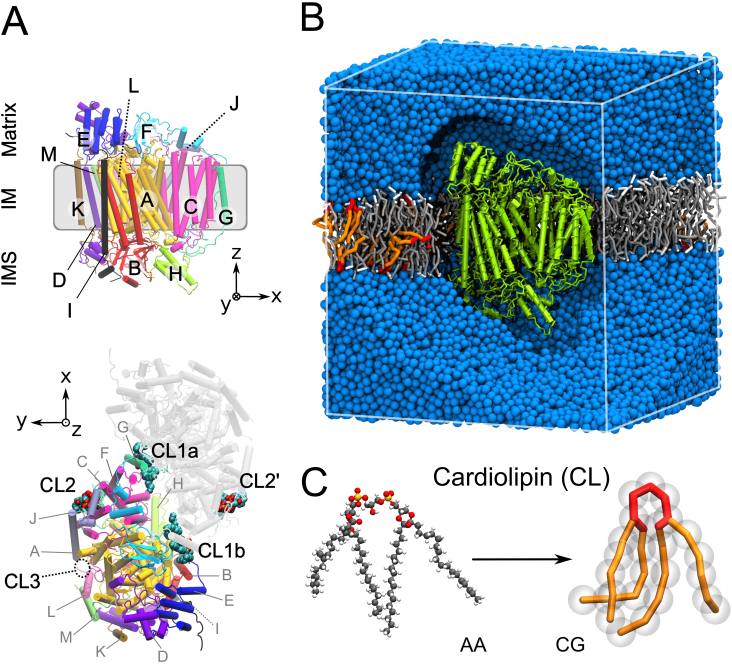
Figure 1. Structural characteristics of bovine heart cytochrome c oxidase (CIV).
(A) Structure of CIV with its thirteen subunits (A–M) shown in a cartoon representation with a chain-based color-code for one monomer. The subunit's nomenclature following a roman number code used in others studies20 is given in Supplementary Information. We kept on the letter code to avoid confusion with the site names. In the bottom view of the dimer structure as found in the X-ray structure CLs are depicted24. There are three CL binding sites per monomer (CL1a, CL1b and CL2), with one facing the matrix side (CL2) and two facing the intermembrane space (IMS; CL1a and CL1b). CL3 indicates the location of an additional binding site suggested by photolabeling experiments19. (B) Simulation box for CIV system, with the protein shown in green, POPC molecules in gray/white, CLs in red/orange and the aqueous phase in blue. (C) Structure of a cardiolipin molecule in an atomistic (AA) and a coarse-grain (CG) representation.
It is only with the most recent crystal structures of CIV23,24 that CLs were found bound to the protein. In the dimeric form of CIV four CLs co-crystallize with the complex, defining three potential CL binding sites per complex (CL1a, b and CL2 in Fig. 1A) after consideration of the dimer's symmetry. The intuitive assignment of the CL binding sites based on their location on the complex was in overall agreement with the in-depth photolabeling study of Sedlak et al.19. The low-affinity CL binding sites are located at the interface of the dimer and interact with subunits G (CL1a/b in Fig. 1), while a high affinity binding site was found on a more membrane exposed protein surface interacting with subunit J (CL2 in Fig. 1). There is however a CL binding site identified by Sedlak et al.19 that is missing in the crystal structure (CL3 in Fig. 1).
Here we explore the binding of CL to complex IV in a monomeric form by means of coarse-grain molecular dynamics (CGMD) simulations. MD simulations have been successfully used to explore the lipid/protein interplay25,26 and more specifically lipid binding to a variety of membrane proteins25,27,28,29,30,31,32. The use of a CG model presents the advantage to investigate the lipid binding to proteins on much longer time scales than conventional atomistic models and thus allows exploring the processes involved from a more realistic dynamic perspective. We recently applied a similar approach to the study of CL binding sites on the cytochrome bc115 in which case we have characterized some key dynamic features of known CL binding sites and revealed a set of new membrane-exposed binding sites potentially involved in the formation of supercomplexes.
The set of CGMD simulations presented here clearly identified the precise positions of the two CLs with high-affinity binding sites on CIV19. The lack of the low-affinity binding sites expected at the interface of the dimer are not observed in the simulations, which confirms that they are strongly associated with the dimer form of CIV and might only exist with the dimer. Five additional CL binding sites with low-affinity are found and may be easily rationalized in light of the position of other co-crystallized lipids24 and their common features. Most remarkably two of the binding sites are found located at the matrix entrance of known proton uptake pathways (D and H). We show how the structure of the protein reveals extensions of these pathways that directly link the CLs in these binding sites to the entrance of the pathways. In the context of the ability of CL to trap protons our data strongly support that CL maximizes CIV electron transport activity by providing protons to the uptake pathways.
