Abstract
Pancreatic cancer is the fourth leading cause of cancer death in both men and women in the United States. However, it has the poorest prognosis of any major tumor type, with a 5-yr survival rate of approximately 5%. Cigarette smoking, increased body mass index, heavy alcohol consumption, and a diagnosis of diabetes mellitus have all been demonstrated to increase risk of pancreatic cancer. A family history of pancreatic cancer has also been associated with increased risk suggesting inherited genetic factors also play an important role, with approximately 5–10% of pancreatic cancer patients reporting family history of pancreatic cancer. While the genetic basis for the majority of the familial clustering of pancreatic cancer remains unclear, several important pancreatic cancer genes have been identified. These consist of high penetrance genes including BRCA2 or PALB2, to more common genetic variation associated with a modest increase risk of pancreatic cancer such as genetic variation at the ABO blood group locus. Recent advances in genotyping and genetic sequencing have accelerated the rate at which novel pancreatic cancer susceptibility genes have been identified with several genes identified within the past few years. This review addresses our current understanding of the familial aggregation of pancreatic cancer, established pancreatic cancer susceptablity genes and how this knowledge informs risk assessment and screening for high-risk families.
Keywords: pancreatic cancer, genetics, familial cancer
INTRODUCTION
There will be an estimated 43,140 new cases of pancreatic cancer diagnosed in the United States in 2010 and 36,800 deaths [1]. In the United States, the age-adjusted incidence of pancreatic cancer is 13.5 per 100,000 for white males and 10.5 per 100,000 for white females. Risk is higher in blacks 17.1 per 100,000 and 14.8 per 100,000 in males and females, respectively [2]. Approximately, 80% of pancreatic cancer has metastasized at diagnosis and 5-yr survival rate is <5% [1]. Cigarette smoking, increased body mass index, heavy alcohol consumption, and a diagnosis of diabetes mellitus have been associated with increased pancreatic cancer risk [3–9]. Active cigarette smoking has been shown to be associated with a 1.74-fold increased risk (95% CI = 1.61–1.87) [4]. However, risk decreases in former smokers 1.2-fold (95% CI = 1.11–1.29) and no evidence of increased risk is observed 20 yr after smoking cessation [4]. Type 2 diabetes has also been associated with an increased risk of pancreatic cancer, overall odds ratio (OR) = 1.8 (95% CI = 1.5–2.1) compared with non-diabetics. Risk is highest in new onset diabetics (duration <2 yr before diagnosis) OR = 2.9 (95% CI = 2.1–3.9) and decreases with duration of diabetes OR = 1.4 (95% CI = 1.0–2.0) for diagnosis more the 15 yr before pancreatic cancer diagnosis [10]. Approximately 1% of new onset diabetics develop pancreatic cancer within 3 yr of their diabetes diagnosis [11]. Risk of pancreatic cancer increases with increasing body mass index OR = 1.55 (95% CI = 1.16–2.07) for individuals with BMI >35 compared to individuals with BMI of 18.9–24.9 [7]. Heavy alcohol consumption (≥6 drinks per day) has also been associated with increased pancreatic cancer risk OR = 1.46 (95% CI = 1.16–1.83) compared with individuals who drank <1 drink per day [12].
Inherited genetic factors also play an important role in pancreatic cancer risk. Pancreatic cancer, like all cancers is a fundamentally genetic disease caused by both inherited and acquired genetic mutations. Inherited genetic variation plays an important role in both the familial and non-familial (sporadic) occurrences of pancreatic cancer. It is estimated that 5–10% of pancreatic cancer patients have a family history of pancreatic cancer [13,14]. While the genetic mutations responsible for the majority of the clustering of pancreatic cancer in families have yet to be identified, several pancreatic cancer genes have been established, including both high-penetrance genes such as BRCA2 [15], STK11 [16], p16/CDKN2 [17], and PALB2 [18] and low-penetrance genes such as the ABO blood group locus [19]. The goal of this review is to provide an understanding of the importance of the clustering of pancreatic cancer, as well as other cancers, in families and genes that have been associated with an increased risk of pancreatic cancer.
FAMILIAL AGGREGATION OF PANCREATIC CANCER
The first studies to suggest that pancreatic cancer has an inherited genetic component were case reports of families in which there were multiple cases of pancreatic cancer [20]. Observational epidemiological studies including, case-control and cohort studies have demonstrated that individuals with a family history of pancreatic cancer are at an increased risk of developing pancreatic cancer themselves. Overall, case-control studies have estimated the odds of having a family history of pancreatic cancer are 1.9- to 13-fold higher in pancreatic cancer patients compared with healthy controls [21,22]. One of the larger studies of 484 pancreatic cancer cases and 2,099 controls, ascertained through population-based registries in three regions of the USA (Atlanta, Detroit, and New Jersey), found pancreatic cancer patients reported a first-degree relative with pancreatic cancer more often than controls OR = 3.2 (95% CI = 1.8–5.6), and the risk was higher OR = 3.6 (95% CI = 1.5–8.7) among those with an affected sibling compared to those with an affected parent OR = 2.6 (095% CI 1.2–5.4) [23]. A recent pooled analysis of data from 5 cohort and one case-control study estimated the odds of pancreatic cancer to be 1.76 higher (95% CI = 1.19–2.61) among individuals with at least one first-degree relative with pancreatic cancer compared to those without a family history of pancreatic cancer [21]. Risk was even higher in individuals with two or more first-degree relatives with pancreatic cancer, OR 4.26 (95% CI = 0.48–37.79) [21]. Prospective studies from the National Familial Pancreatic Tumor Registry (NFPTR, www.NFPTR.org) have demonstrated that incidence of pancreatic cancer is greater in children, parents and siblings of patients with familial pancreatic cancer (defined as a pair of first-degree relatives in the kindred with pancreatic cancer), standardized incidence ratio (SIR) 6.79, 95% CI 4.54–9.75, as compared to children, parents and siblings of patients with sporadic pancreatic cancer (families without a pair of first-degree relatives with pancreatic cancer), SIR = 2.41, (95% CI = 1.04–4.74).