Results
Identification of seven CL binding sites on bovine heart cytochrome c oxidase
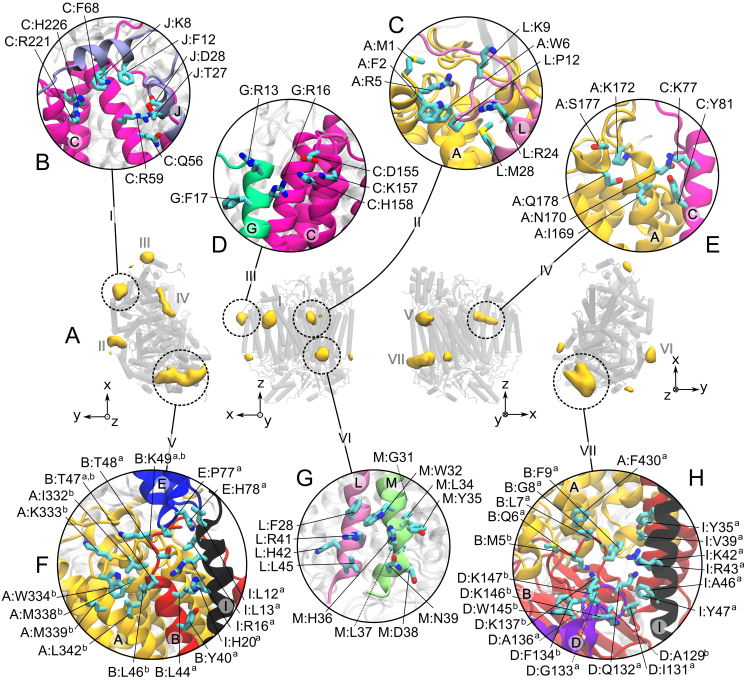
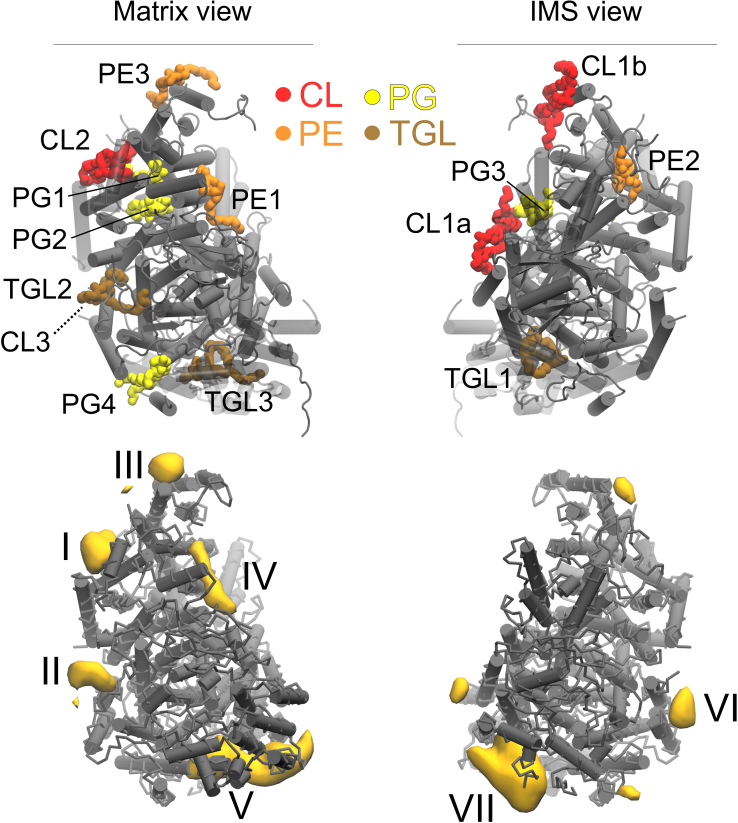
We characterized the CL density around the protein from a 100 μs CGMD simulation of this complex embedded in a POPC/CL (20/1 molar ratio) membrane. The average densities of CLs (Fig. 2A) demonstrate the existence of several preferential sites of interaction of CL with the protein. We define CL binding sites as the locations having a CL density more than five times the bulk value. Accordingly, seven binding sites are found on either leaflet of the membrane and are labeled I to VII (Fig. 2A). Sites I–V are on the matrix side of the membrane and sites VI and VII on the side of the IMS. Sites V and VII may be occupied by two CLs and are therefore subdivided into Va/Vb and VIIa/VIIb. A hierarchy of binding strength was observed, starting with many CL binding/unbinding events for some sites (e.g. 380 exchanges for site VIIb) but only few exchanges for others (e.g. few unbinding events for site I) during the same amount of simulation time. For all sites but site I, binding events by at least 4 CLs were observed (see Supplementary Movie), allowing us to extract meaningful statistical averaged occupations and CL residence times listed in Table 1. The detailed structural characterization of the binding sites is presented in Figure 2B-H.
Figure 2. Cardiolipin (CL) binding sites on cytochrome c oxidase (CIV).
Binding sites are extracted from a 100 μs of CGMD simulation of the complex embedded in a CL/POPC membrane bilayer. (A) From left to right: matrix, membrane (two orientations) and inter membrane space (IMS) views of CIV with the CL densities shown in yellow volume maps at an isovalue corresponding to at least 5 times the bulk density. The protein is shown as shaded grey cylinders with the CL densities projected onto them. (B–H) Detailed description of the CL binding sites I to VII, respectively. The residues are numbered as follows: “chain:residuesub-site”. For each site, the subunits involved in the interactions with the CLs are depicted as colored cartoon as in Fig. 1. The rest of the protein is shown in a transparent gray cartoon.
Table 1. Occupation (Ξ) and residence time (θ, μs) of cardiolipin binding sites on cytochrome c oxydase (CIV). The values are averaged over a 100 μs CGMD simulation. The accuracy of the occupation levels is ±0.02 at most and of the residence times within ±0.1 μs. See the Extended Methods given as Supplementary Information for details. #CLs is the number of different CLs getting in contact with a site and #events the number of binding/unbinding events. The lipids described by Shinzawa-Itoh et al.24 (see Fig. 4) at the locations corresponding to the CL binding sites found in the simulations are indicated in the bottom row. The CL binding site predicted by Sedlák et al.19 at the site II is also indicated.
| Site | I | II | III | IV | Va | Vb | VI | VIIa | VIIb |
|---|---|---|---|---|---|---|---|---|---|
| Ξ | 0.99 | 0.60 | 0.57 | 0.36 | 0.74 | 0.68 | 0.52 | 0.88 | 0.50 |
| θ | >50* | >60* | 1.0 | 0.5 | 0.4 | 0.6 | 0.4 | 10.3 | 0.2 |
| #CLs | 1 | 4 | 17 | 11 | 20 | 10 | 13 | 4 | 20 |
| #events | 18 | 11 | 104 | 109 | 235 | 122 | 147 | 41 | 380 |
| ref 19 | – | CL | – | – | – | – | — | – | – |
| 2DYR24 | CL | TGL2 | PE3 | PE1 | TGL3 | TGL3 | — | TGL1 | TGL1 |
*The strong binding of CLs in sites I and II led to poor binding/unbinding events statistics and prevented us from determining accurate residence time. Instead we indicate a rough estimate obtained from the binding behavior. See Figure S1 in Supplementary Information.
Site I quickly bound a single CL that remained bound during the entire simulation although reorientation of the CL led to multiple short unbinding events (see Supplementary Figure S1). These events complicated the computation of the lifetime estimated at more than 50 μs (Table 1). In this site the CL mainly interacts with the subunit C and with a couple of residues of subunit J (Fig. 2B). Sites II and III are adjacent to site I and located at the junction of subunits A and L and subunits C and G, respectively (Fig. 2C–D). They both bind one CL at a time and have slightly lower occupancies (60%) than site I. Although it is difficult to precisely determine the time scale of CLs binding to site II (see Supplementary Fig. S1; estimate ~60 μs) it is significantly longer than in site III in which CLs exchange on a time scale of ~1 μs. Site IV is located on the face of CIV that corresponds to the dimer interface in the crystal structure. This site, involving subunits A and C, is occupied only 36% of the time and CLs bind on a sub-microsecond timescale. Site V is located at the other extremity of CIV as compared to site I. CLs in site Va and Vb interact with subunits B, E and I, and A and B, respectively. They have similar occupancies, ~70%, and similar residence times, ~0.5 μs. Site VI is located on the IMS side of CIV. It involves the C-termini of subunits L and M, has a low CL occupancy, 50%, and a small CL residence time, ~0.4 μs. Site VII is located on the same protein surface as site V, but on the opposite leaflet (IMS). CLs in site VIIa and VIIb primarily interact with subunits A, B, D and I on the IMS side of the protein. Site VIIa is significantly more occupied (88%) than site VIIb (50%) and has a relatively long residence time of ~10 μs. The difference between sites VIIa and VIIb might reflect that site VIIa is buried deeper in a cavity formed by helices of subunits B and I, whereas site VIIb is more exposed to the membrane.
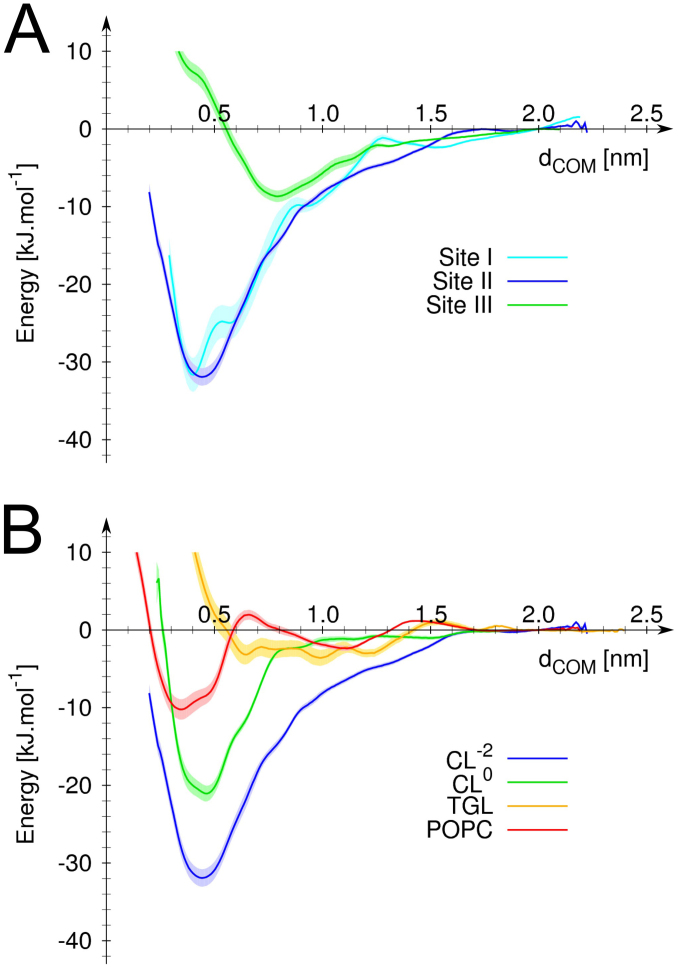
We further characterized the binding strength of CL to CIV by determining the potentials of mean force (PMFs) of CL's binding to a selection of the sites (Fig. 3). The data clearly indicates that CL's binding to site I and II is stronger than to the other sites. Site III was taken as a reference site because it is relatively well occupied, accessible to the membrane and explored by many CLs so that the statistics obtained is reliable (Table 1). The data also indicate that CL binds similarly to site II than to site I. The comparison of the PMFs of a CL binding to site II with or without the −2 e charge of its headgroup indicates that although the charge of the headgroup bears the most significant contribution to the binding, the tails also contribute to CL's binding strength. The contribution from the CL's bulky tail is by itself larger than the binding strength of a POPC molecule.