Several studies have examined if the risk of pancreatic cancer varies by age of onset of pancreatic cancer in the family, however, these studies have yielded inconsistent results, with some studies shown no association between age-of-onset and risk while others showing that the age-of-onset of pancreatic cancer may be slightly younger in patient with a family history. In a series of patients reported by James et al. 36.7% of patients with a family history of pancreatic cancer developed disease before the age of 50 compared with 18.3% of sporadic pancreatic cancer patients (P < 0.017). Conversely, several case-control studies have demonstrated that the age-of-onset of pancreatic cancer in the proband was not associated with having a family history of pancreatic cancer [23,24]. The PacGENE consortium, a consortia of familial pancreatic cancer registries compared the mean age-at-onset of 466 familial pancreatic cancer probands (64.1 ± 11.8 yr) and 670 affected relatives (65.4 ± 11.6 yr) to that in the general US SEER population (70.0 ± 12.1 yr, P < 0.001, for both groups). However, a portion of this difference may reflect ascertainment bias because families self-enroll in these high-risk family registries. Prospective data from the NFPTR, demonstrated that having a young-onset patient (<50 yr) in the family did not alter the risk of pancreatic cancer for sporadic pancreatic cancer kindred members, however, pancreatic cancer risk was higher among members of familial pancreatic cancer kindreds with a young-onset case (<50) in the kindred SIR = 9.31 (95% CI = 3.42–20.28) than those without a young-onset case in the kindred SIR 6.34 = (95% CI = 4.02–9.51) [25].
Segregation analysis provided statistical evidence that there is a major gene(s) responsible for the clustering of pancreatic cancer in families. Analysis of 287 pancreatic cancer families ascertained through a Johns Hopkins Hospital index case estimated that 6/1,000 individuals were estimated to carry a high-risk pancreatic cancer genotype and the lifetime risk of pancreatic cancer (age 85) was 32% [26].
In addition to the clustering of pancreatic cancer within a family, co-aggregation in families of cancers at other sites along with pancreatic cancer suggests the presence of a predisposition gene that is responsible for a cancer syndrome. Analysis of data from several large cohort studies and the Mayo clinic case-control study estimated odds of prostate cancer (OR 1.45, 95% CI 1.12–1.89) was higher among individuals with a family history of pancreatic cancer [21]. In addition, data from a large series of pancreatic cancer patients at the Mayo clinic suggested the relatives of pancreatic cancer patients were at an increased risk of liver carcinoma (SIR = 2.70, 95% CI = 1.51–4.46). Cote et al. reported an excess risk of lymphoma (OR = 2.83, 95% CI = 1.02–7.86) in the relatives of 247 pancreatic cancer patients compared to that in 420 controls. Data from the NFTPR at Johns Hopkins supported the finding of an increased risk of colon cancer among the relatives of young onset (<age 50) pancreatic cancer probands, weighted standardized mortality ratio (wSMR) = 2.31 95% CI = 1.30–3.81. In addition, they reported that relatives of young onset pancreatic cancer patients were at higher risk of dying from cancers of the breast (wSMR = 1.98, 95% CI = 1.01–3.52), and prostate (wSMR = 2.31, 95% CI = 1.14–4.20). Additionally, in the NFPTR the relatives of familial pancreatic cancer probands had a significantly increased risk of dying from breast (wSMR 1.66, 95% CI 1.15–2.34), ovarian (wSMR = 2.05, 95% CI 1.10–3.49), and bile duct cancers (wSMR = 2.89, 95% CI 1.04–6.39), whereas the relatives of sporadic probands were at increased risk of dying from bile duct cancer (wSMR = 3.01, 95% CI 1.09–6.67) [27].
PANCREATIC CANCER SUSCEPTIBILITY GENES
While the genetic basis of the majority of the familial clustering of pancreatic cancer has yet to be elucidated, several important pancreatic cancer susceptibility genes have been identified. Recent advances in genome sequencing and array genotyping have accelerated the pace at which new pancreatic cancer susceptibility genes are being identified and have lead to the discovery of several new pancreatic cancer genes in the past 2 years and ongoing studies using these new methods are likely to identify many more pancreatic cancer genes in the coming years.
There is a wide spectrum of genetic variation involved in pancreatic cancer. This is similar to what is observed for most complex diseases (diseases due to both environmental and genetic basis). On one end of the spectrum is rare genetic variation that is often associated with a very high lifetime risk of developing diseases, high penetrance genes. On the opposite end of the spectrum is common genetic variation that typically confers only a minor or modest increase risk of disease, low penetrance genes. Genome-wide association studies in which the prevalence of common genetic variants is compared among cases and controls have great potential to identify these common genetic changes associated with pancreatic cancer risk. Conversely, family-based studies using linkage or genome-sequencing approaches are better suited to the identification of rare high penetrance genes. A list of established pancreatic cancer susceptibility genes is presented in Table 1.
Table 1.
Established and Suggested Pancreatic Cancer Susceptibility Gene
| Gene(s) | Genetic syndrome | Risk of pancreatic cancer | |
|---|---|---|---|
| High penetrance genes | BRCA2 | Hereditary breast and ovarian cancer | OR = 3.5 (95% CI 1.87–6.58) [28] |
| STK11/LKB1 | Peutz-Jeghers | SIR = 132 (95% CI 44–261) [75] | |
| PALB2 | Familial breast cancer | Increased [18] | |
| PRSS1 | Hereditary pancreatitis | SIR = 53 (95% CI 23–105) [50] | |
| SPINK1 | |||
| CDKN2A | Familial melanoma | SIR = 13–38 [76] | |
| Unknown | Familial pancreatic cancer | SIR = 6–32 [25,77] | |
| Possible high to moderate penetrance genes | Mismatch repair genes | Hereditary non-polyposis colorectal cancer | No effect up to SIR 8.6 (95% CI, 4.7–15.7) [42–44] |
| BRCA1 | Hereditary breast and ovarian cancer | No effect up to OR = 2.26 (95% CI 1.26 to 4.06) [30,34,35] | |
| Low penetrance genes | ABO | OR = 1.20 (95% CI1.12–1.28, per allele) [19] | |
| CFTR | OR = 1.40; 95% CI, 1.04–1.89 [78] | ||
| Genomic regions with genome-wide significant evidence of association (P < 5 × 10−8) | 1q32.1 | OR = 0.77, 95% CI 0.71–0.84 [58] | |
| 13q22.1 | OR = 1.26, 95% CI 1.18–1.35 [58] |
HIGH PENETRANCE GENES
BRCA2
Mutations in the BRCA2 have been shown to be associated with an increased risk of breast, ovarian, prostate, and pancreatic cancer. Analysis of a large series of BRCA2 mutation positive families ascertained for young onset breast and/or ovarian cancer estimated that BRCA2 mutations carriers have a 3.5-fold (95% CI 1.9–6.6) increased risk of pancreatic cancer compared with non-carriers [28]. Several studies have estimated the prevalence of mutations in the BRCA2 gene among pancreatic cancer patients. The first report of mutation in the BRCA2 gene among patients with pancreatic cancer was by Goggins et al. who reported that 7% (4/41) Johns Hopkins Hospital surgical patients with adenocarcinomas of the pancreas were found to have a germline deleterious mutation in the BRCA2 gene [29]. Subsequent studies indicate the prevalence of BRCA2 mutations in unselected pancreatic cancer patients may in fact be lower, ranging up to 4% among pancreatic cancer patients of Ashkenazi Jewish descent [30]. However, the prevelance of mutations may be significantly higher among individuals with a family history of pancreatic cancer. Murphy et al. [15] reported that 16% of patients with familial pancreatic cancer from kindreds where there were 3 or more pancreatic cancers harbor germline mutations in BRCA2. Similarly, Hahn et al. reported 12% of patients from familial pancreatic cancer kindreds had deleterious BRCA2 mutations and Couch et al. reported a large series of 180 pancreatic cancer patients who reported pancreatic cancer in a first- or second-degree relative identified 10 (6%) germline deleterious BRCA2 mutations. Of note, not all pancreatic cancer families with germline BRCA2 mutations also report a family history of breast and/or ovarian cancer and some sporadic pancreatic cancer kindreds with germline BRCA2 defects report no family cancer history at all.
PALB2
Mutations in the PALB2 gene, partner and localizer of BRCA2, have been shown to confer and increased risk of breast and pancreatic cancer. The PALB2 protein binds with BRCA2 protein stabilizing it in the nucleus, the BRCA2/PALB2 complex is part of the Fanconi Anemia DNA repair pathway and acts in double-stranded DNA repair. Approximately 1% of non-BRCA1/BRCA2 deficient familial breast cancers are caused by germline defects in PALB2 [31]. In 2009, using a whole-exome sequencing approach Jones et al. identified a truncating mutation in PALB2 in a patient with familial pancreatic cancer. Analysis of 96 additional familial pancreatic cancer patients from families with two or more pancreatic cancers identified three additional truncating mutations in the PALB2 gene [18]. Subsequent studies have identified additional PALB2 mutations in 1–3% of familial pancreatic cancer kindreds [32,33].
BRCA1
While there is strong evidence supporting an increased risk of pancreatic cancer among carriers of the BRCA2 gene, the role of BRCA1 gene mutation in pancreatic cancer susceptibility is less clear. While some large studies have estimated that there is a 2.26-fold (95% CI 1.26–4.06) increased risk of pancreatic cancer in BRCA1 mutation carriers [34], other studies have reported no increased in the prevalence of BRCA1 mutations in pancreatic cancer patients [30]. A study of 145 Ashkenazi Jewish pancreatic cancer patients did not detect an excess frequency of BRCA1 mutations, whereas the risk of BRCA2 mutations was significantly elevated OR = 3.85; 95% CI = 2.1–10.8 [30]. In addition analysis of 66 familial pancreatic cancer patients from NFPTR kindreds with three or more pancreatic cancer failed to identify any deleterious BRCA1 mutations despite the fact approximately half of the kindreds reported a family history of breast of ovarian cancer in addition to pancreatic cancer [35].
p16/CDKN2A
Germline mutations in the p16/CDKN2A gene are most commonly associated with familial melanoma. While only a portion of the familial aggregation of melanoma is due to germline defects in p16/CDKN2A, individuals with germline defects in p16/CDKN2A have been shown to have an increased risk of pancreatic cancer. It has been estimated that individuals from familial melanoma kindreds have a 13- to 22-fold increased risk of developing pancreatic cancer [36] and individuals who carry p16/CDKN2A mutations have a 38-fold increased risk of developing pancreatic cancer compared to the general population [37]. Pedigree analysis of p16/CDKN2A gene in 27 Dutch familial melanoma kindreds demonstrated that 19 of the 27 families harbored specific mutation, the p16-Leiden founder mutation, which is a 19 bp deletion in exon 2 of the p16/CDKN2A gene. The lifetime risk of pancreatic cancer in carriers of the p16-leiden founder mutation was estimated to be 17% by the age of 75 [38]. In addition to pancreatic cancer and melanoma, a variety of cancers have been reported to be increased familial melanoma kindreds, including carcinoma of the lung, and breast as well as sarcoma [17,36].
LYNCH SYNDROME
Lynch syndrome is an autosomal dominant hereditary disease characterized by early onset of colorectal cancer [39]. Patients with Lynch syndrome have germline mutations in genes coding for proteins associated with DNA mismatch repair genes hMSH2, hMLH1, hPMS1, hPMS2, and hMSH6/GTBP [39]. The lifetime risk (age 70) of colon cancer among mutation carriers is estimated to be 68.7% (95% CI = 58.6–78.9%) for men and 52.2% (95% CI = 37.6–66.9%) for women. Women who carry mutations in these genes are also at a very high lifetime risk of developing endometrial cancer, 54% (95% CI = 41.9–66.1%) [40]. In addition to colorectal and endometrial cancer, mutation carriers are also at an increased risk of developing cancers of the ovary, stomach, bile duct, kidney, bladder, ureter, and skin [39].
Lynch et al. [41] first reported that families with Lynch syndrome have an increased risk of developing pancreatic cancer. A recent study by Kastrinos et al., examined 147 families that carried a mutation a mismatch repair gene. These families had an 8.6-fold (95% CI = 4.7–15.7) increased risk of pancreatic cancer compared with the general population and an estimated 3.68% (95% CI = 1.45–5.88%) lifetime (age 70) risk of pancreatic cancer [42]. While Geary et al. [43] also reported an increased rate of pancreatic cancer HNPCC families another study by Barrow et al. [44] observed no excess risk. Adenocarcinomas of the pancreas in patients with HNPCC typically show microsatellite instability (MSI+) and a distinct medullary histopathology [45].
HEREDITARY PANCREATITIS
Hereditary pancreatitis, which typically manifests as repeated attacks of acute pancreatitis beginning in childhood and often leads to pancreatic insufficiency, is a rare inherited form of chronic pancreatitis [46]. Mutations in two genes have been found to cause hereditary pancreatitis. Mutations in the cationic trypsinogen (PRSS1) gene, causes an autosomal dominant form of hereditary pancreatitis [47,48] while mutations in the serine protease inhibitor gene (SPINK1) causes an autosomal recessive form of hereditary pancreatitis [49].
Individuals with hereditary pancreatitis have been shown to have a 53-fold (95% CI 23–105) increased risk for developing pancreatic cancer [50] and a lifetime risk (age 70) of pancreatic cancer of 30–40% [50,51]. The risk is even higher among smokers with hereditary pancreatitis who tend to develop disease 20 yr before non-smokers [51].