Figure 3. Potential of mean force for binding of various lipids to sites I, II and III.
(A) Comparison of CL's binding strength to site I (cyan), II (blue) and III (green). (B) Comparison of binding strength of TGL (yellow), POPC (orange) and CL to site II. Two CLs were tested; double charged (−2 e) and neutral, blue and green curves, respectively. In both panel the relative free energy of the system is expressed as a function of the distance, dCOM, between the center of masse of the lipid headgroup and of the binding site as defined in Fig. 2. The error on the measure (estimated using the Bayesian bootstrapping method) is shown by the shaded area behind the curves.
Residue content of the binding sites
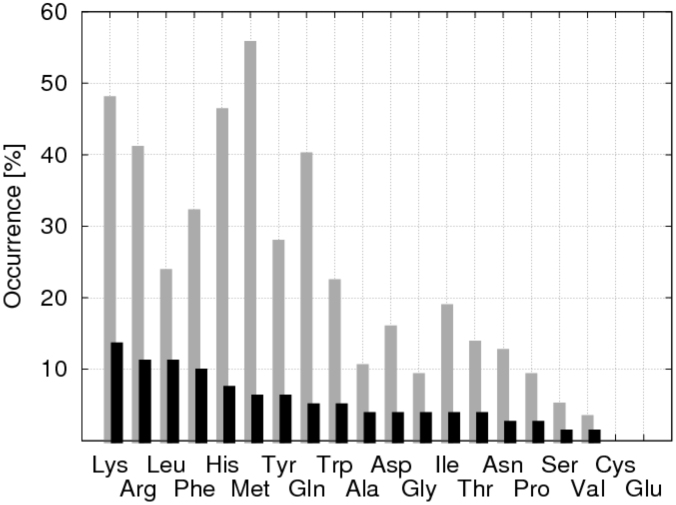
Interesting features emerged from the analysis of the residues in contact with CLs during the 100 µs simulation (Fig. 4). First, all the residues that made at least one contact with a CL molecule virtually span the entire transmembrane region of the protein, indicating that the CLs explored the complete transmembrane protein surface. As expected, the distribution of contacts as a function of the residue type (grey bars in Fig. 4) is in general agreement with the amino acid distribution in integral membrane proteins33 keeping in mind that a residue would need to be exposed to the membrane to be in contact with a CL. Furthermore, focusing on the composition of the CL binding sites (black bars in Fig. 4), the positively charged amino acids (lysine and arginine) were found to account for ~25% of the CL ligands. The large contribution from positively charged residues might be expected since CL carries a −2 e charge and is illustrated by the strong correlation between the locations of the CL's binding sites with the positive regions of electrostatic potential on the protein's surface (Fig. S2). Lysine is slightly favored over arginine (14 vs 11%), which contrasts with the results obtained previously for CIII15. In that case arginine was significantly more represented in the binding sites than lysine, which then represented only ~5% of the CL's ligands against 18% for arginine. Phenylalanine and leucine were found to be a relatively important contribution to the CL binding sites (> 10%) in both CIII and CIV.
Figure 4. Residue content of the CL binding sites of cytochrome c oxidase (CIV).
The gray sticks indicate the percentage of each residue type at least once in contact with a CL; e.g. present in the section of the CIV accessible to CLs. The black sticks give for each residue type the percentage of its participation to the CL binding sites.
Discussion
A thorough analysis of the lipids bound to CIV was presented by Shinzawa-Itoh et al.24 combining a crystal structure of the complex with mass spectroscopy. They discussed up to seven species of lipids per monomer of CIV among which three CLs, three phosphatidylethanolamines (PEs), four phosphatidylglycerols (PGs) and three triglycerides (TGLs). These lipid types are found in the native mitochondrial inner membrane3 (TGL only recently24) and their presence in the crystal is therefore not expected to be an artifact of the crystallization process. Their positions are depicted in Figure 5 together with the CL binding sites found in the simulations. The comparison of the two sets of molecules allows to unambiguously identifying the two CL binding sites with high-affinity and involved in CIV's proton transport activity, and suggesting the most likely candidates for the low-affinity binding sites that have been linked to CIV's structural integrity. Our interpretation is in complete agreement with earlier predictions of Robinson and co-workers13,19,22.