PEUTZ-JEGHERS SYNDROME [STK11]
Peutz-Jeghers syndrome, an autosomal dominant disorder characterized by hamartomatous polyps in the gastrointestinal tract and pigmented macules of the lips, buccal mucosa, and digits. Most cases of Peutz-Jeghers syndrome (>80%) are caused by mutations in the STK11 tumor suppressor gene, on chromosome 19p13, which encodes for a serine-threonine kinase. Individuals with Peutz-Jeghers syndrome have been shown to be at an increased risk of several gastrointestinal cancers including esophageal, stomach, small intestine, colon, lung, breast, uterine, ovarian, and pancreatic cancer [52]. Overall, individuals with Peutz-Jeghers syndrome have a lifetime (age 70) risk of cancer of 93% [53]. Giardiello et al. examined 210 individuals with Peutz-Jeghers syndrome and observed a 132-fold (95% CI 44–261) increased risk of pancreatic cancer compared to the general population. The lifetime risk of pancreatic cancer in Peutz-Jeghers patients had been estimated to be 11–32% [53].
Recent studies have suggested that pancreatic cancers that arise in Peutz-Jeghers patients may due to through the intraductal papillary mucinous neoplasm (IPMN) precursor pathway. IMPNs are cystic lesions of pancreas and therefore are detectable via computed tomography (CT) or endoscopic ultrasonography (EUS).
SUSCEPTIBILITY VARIANTS
Genome-wide association studies (GWAS) are powerful tools aimed at identifying relatively common variants that may increase risk of disease. Unlike candidate gene studies, in which a priori knowledge is used to identify plausible cancer susceptibility genes, GWAS have the advantage of examining common genetic variation (variation the occurs at frequencies >5–10%) in an unbiased manner. Through this approach we have been able to identify susceptibility genes for a wide variety of diseases including breast cancer, colon cancer, and pancreatic cancer [19,54]. The first GWAS for pancreatic cancer have just been completed, the PanScanI and II studies which jointly examined 550 K variants in 3,851 affected individuals (cases) and 3,934 unaffected controls drawn from 12 prospective cohort studies and eight case-control studies, with the majority of PanScan sites from North American or Europe and a smaller GWAS of 991 pancreatic cancer patients and 5,209 controls from Japan. It is important to note that the associated variants identified in association studies may not be causal, but only in linkage disequilibrium with the causal genetic variation. Additional functional and fine-mapping studies are necessary to identify the causal variants. In the sections below, the results of the GWAS as well as some candidate gene studies are detailed.
ABO
The ABO gene encodes α(1,3)-N-acetylgalactosaminyl transferase α catalyzing attachment of the A antigen and α(1,3)-galactosyl transferase attachment of the B antigen on oligosaccharide chains on the surface of erythrocytes, gastrointestinal mucosa and elsewhere. In the PanScanI study, 1,896 cases and 1,939 controls from 12 cohort studies and one case-control study were genotyped using an Illumina 550 K SNP panel and genome-wide significant evidence of association was reported to rs505922, a SNP in the ABO gene. This finding was replicated in an independent sample of 2,457 affected individuals and 2,654 controls from eight case-control studies from the PanScan II study. Joint analysis of these two phases yielded multiplicative per-allele OR 1.20 (95% CI = 1.12–1.28, combined P = 5.37 × 10−8) [19]. The SNP rs505922 is in complete linkage disequilibrium with the O/non-O blood group allele, such that individuals with non-O blood groups are at an increased risk of developing pancreatic cancer. Additional epidemiological studies, using reported ABO blood group and ABO serotype support the associated between ABO blood group and pancreatic cancer risk pancreatic cancer risk. Using SNP genotype data to determine serotype status, Wolpin et al. reported that compared to individuals with O blood type, the OR of pancreatic cancer for individuals with A, AB, and B blood type were 1.38 (95% CI = 1.18–1.62), 1.47 (95% CI = 1.07–2.02), and 1.53 (95% CI = 1.21–1.92), respectively [55]. In addition, a recent study by Risch et al. reported an association between non-O blood group and pancreatic risk. Furthermore, risk among non-O individuals was even higher if they were also seropositive for CagA-negative H. pylori (OR 2.78: 95% CI 1.49–5.20). Interestingly, no increased risk was observed for CagA-negative H. pylori individuals of O blood type (OR 1.28, 95% CI 0.62–2.64) [56]. ABO blood group has also been shown to be associated the several other GI diseases, most notably gastric cancer.
1q32.1
Combined analysis ~550 K SNPS in the PanScan I and II studies provided evidence of association to five SNPs in the NR5A2 region on 1q32.1, including one intronic SNP in this gene [57]. The SNP with the strongest association was rs3790844 multiplicative per-allele OR = 0.77, 95% CI 0.71–0.84 (P = 2.45 × 10−10) [58]. NR5A2 is a nuclear receptor subfamily 5, group A, member 2, also known as liver receptor homolog 1. Studies have suggested LRH-1/NR5A2 is involved in the initiation of intestinal tumors by influencing inflammation and cell cycle regulation [59]. Furthermore, decreased LRH-1 expression is related to a decrease in TNF-alpha levels [59].
13q22.1
In a large gene desert located on chromosome 13q22.1, two SNPs rs9543325 and rs9564966 with strong inter-marker LD R2 = 0.85 and R2 = 0.6 provided evidence of association in joint analysis of the PanScanI and II data. The multiplicative per-allele OR were 1.26, (95% CI = 1.18–1.35, P = 3.27 × 10−11) for rs9543325 and 1.21 (95% CI = 1.13–1.30, P = 5.86 × 10−8) [58].
CLPTM1/TERT
The CLPTM1/TERT region on 5p15.33 has been shown to be important in several cancers. In particular, recent genome-wide association studies of lung and other cancers provide strong evidence of association in this region [60,61]. In this region, there was evidence of association to rs401681 multiplicative per-allele OR 1.19 (95% CI = 1.11–1.27, P = 3.66 × 10−7) in the joint analysis of the PanScanI and II data [58]. TERT is a telomere maintenance gene, and abnormal telomere length has been demonstrated in many cancers including pancreatic cancer. In particular, the majority of pancreatic adenocarcinomas show chromosome ends lacking telomeric repeat sequences [62].
FOXQ1
Genome-wide association analysis of 991 pancreatic cancer patients and 5,209 controls from Japan provided evidence of association to rs9502893, multiplicative per-allele OR 1.29 (95% CI 1.17–1.43, P = 3.30 × 10−7) [63]. This SNP is located on 6q25.3 and is in a large LD block with the FOXQ1 gene. FOXQ1 is one of the 43 members of the Fox family of transcription factor genes. FOXQ1 protein is under-expressed in pancreatic cancers.
BICD1
Evidence of association was also reported for rs708224 on 12p11 multiplicative per-allele OR 3.30 (95% CI = 1.19–1.47, P = 3.30 × 10−7) in the Japanese genome-wide association study [63]. This SNP is located in the second intron of the BICD1 gene. Some studies have suggested a role for BICD1 in telomere length.
DPP6
DPP6 gene had been shown to be somatically altered in pancreatic cancers and it has been suggested that this genes plays a role in pancreatic cancer invasion. Several SNPs in the DPP6 gene provided some evidence of association in the Japanese population, the strongest evidence was for rs6464375 when examined using a recessive model of inheritance, OR 3.73 (95% CI = 2.24–6.21, P = 4.41 × 10−7). However, it is important to note there was limited evidence of association under an additive genetic model (P = 0.01) and given the small size of the study population, addition studies are needed to replicate these findings [63].
CFTR
Cystic fibrosis (CF) is an autosomal recessive condition caused by a defect in protein cystic fibrosis transmembrane conductance regulator (CFTR) which regulates mucus in the lung and gastrointestinal tract. Most patients with CF develop pancreatic insufficiency. There is some evidence supporting an increased risk of pancreatic cancer among CFTR mutation carriers. A study of 949 pancreatic cancer patients and 13,340 controls reported 5.3% of patients and 3.8% of controls carried a mutation in the CFTR gene (OR = 1.40, 95% CI = 1.04–1.89) [64]. However, other smaller studies have failed to detect and association between CFTR mutation and pancreatic cancer risk [65] (Table 1).
CANDIDATE SUSCEPTIBILITY VARIANTS
In addition to the findings from genome-wide associations studies, there have been numerous candidate gene studies conducted to identify pancreatic cancer susceptibility variants. Many of these studies focus on biological pathways known to be important in pancreatic cancer risk such as tobacco metabolism, DNA repair, inflammation, and folate metabolism as well as others. In addition, to the main effects of these genes, variation in these genes may also interact with environmental exposures to influence risk. While some of these studies have reported positive findings, the results are inconsistent across studies and further replication is needed to determine if variation in these genes influence pancreatic cancer risk. For example, cigarette exposure accounts for an estimated 26% of pancreatic cancers [4] therefore the genes involved in tobacco metabolism may be involved in pancreatic cancer susceptibility. Variation in the NAT1, NAT2, GSTM1, GSTT1, and GSTP1 genes have been studied to examine there association with pancreatic cancer, while some studies have suggested variation in the NAT1 many be involved in pancreatic cancer risk other have reported no association [66–68].
MULTIGENETIC CAUSALITY AND GENE/ENVIRONMENT INTERACTION
In addition to the direct effect of genetic factors on pancreatic cancer, it is likely that gene–gene interactions and gene–environment interactions may also play an important role in pancreatic cancer risk. While many studies have looked for interactions between candidate genes or between candidate genes and environmental factors, the results of these studies have been inconsistent. Given the individual effects of these genes and/or environmental factors on pancreatic cancer risk may be quite modest (OR ~1.5 or less) large studies are needed with both genetic data and detailed environmental risk factor profiles.
RISK ASSESSMENT AND SCREENING IN HIGH RISK POPULATIONS
Family history is a strong predictor of pancreatic cancer risk because it is suggestive of the presence of a genetic predisposition to pancreatic cancer, although shared environmental factors also play a role. As we have discussed above, risk increases with the number of affected relatives, however, family size, current age, or age of onset of pancreatic cancer and the exact relationship between affected family members are also important indicators of risk. Quantitative risk models have been developed for many types of cancers, including the Gail [69] and BRCAPRO [70] models for breast cancer. In April 2007, the first risk prediction tool for pancreatic cancer, PancPRO was released [71]. PancPRO builds on the foundation developed in BRCAPRO. In brief, this model predicts the probability that an individual has a pancreatic cancer susceptibility gene and their associated lifetime risk of pancreatic using family history information provided by the proband and family. The PancPRO model has been shown to provide accurate risk assessment for familial pancreatic cancer kindreds with an observed to predicted pancreatic cancer ratio of 0.83 (95% CI = 0.52–1.20) and high discriminatory ability, with an area under the curve of 0.75 (95% CI = 0.68–0.81) [71] and a user-friendly version is freely available as part of the CaGene package (www4.utsouthwestern.edu/breasthealth/cagene) and the R software implementation at (http://astor.som.jhmi.edu/BayesMendel).
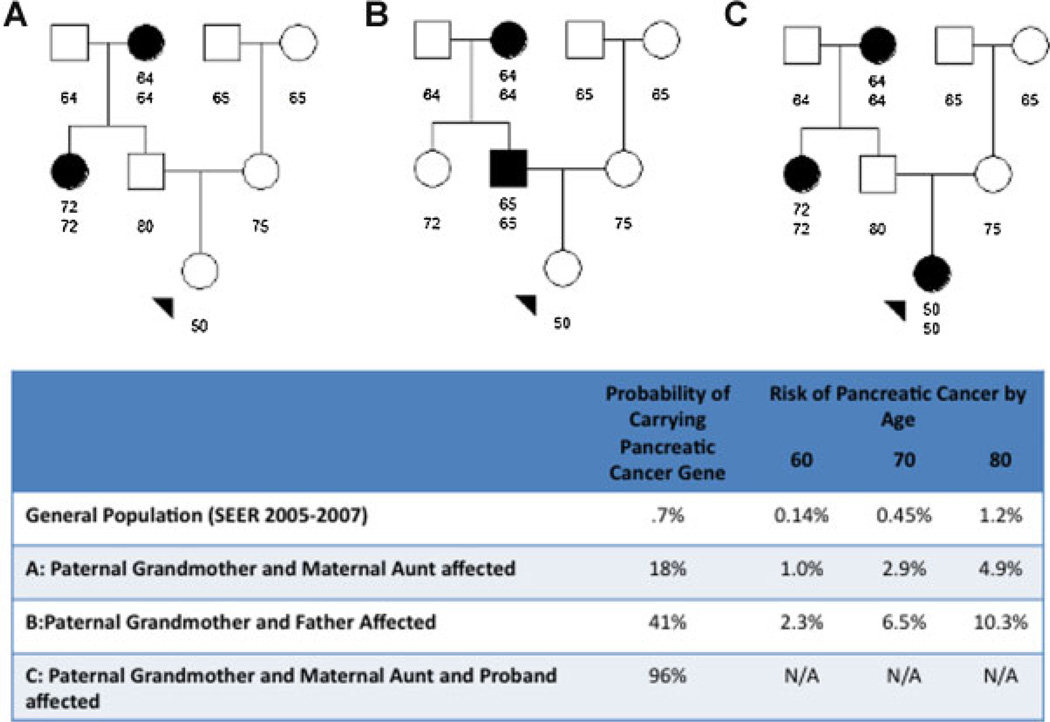
An example risk assessment using the PancPRO model is presented in Figure 1. Panel 1A displays a familial pancreatic cancer kindred. The proband is a 50-yr-old female whose grandfather developed pancreatic cancer at age 64 and paternal aunt at age 72, the other family members are pancreatic cancer free at the ages shown. Using the family history data provided, the PancPRO model estimates the probability of carrying a high-penetrance pancreatic cancer susceptibility gene of 18% and their risk of developing pancreatic cancer is 1.0%, 2.9%, and 4.9% by ages 60, 70, and 80, respectively. In contrast, general population age-averaged SEER rates for whites, a pancreatic cancer-free 50-yr-old has a risk of 0.14%, 0.45%, and 1.2% of developing pancreatic cancer by age 60, 70, and 80, respectively. If we permute the family history of the proband such that the father is affected with pancreatic cancer at age 65 and the paternal aunt is unaffected at age 72, the proband’s carrier probability increases to 41% and their risk of pancreatic cancer increases to 2.3%, 6.5%, and 10.3% by age 60, 70, and 80, respectively, as shown in panel 1B. Similarly, the proband in the original pedigree was diagnosed with pancreatic cancer at age 50, as shown in panel 1C, the proband’s probability of carrying a pancreatic cancer gene is estimated to be 96%.