Figure 5. Location of the co-crystallized phospholipid positions and densities computed with our CGMD simulation.
The experimental positions (upper part) were extracted from the PBD entry 2dyr24. The densities (lower part) are extracted from the 100 μs CGMD simulation. Views from the matrix (left) and the IMS (right) sides of CIV are shown. CL: cardiolipin; PC: phosphatidylcholine; PE: phosphatidylethanolamine; PG: phosphatidylglycerol; TGL: triglyceride.
Among the three CL binding sites per monomer found in the dimer structure of CIV23,24 (CL1a, b and CL2 in Fig. 5), only CL2 is observed in the simulations. It is located at the site I. This site is filled early in our simulation and remains occupied during the entire simulation (Table 1 and Supplementary Fig. S1), which gives it one of the highest binding affinities observed in the simulations. This result is in agreement with its presence in the crystal structure and the PMFs described above, and confirms that it corresponds to one of the high-affinity sites as predicted by Sedlák et al.19. CL1a/b, located at the dimer interface in the crystal structures23,24, are not stable in our simulations of CIV as a monomer. These two CLs have been suggested to be the ones loosely bound to CIV and strongly connected to the existence of the dimeric form of CIV by their stabilization of subunits G and H18,19. Our simulations indicate that the tight association of CIVs in the dimer is a determinant factor for the stability of CL1a and CL1b. To test this hypothesis, additional simulation of the dimeric structure of CIV including the experimental CL1a/CL1b were performed. Both sites were found stable on the μs time scale.
The site II defined by the simulations (Fig. 2 and 5) confirms the location of the second high-affinity CL binding site predicted by Sedlák et al.19 using photolabeling and missing in the crystal structures (CL3 in Fig. 1). The relatively low occupancy and residence time (Table 1) might be surprising at first but reflect that it took ~40 μs for a CL to enter the protein cavity forming the binding site (see Supplementary Fig. S1). Once the CL made the proper contacts the site is fully occupied and therefore corresponds to a high-affinity binding site. This result is corroborated by the PMF of CL binding to site II, which shows a similar bonding strength to site I (Fig. 3A). It is interesting to note that a TGL (TGL2 in Fig. 5) occupies the site II in the crystal structure24. From our understanding of the system it is extremely unlikely that both CL and TGL molecules occupy site II. The PMFs of the binding of a CL vs. a TGL (Fig. 3B) demonstrate not only that CL binds much stronger but also that TGL is no stable in site II. Unrestrained simulations confirm this observation: a TGL placed in site II left it quickly and went to mix with the bulk after exploring the surrounding of the protein surface around the site. The PMFs showed that an important part of the CL's binding strength is due to coulombic interactions, which might rationalize the weak binding of TGL; it is a neutral molecule. It is not clear at this point why a TGL molecule is present in the crystal although it was argued not to be an artifact of the purification24. One might speculate that the TGL molecule had the capability to replace the CL using the similarity of their bulky tails. This scenario implies however that the CL was first destabilized, which might occur by disruption of the interaction of its headgroup during the crystallization process. Note the PMFs showed a neutral CL still binds site II stronger than TGL suggesting that TGL's bulky tails are not sufficient to stabilize it. See the Supplementary Information for additional discussion on the presence of TGL molecules in CL's binding sites.
The remaining CL binding sites (III–VII) found in the simulations are relatively weak binders and unless they are buried within a large protein cavity (sites Va and VIIa) they have a low occupancy. They all correspond to the location of binding of another lipid type in the crystal, which is in line with previous studies showing the conservation of binding sites by different lipid types14. We review the several cases observed in CIV and show that it is quite straightforward to rationalize the discrepancies from the similarities of CLs with PGs and TGLs (CL and PG share a negatively charged headgroup, while CL and TGL bulky tails), see Supplementary Information for more detailed discussion.
The high degree of conservation of lipid binding sites on CIV from various organisms14 suggests an important functional role of these bound lipids. CLs have been shown to affect CIV in two ways. First, there is extensive data demonstrating that the presence of two CLs with high-affinity binding to CIV, which we have shown are bound in site I and II, is mandatory for a fully functional CIV22. It is however unknown by which mechanism they operate. CIV enzymatic activity is often summarized by its electron transport activity (up take from cytochrome c) but CIV uses as many protons as electrons to transform a dioxygen molecule into two water molecules. CLs have the ability to trap protons34 and thereby can facilitate proton translocation along the membrane surface35. This particular feature of CL has been suggested to be an important aspect of CIII mechanism for proton transfer16,36,37 and might extend to CIV by providing proton sources to either of the D-, K- or H-pathways38,39,40,41. The proximity of CL3 to the D-pathway entrance has suggested that CL might be acting as a proton antenna to this pathway19 although only limited structural insight is available since this site is not occupied in the crystal structures23,24.