Figure 1.
Example of risk assessment in a pancreatic cancer family using the PancPRO model.
SCREENING IN HIGH-RISK FAMILIES
The development of a successful early detection screening program for pancreatic cancer is needed to reduce the mortality of this disease. With current technologies, screening of the general population given the low incidence of pancreatic cancer is not practicable. However, there are currently several ongoing clinical trails evaluating the usefulness of screening of high-risk populations, including individuals with a strong family history of pancreatic cancer or individuals who carry a mutation in a established high-penetrance pancreatic cancer susceptibility gene (such as STK11, BRCA2, etc.). The majority of these studies have evaluated the use of EUS, CT, and/or magnetic resonance imaging (MRI) to detect small lesions in the pancreas. In the second phase of the “Cancer of the Pancreatic Screening Study (CAPS)” study Canto et al. examined seventy-eight patients with a strong family history of pancreatic cancer or with Peutz-Jeghers Syndrome, using EUS and CT, 8 of which were found to have a confirmed pancreatic neoplasia [72]. Vasen et al. [73] screened 79 patients with the p16-Lieden mutation with MRI and magnetic cholangiopancreatography (MRCP), after a median of 4 yr follow-up (range 0–10), seven individuals were found to have pancreatic cancer. These studies along with others [74] demonstrate that the early detection of pancreatic cancer is possible. Identification of pancreatic cancer susceptibility genes can help identify high-risk population who may benefit from the early detection screening tests currently under evaluation.
DISCUSSION
Over the past decade many important pancreatic cancer susceptibility genes have been identified, these include high-penetrance genes BRCA2, PALB2, PRSS1, SPINK1, STK11, and DNA mis-match repair genes and low penetrance genes CFTR, ABO. However, the genetic basis of the majority of pancreatic cancer remains unclear, as these established high-penetrance genes explain only 10–15% of the familial aggregation of pancreatic cancer. Recent technological improvements in genotyping and genome sequencing have accelerated the pace at which new pancreatic cancer genes have been discovered, including the recent discovery of PALB2 as a pancreatic cancer susceptibility gene. These new technologies will facilitate the discovery of additional pancreatic cancer genes in the coming years.
Identification of the inherited genetic basis of pancreatic cancer can not only lead to a better understanding of the etiology of this disease but can aid in the early detection of pancreatic cancer through the identification of high-risk populations who are most likely to benefit from the early detection screening programs currently under development.
ACKNOWLEDGMENTS
This work was supported by the Sol Goldman Pancreatic Cancer Research Center, NCI SPORE in Gastrointestinal Cancer CA62924.
Grant sponsor: Sol Goldman Pancreatic Cancer Research Center; Grant sponsor: NCI SPORE; Grant number: CA62924.
Abbreviations
- OR
odds ratio
- NFPTR
National Familial Pancreatic Tumor Registry
- SIR
standardized incidence ratio
- wSMR
weighted standardized mortality ratio
- MSI+
microsatellite instability
- IPMN
intraductal papillary mucinous neoplasm
- CT
computed tomography
- EUS
endoscopic ultrasonography
- GWAS
genome-wide association studies
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- EUS
endoscopic ultrasonography
- MRI
magnetic resonance imaging
- CAPS
Cancer of the Pancreatic Screening Study
- MRCP
magnetic cholangiopancreatography
REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Surveillance EaERP. National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch; 2003. SEER*Stat Database: Incidence SEER 9 Regs, Nov 2002 Sub (1973–2000) released April 2003, based on the November 2002 submission: [Google Scholar]
- 3.Lynch SM, Vrieling A, Lubin JH, et al. Cigarette smoking and pancreatic cancer: A pooled analysis from the pancreatic cancer cohort consortium. Am J Epidemiol. 2009;170:403–413. doi: 10.1093/aje/kwp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iodice S, Gandini S, Maisonneuve P, Lowenfels AB. Tobacco and the risk of pancreatic cancer: A review and meta-analysis. Langenbecks Arch Surg. 2008;393:535–545. doi: 10.1007/s00423-007-0266-2. [DOI] [PubMed] [Google Scholar]
- 5.Hassan MM, Bondy ML, Wolff RA, et al. Risk factors for pancreatic cancer: Case-control study. Am J Gastroenterol. 2007;102:2696–2707. doi: 10.1111/j.1572-0241.2007.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michaud DS, Vrieling A, Jiao L, et al. Alcohol intake and pancreatic cancer: A pooled analysis from the pancreatic cancer cohort consortium (PanScan) Cancer Causes Control. 21:1213–1225. doi: 10.1007/s10552-010-9548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arslan AA, Helzlsouer KJ, Kooperberg C, et al. Anthropometric measures, body mass index, and pancreatic cancer: A pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan) Arch Intern Med. 170:791–802. doi: 10.1001/archinternmed.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiao L, Berrington de Gonzalez A, Hartge P, et al. Body mass index, effect modifiers, and risk of pancreatic cancer: A pooled study of seven prospective cohorts. Cancer Causes Control. 21:1305–1314. doi: 10.1007/s10552-010-9558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everhart J, Wright D. Diabetes mellitus as a risk factor for pancreatic cancer. A meta-analysis. JAMA. 1995;273:1605–1609. [PubMed] [Google Scholar]
- 10.Li D, Tang H, Hassan MM, Holly EA, Bracci PM, Silverman DT. Diabetes and risk of pancreatic cancer: A pooled analysis of three large case-control studies. Cancer Causes Control. 22:189–197. doi: 10.1007/s10552-010-9686-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chari ST, Leibson CL, Rabe KG, Ransom J, de AM, Petersen GM. Probability of pancreatic cancer following diabetes: A population-based study. Gastroenterology. 2005;129:504–511. doi: 10.1053/j.gastro.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucenteforte E, La Vecchia C, Silverman D, et al. Alcohol consumption and pancreatic cancer: A pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4) Ann Oncol. 2011 doi: 10.1093/annonc/mdr120. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch HT, Smyrk T, Kern SE, et al. Familial pancreatic cancer: A review. Semin Oncol. 1996;23:251–275. [PubMed] [Google Scholar]
- 14.Hruban RH, Canto MI, Goggins M, Schulick R, Klein AP. Update on familial pancreatic cancer. Adv Surg. 44:293–311. doi: 10.1016/j.yasu.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy KM, Brune KA, Griffin C, et al. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: Deleterious BRCA2 mutations in 17% Cancer Res. 2002;62:3789–3793. [PubMed] [Google Scholar]
- 16.Su GH, Hruban RH, Bansal RK, et al. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am J Pathol. 1999;154:1835–1840. doi: 10.1016/S0002-9440(10)65440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch HT, Brand RE, Hogg D, et al. Phenotypic variation in eight extended CDKN2A germline mutation familial atypical multiple mole melanoma-pancreatic carcinoma-prone families: The familial atypical mole melanoma-pancreatic carcinoma syndrome. Cancer. 2002;94:84–96. doi: 10.1002/cncr.10159. [DOI] [PubMed] [Google Scholar]
- 18.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein AP, Hruban RH, Brune KA, Petersen GM, Goggins M. Familial pancreatic cancer. Cancer J. 2001;7:266–273. [PubMed] [Google Scholar]
- 21.Jacobs EJ, Chanock SJ, Fuchs CS, et al. Family history of cancer and risk of pancreatic cancer: A pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan) Int J Cancer. 127:1421–1428. doi: 10.1002/ijc.25148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghadirian P, Boyle P, Simard A, Baillargeon J, Maisonneuve P, Perret C. Reported family aggregation of pancreatic cancer within a population- based case-control study in the Francophone community in Montreal, Canada. Int J Pancreatol. 1991;10:183–196. doi: 10.1007/BF02924156. [DOI] [PubMed] [Google Scholar]
- 23.Silverman DT, Schiffman M, Everhart J, et al. Diabetes mellitus, other medical conditions and familial history of cancer as risk factors for pancreatic cancer. Br J Cancer. 1999;80:1830–1837. doi: 10.1038/sj.bjc.6690607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schenk M, Schwartz AG, O’Neal E, et al. Familial risk of pancreatic cancer. J Natl Cancer Inst. 2001;93:640–644. doi: 10.1093/jnci/93.8.640. [DOI] [PubMed] [Google Scholar]
- 25.Brune KA, Lau B, Palmisano E, et al. Importance of age of onset in pancreatic cancer kindreds. J Natl Cancer Inst. 2010;102:119–126. doi: 10.1093/jnci/djp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein AP, Beaty TH, Bailey-Wilson JE, Brune KA, Hruban RH, Petersen GM. Evidence for a major gene influencing risk of pancreatic cancer. Genet Epidemiol. 2002;23:133–149. doi: 10.1002/gepi.1102. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Brune KA, Visvanathan K, et al. Elevated cancer mortality in the relatives of patients with pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2829–2834. doi: 10.1158/1055-9965.EPI-09-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 29.Goggins M, Schutte M, Lu J, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 30.Ferrone CR, Levine DA, Tang LH, et al. BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J Clin Oncol. 2009;27:433–438. doi: 10.1200/JCO.2008.18.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tischkowitz MD, Sabbaghian N, Hamel N, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137:1183–1186. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slater EP, Langer P, Niemczyk E, et al. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 78:490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 34.Thompson D, Easton DF. Cancer incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 35.Axilbund JE, Argani P, Kamiyama M, et al. Absence of germline BRCA1 mutations in familial pancreatic cancer patients. Cancer Biol Ther. 2009;8:1–5. doi: 10.4161/cbt.8.2.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch HT, Fusaro RM, Lynch JF, Brand R. Pancreatic cancer and the FAMMM syndrome. Fam Cancer. 2008;7:103–112. doi: 10.1007/s10689-007-9166-4. [DOI] [PubMed] [Google Scholar]
- 37.Rutter JL, Bromley CM, Goldstein AM, et al. Heterogeneity of risk for melanoma and pancreatic and digestive malignancies: A melanoma case-control study. Cancer. 2004;101:2809–2816. doi: 10.1002/cncr.20669. [DOI] [PubMed] [Google Scholar]
- 38.Vasen HF, Gruis NA, Frants RR, van dV, Hille ET, Bergman W. Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden) Int J Cancer. 2000;87:809–811. [PubMed] [Google Scholar]
- 39.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 40.Hampel H, Stephens JA, Pukkala E, et al. Cancer risk in hereditary nonpolyposis colorectal cancer syndrome: Later age of onset. Gastroenterology. 2005;129:415–421. doi: 10.1016/j.gastro.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Lynch HT, Voorhees GJ, Lanspa SJ, McGreevy PS, Lynch JF. Pancreatic carcinoma and hereditary nonpolyposis colorectal cancer: A family study. Br J Cancer. 1985;52:271–273. doi: 10.1038/bjc.1985.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kastrinos F, Mukherjee B, Tayob N, et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA. 2009;302:1790–1795. doi: 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geary J, Sasieni P, Houlston R, et al. Gene-related cancer spectrum in families with hereditary non-polyposis colorectal cancer (HNPCC) Fam Cancer. 2008;7:163–172. doi: 10.1007/s10689-007-9164-6. [DOI] [PubMed] [Google Scholar]
- 44.Barrow E, Robinson L, Alduaij W, et al. Cumulative life-time incidence of extracolonic cancers in Lynch syndrome: A report of 121 families with proven mutations. Clin Genet. 2009;75:141–149. doi: 10.1111/j.1399-0004.2008.01125.x. [DOI] [PubMed] [Google Scholar]
- 45.Alexander J, Watanabe T, Wu TT, Rashid A, Li S, Hamilton SR. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol. 2001;158:527–535. doi: 10.1016/S0002-9440(10)63994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Bodic L, Bignon JD, Raguenes O, et al. The hereditary pancreatitis gene maps to long arm of chromosome 7. Hum Mol Genet. 1996;5:549–554. doi: 10.1093/hmg/5.4.549. [DOI] [PubMed] [Google Scholar]