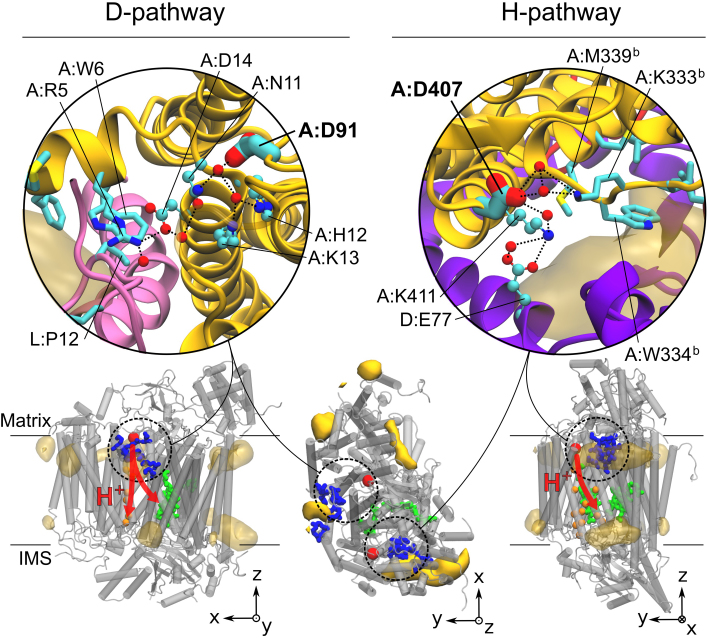
Our data confirm that this ability of CL to carry protons is indeed relevant to site II but might also be to site Vb. Both sites are located on the matrix side of the IM where the protons are taken up. Site II corresponds to CL3 (Fig. 1, 5) and is very close to the D-pathway entrance (~1.1 nm; Fig. 6), while site Vb is close to the entrance of the H-pathway (~0.8 nm; Fig. 6). Up to now residues A:D91 (A:D132 in Rodobacter sphaeroides) and A:D407 define the matrix side entrances to the D- and H-pathways, respectively. Only little is known on the proton path before these residues. A close inspection of the protein structure revealed in both cases the existence of a strong H-bond network starting at the CL binding sites II and Vb and leading to the entrances of the D- and H-pathways, respectively (Fig. 6). These networks form clear extensions of the D- and H-pathways towards the exterior of the protein on the matrix side. They directly connect the CL binding sites to A:D91 (A:D132 in Rodobacter sphaeroides) and A:D407, respectively, and thereby strongly support the idea that CLs provide a source of protons at the surface of the membrane facilitating CIV electron transport activity.
Figure 6. H-bond networks leading from the cardiolipin (CL) binding sites to the entrance of D- and H-pathways, A:D91 and A:D407, respectively.
The CL binding sites are shown in stick representation. The yellow surface maps depict the CL densities shown in Fig. 2. A:D91 and A:D407 are shown in large sticks. The residues and water molecules (red spheres) participating to the networks are shown in a ball-and-stick representation and numbered as in Fig. 2. The bottom row shows side and top views of CIV with the CL densities, the residues involved in the sites in blue, the residues involved in the transmembrane section of the proton pathways (red arrows) in orange spheres and the heme molecules in green. The large red spheres position the entrances (A:D91 and A:D407) of the pathways.
It is also notable that up to four CLs occupy the protein cavity close to the H-pathway; two lipids in sites Va/b and VIIa/b on the matrix and IMS side of the membrane, respectively. This aggregation of CLs might be relevant to the ‘dielectric channel' activity of CIV proposed by Rich42.
The second way by which CLs affect CIV is by providing structural integrity. Two additional CLs, which we propose being CL1a and CL1b in the crystal structures (Fig. 1, 5), serve the structural integrity of CIV by stabilizing subunits G and H, and thereby its dimeric form17. These two CLs have been shown to bind CIV with a low-affinity19 and are not observed in the simulation of the monomeric form of CIV (Fig. 2) but are stable in the dimeric structure. This ability of CLs to stabilize CIV dimers can be extended to high-order oligomers of the respiratory chain complexes as it was reported in the context of the formation of supercomplexes. This behavior is most notable for the formation of supercomplexes CIII2-CIV and CIII2-CIV29,12. Our recent CGMD simulations of the CIII15 suggested that CL binding sites on the surface of the complex might define the location of protein-protein contact with CIV. The CL binding sites found on CIV might share a similar function.
In summary, the CGMD simulations used in this study to investigate the CL binding sites on the respiratory chain complex IV, cytochrome c oxidase, provide new and valuable insights on the way CLs participate to the function of proteins. The locations of the CL binding sites that most likely correspond to both the high and low-affinity CL binding to CIV are identified with high level of confidence. They agree with and reconcile all known experimental data. CL binding sites on the surface of the protein are found at the proximity of two of the three known proton-uptake pathways, the D- and H-pathways. Two clear interaction networks connecting the CL binding sites to the entrances of these two pathways are uncovered. To our knowledge they provide the first complete proton pathway between the membrane surface (CL binding sites) and the D- and H-pathways in cytochrome c oxidase. This data strongly supports that CLs take active part in the proton delivery mechanism. Given the wide impact of CL in the respiratory chain machinery the role suggested by our data should extend to other components of the respiratory chain.
Methods
Molecular models
The Protein Data Bank (PDB) entries 1occ and 2occ38 were used to built a complete atomistic model of CIV, which was simulated as a monomer, its functional form. In the experimental dimeric structure of CIV24 four CLs co-crystallize with the protein (Fig. 1A). In a monomeric configuration, the experimental CL positions are all exposed to the membrane bulk and were not included in the initial conformation. In the simulations, the complex lipid composition of the mitochondrial membrane was modeled by a two components mixture of CL:POPC with a molecular ratio of 1:20 (5% of CL, 10% of total phosphorus content). This value is close to the experimental molecular ratio observed for bovine heart mitochondria (15 to 20% of the phosphorus content3).
The systems were described with the Martini CG force field for biomolecules (version 2.043) and its extension to proteins (version 2.144) together with the ElNeDyn approach45. The Martini force field defines chemical groups as CG beads (or super-atoms) parameterized to reproduce known thermodynamical observables associated to these groups. Molecules are constructed from these super-atoms using conventional bond, angle and dihedral potentials such that they reproduce the structure and flexibility of atomistic models43,44. ElNeDyn maintains the secondary and tertiary structures of CG proteins through an elastic network. In our model, each subunit is maintained independently, so that domain motions (reorientation of subunits) are possible. CG parameters for CLs and TGLs were taken from the works of Dahlberg et al.46 and Vuorela et al.47.
Simulation details
All CG simulations were performed using the GROMACS simulation package version 4.048. Conventional simulation setups associated with the use of Martini and ElNeDyn were used. The protein, membrane bilayer (POPC and CL) and aqueous phase (water and Na+ ions) were coupled independently to an external temperature bath49 at 300 K and the pressure was weakly coupled to an external bath49 at 1 bar using a semi-isotropic pressure scheme. Further details of the models, simulation protocols and limitations, as well as the methods used for analysis, are published as Supplementary Information.
The CG protein structure was inserted into a pre-equilibrated CL:POPC membrane patch. The system was energy minimized and simulated for 10 ns with position restraints on the backbone beads of the protein to relax the solvent and side-chains before starting production runs. The system shown in Fig. 1B contains the protein (1780 residues; 4117 beads), a POPC bilayer (966 lipids; 12,558 beads) including CLs (48 CLs; 1296 beads) and the aqueous phase (51,549 water beads and 103 sodium ions).
The potentials of mean force (PMF) of lipid molecules binding to CIV binding sites were computed using an umbrella sampling approach with the distance to the protein surface as reaction coordinate. Details are given as Supplementary Information.
Author Contributions
C.A. conducted the simulations. X.P. and C.A. built the models and analysed the trajectories. X.P., C.A. and S.J.M. designed the models and simulations. X.P., C.A. and S.J.M. interpreted the data and wrote the paper.
Supplementary Material
Supplementary material
Acknowledgments
The authors would like to thank Peter Rich, Jean-Pierre Mazat and Juan Elezgaray for insightful discussions, and Jaakko Uusitalo for helping performing the PMFs. This work was supported by the Netherlands Organisation for Scientific Research (NWO) (to X.P. and S.J.M., ECHO.08.BM.041).
References
- Claypool S. M. & Koehler C. M. The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 37, 32–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Voelker D. R. & Langer T. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192, 7–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G. Lipids of mitochondria. Biochim. Biophys. Acta 822, 1–42 (1985). [DOI] [PubMed] [Google Scholar]
- van Meer G., Voelker D. R. & Feigenson G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell. Biol. 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicco A. J. & Sparagna G. C. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. - Cell Physiol. 292, 33–44 (2007). [DOI] [PubMed] [Google Scholar]
- Schlame M. & Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 580, 5450–5455 (2006). [DOI] [PubMed] [Google Scholar]
- Saini-Chohan H. K. et al. Cardiolipin biosynthesis and remodeling enzymes are altered during development of heart failure. J. Lipid. Res. 50, 1600–1608 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. & Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Mileykovskaya E. & Dowhan W. Gluing the respiratory chain together. J. Biol. Chem. 277, 43553–43556 (2002). [DOI] [PubMed] [Google Scholar]
- Pfeiffer K. et al. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 (2003). [DOI] [PubMed] [Google Scholar]
- Gohil V. et al. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J. Biol. Chem. 279, 42612–42618 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang M., Mileykovskaya E. & Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 280, 29403–29408 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N. C., Zborowski J. & Talbert L. H. Cardiolipin-depleted bovine heart cytochrome-c-oxidase - binding stoichiometry and affinity for cardiolipin derivatives. Biochemistry 29, 8962–8969 (1990). [DOI] [PubMed] [Google Scholar]
- Qin L., Hiser C., Mulichak A., Garavito R. M. & Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 103, 16117–16122 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnarez C., Mazat J.-P., Elezgaray J., Marrink S.-J. & Periole X. Evidence for cardiolipin binding sites on the membrane-exposed surface of the respiratory chain complex III. J. Am. Chem. Soc., accepted (DOI: 10.1021/ja310577u). [DOI] [PubMed] [Google Scholar]
- Lange C., Nett J. H., Trumpower B. L. & Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 20, 6591–6600 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlák E. & Robinson N. C. Phospholipase A2 digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry 38, 14966–14972 (1999). [DOI] [PubMed] [Google Scholar]
- Musatov A. & Robinson N. C. Cholate-induced dimerization of detergent- or phospholipid-solubilized bovine cytochrome c oxidase. Biochemistry 41, 4371–4376 (2002). [DOI] [PubMed] [Google Scholar]