- 47.Whitcomb DC, Preston RA, Aston CE, et al. A gene for hereditary pancreatitis maps to chromosome 7q35. Gastroenterology. 1996;110:1975–1980. doi: 10.1053/gast.1996.v110.pm8964426. [DOI] [PubMed] [Google Scholar]
- 48.Whitcomb DC, Gorry MC, Preston RA, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene [see comments] Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 49.Witt H, Luck W, Hennies HC, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 50.Lowenfels AB, Maisonneuve P, DiMagno EP, et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst. 1997;89:442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 51.Lowenfels AB, Maisonneuve P, Whitcomb DC, Lerch MM, DiMagno EP. Cigarette smoking as a risk factor for pancreatic cancer in patients with hereditary pancreatitis. JAMA. 2001;286:169–170. doi: 10.1001/jama.286.2.169. [DOI] [PubMed] [Google Scholar]
- 52.Giardiello FM, Welsh SB, Hamilton SR, et al. Increased risk of cancer in the Peutz-Jeghers syndrome. N Engl J Med. 1987;316:1511–1514. doi: 10.1056/NEJM198706113162404. [DOI] [PubMed] [Google Scholar]
- 53.van Lier MG, Wagner A, Mathus-Vliegen EM, Kuipers EJ, Steyerberg EW, van Leerdam ME. High cancer risk in Peutz-Jeghers syndrome: A systematic review and surveillance recommendations. Am J Gastroenterol. 105:1258–1264. doi: 10.1038/ajg.2009.725. (author reply 1265). [DOI] [PubMed] [Google Scholar]
- 54.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolpin BM, Chan AT, Hartge P, et al. ABO blood group and the risk of pancreatic cancer. J Natl Cancer Inst. 2009;101:424–431. doi: 10.1093/jnci/djp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Risch HA, Yu H, Lu L, Kidd MS. ABO blood group, Helicobacter pylori seropositivity, and risk of pancreatic cancer: A case-control study. J Natl Cancer Inst. 2010;102:502–505. doi: 10.1093/jnci/djq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petersen GMAL, Fuchs CS, Kraft Peter, Stolzenberg-Solomon RZ. A genome-wide association study identified pancreatic cancer susceptability loci on chromosomes 13q22.1, 1q31.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoonjans K, Dubuquoy L, Mebis J, et al. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc Natl Acad Sci USA. 2005;102:2058–2062. doi: 10.1073/pnas.0409756102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gisselsson D, Jonson T, Petersen A, et al. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Low SK, Kuchiba A, Zembutsu H, et al. Genome-wide association study of pancreatic cancer in Japanese population. PLoS One. 5:e11824. doi: 10.1371/journal.pone.0011824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McWilliams RR, Petersen GM, Rabe KG, et al. Cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations and risk for pancreatic adenocarcinoma. Cancer. 116:203–209. doi: 10.1002/cncr.24697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsubayashi H, Fukushima N, Sato N, et al. Polymorphisms of SPINK1 N34S and CFTR in patients with sporadic and familial pancreatic cancer. Cancer Biol Ther. 2003;2:652–655. [PubMed] [Google Scholar]
- 66.Lin Y, Yagyu K, Egawa N, et al. An Overview of Genetic polymorphisms and pancreatic cancer risk in molecular epidemiologic studies. J Epidemiol. 2011;21:2–12. doi: 10.2188/jea.JE20100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D, Jiao L, Li Y, et al. Polymorphisms of cytochrome P4501A2 and N-acetyltransferase genes, smoking, and risk of pancreatic cancer. Carcinogenesis. 2006;27:103–111. doi: 10.1093/carcin/bgi171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duell EJ, Holly EA, Bracci PM, Liu M, Wiencke JK, Kelsey KT. A population-based, case-control study of polymorphisms in carcinogen-metabolizing genes, smoking, and pancreatic adenocarcinoma risk. J Natl Cancer Inst. 2002;94:297–306. doi: 10.1093/jnci/94.4.297. [DOI] [PubMed] [Google Scholar]
- 69.Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 70.Berry DA, Parmigiani G, Sanchez J, Schildkraut J, Winer E. Probability of carrying a mutation of breast-ovarian cancer gene BRCA1 based on family history. J Natl Cancer Inst. 1997;89:227–238. doi: 10.1093/jnci/89.3.227. [DOI] [PubMed] [Google Scholar]
- 71.Wang W, Chen S, Brune KA, Hruban RH, Parmigiani G, Klein AP. PancPRO: Risk assessment for individuals with a family history of pancreatic cancer. J Clin Oncol. 2007;25:1417–1422. doi: 10.1200/JCO.2006.09.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Canto M, Goggins M, Hruban R, et al. Screening for early pancreatic neoplasia in high-risk individuals: A prospective controlled study. Clin Gast Hepatol. 2006;4:766–781. doi: 10.1016/j.cgh.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 73.Vasen HF, Wasser M, van Mil A, et al. Magnetic resonance imaging surveillance detects early-stage pancreatic cancer in carriers of a p16-Leiden mutation. Gastroenterology. 140:850–856. doi: 10.1053/j.gastro.2010.11.048. [DOI] [PubMed] [Google Scholar]
- 74.Brentnall TA, Bronner MP, Byrd DR, Haggitt RC, Kimmey MB. Early diagnosis and treatment of pancreatic dysplasia in patients with a family history of pancreatic cancer. Ann Intern Med. 1999;131:247–255. doi: 10.7326/0003-4819-131-4-199908170-00003. [DOI] [PubMed] [Google Scholar]
- 75.Giardiello FM, Brensinger JD, Tersmette AC, et al. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000;119:1447–1453. doi: 10.1053/gast.2000.20228. [DOI] [PubMed] [Google Scholar]
- 76.Goldstein AM, Fraser MC, Struewing JP, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med. 1995;333:970–974. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 77.Klein AP, Brune KA, Petersen GM, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–2638. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 78.McWilliams R, Highsmith WE, Rabe KG, et al. Cystic fibrosis transmembrane regulator gene carrier status is a risk factor for young onset pancreatic adenocarcinoma. Gut. 2005;54:1661–1662. doi: 10.1136/gut.2005.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]