- Sedlák E., Panda M., Dale M. P., Weintraub S. T. & Robinson N. C. Photolabeling of cardiolipin binding subunits within bovine heart cytochrome c oxidase. Biochemistry 45, 746–754 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadenbach B., Jarausch J., Hartmann R. & Merle P. Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecyl sulfate-gel electrophoretic procedure. Anal Biochem 129, 517–521 (1983). [DOI] [PubMed] [Google Scholar]
- Robinson N. C., Strey F. & Talbert L. H. Investigation of the essential boundary layer phospholipids of cytochrome c oxidase using Triton X-100 delipidation. Biochemistry (1980). [DOI] [PubMed] [Google Scholar]
- Robinson N. C. Functional binding of cardiolipin to cytochrome c oxidase. J. Bioenerg. Biomembr. 25, 153–163 (1993). [DOI] [PubMed] [Google Scholar]
- Tsukihara T. et al. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl. Acad. Sci. U.S.A. 100, 15304–15309 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa-Itoh K. et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713–1725 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periole X., Huber T., Marrink S.-J. & Sakmar T. P. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J. Am. Chem. Soc. 129, 10126–10132 (2007). [DOI] [PubMed] [Google Scholar]
- Louhivuori M., Risselada H. J., van der Giessen E. & Marrink S.-J. Release of content through mechano-sensitive gates in pressurized liposomes. 107, 19856–19860 (2010). [DOI] [PMC free article] [PubMed]
- Grossfield A., Feller S. E. & Pitman M. C. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc. Natl. Acad. Sci. U.S.A. 103, 4888–4893 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfeld P. J., Hopkinson R., Ashcroft F. M. & Sansom M. S. P. PIP2-binding site in Kir channels: definition by multiscale biomolecular simulations. Biochemistry 48, 10926–10933 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aponte-Santamaria C., Briones R., Schenk A. D., Walz T. & de Groot B. L. Molecular driving forces defining lipid positions around aquaporin-0. Proc. Natl. Acad. Sci. U.S.A. 109, 9887–9892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer L. V. et al. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc. Natl. Acad. Sci. U.S.A. 108, 1343–1348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domański J., Marrink S.-J. & Schäfer L. V. Transmembrane helices can induce domain formation in crowded model membranes. BBA - Biomembranes 1818, 984–994 (2012). [DOI] [PubMed] [Google Scholar]
- Koivuniemi A., Vuorela T., Kovanen P. T., Vattulainen I. & Hyvönen M. T. Lipid exchange mechanism of the cholesteryl ester transfer protein clarified by atomistic and coarse-grained simulations. PLoS Comput. Biol. 8, e1002299– (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmschneider M. & Sansom M. S. P. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta 1512, 1–14 (2001). [DOI] [PubMed] [Google Scholar]
- Kates M., Syz J. Y., Gosser D. & Haines T. H. pH-dissociation characteristics of cardiolipin and its 2'-deoxy analogue. Lipids 28, 877–882 (1993). [DOI] [PubMed] [Google Scholar]
- Haines T. H. Anionic lipid headgroups as a proton-conducting pathway along the surface of membranes: a hypothesis. Proc. Natl. Acad. Sci. U.S.A. 80, 160–164 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines T. H. & Dencher N. A. Cardiolipin: a proton trap for oxidative phosphorylation. FEBS Lett. 528, 35–39 (2002). [DOI] [PubMed] [Google Scholar]
- Haines T. H. A new look at cardiolipin. Biochim. Biophys. Acta 1788, 1997–2002 (2009). [DOI] [PubMed] [Google Scholar]
- Tsukihara T. et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 (1996). [DOI] [PubMed] [Google Scholar]
- Gennis R. B. Multiple proton-conducting pathways in cytochrome c oxidase and a proposed role for the active-site tyrosine. BBA - Bioenergetics (1998). [Google Scholar]
- Namslauer A. & Brzezinski P. Structural elements involved in electron-coupled proton transfer in cytochrome c oxidase. FEBS Lett. 567, 103–110 (2004). [DOI] [PubMed] [Google Scholar]
- Brändén G., Gennis R. B. & Brzezinski P. Transmembrane proton translocation by cytochrome c oxidase. Biochim. Biophys. Acta 1757, 1052–1063 (2006). [DOI] [PubMed] [Google Scholar]
- Rich P. R. Protein Electron Transfer. 217–248 (BIOS Scientific Publishers Ltd., 1996). [Google Scholar]
- Marrink S.-J., Risselada H. J., Yefimov S., Tieleman D. P. & de Vries A. H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824 (2007). [DOI] [PubMed] [Google Scholar]
- Monticelli L. et al. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 4, 819–834 (2008). [DOI] [PubMed] [Google Scholar]
- Periole X., Cavalli M., Marrink S.-J. & Ceruso M. A. Combining an elastic network with a coarse-grained molecular force field: structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 5, 2531–2543 (2009). [DOI] [PubMed] [Google Scholar]
- Dahlberg M. Polymorphic phase behavior of cardiolipin derivatives studied by coarse-grained molecular dynamics. J. Phys. Chem. B 111, 7194–7200 (2007). [DOI] [PubMed] [Google Scholar]
- Vuorela T. et al. Role of lipids in spheroidal high density lipoproteins. PLoS Comput. Biol. 6, e1000964 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D. & Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 (2008). [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A. & Haak J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material