Abstract
The TREX (transcription/export) complex couples transcription elongation to the nuclear export of mRNAs. In this article, we show that the poly(A)+ RNA-binding proteins Gbp2 and Hrb1, which resemble the serine-arginine-rich (SR) family of splicing factors found in higher eukaryotes, are specifically associated with the yeast TREX complex. We also show that Gbp2 and Hrb1 interact with Ctk1, a kinase that phosphorylates the C-terminal domain of RNA polymerase II during transcription elongation. Consistent with these findings, Gbp2 and Hrb1 associate with actively transcribed genes throughout their entire lengths. By using an RNA immunoprecipitation assay, we show that Gbp2 and Hrb1 also are bound to transcripts that are derived from these genes. We conclude that recruitment of the SR-like proteins Gbp2 and Hrb1 to mRNA occurs cotranscriptionally by means of association with the TREX complex and/or Ctk1.
Gene expression encompasses the transcription of the gene into mRNA, mRNA processing, and the export of the mature messenger ribonucleoprotein (mRNP) to the cytoplasm, where it directs the synthesis of proteins. In recent years, it has become clear that the different steps in gene expression are extensively coupled (1–3). The TREX (transcription/export) complex, which links transcription and mRNA export, is a key player in this coupling (4).
The TREX complex contains the heterotetrameric THO complex (Tho2, Hpr1, Mft1, Thp2) and the mRNA export factors Sub2 and Yra1. TREX complex components are also associated with Gbp2 and Hrb1 (4, 5). These proteins are highly homologous and contain serine-arginine-rich (SR) and RNA recognition motif (RRM) domains, both of which are hallmarks of the SR family of splicing factors in metazoans. Gbp2 and Hrb1 are also phosphorylated by the SR protein kinase Sky1 and are imported into the nucleus by means of Mtr10, the homologue of the metazoan SR protein import receptor (6). Together, these observations suggest that Gbp2 and Hrb1 are related to the metazoan SR protein family. However, it is not yet known why Gbp2 and Hrb1 are associated with TREX complex components.
The THO complex plays a role in transcription-dependent recombination and in transcription itself (7). Recently, it has been shown that the THO complex is required for efficient transcription elongation (8). Consistent with a function in transcription elongation, the THO complex is recruited to actively transcribed genes and moves along the ORF together with RNA polymerase II (4). Deletion of any one of the components of the THO complex leads to an mRNA export defect, indicating a function in mRNA export as well (4, 9). In addition, the THO complex is thought to play a role in packaging the newly synthesized mRNA. In Δhpr1 mutants, the nascent mRNA can both impair the efficiency of transcription elongation and promote recombination (10). In addition, an RNA:DNA hybrid forms in these cells (10). Thus, Hpr1 and the other components of the TREX complex could take over the nascent mRNA to keep elongation efficient. Along these lines, in THO deletion mutants the mRNA is more quickly degraded than in wild-type cells (9). This degradation of mRNA can be diminished by deletion of RRP6, a component of the nuclear exosome (9). Thus, the THO complex plays a role in transcription elongation as well as in the further formation and assembly of the mRNP.
The THO complex binds to the mRNA export factors Sub2 and Yra1 and is thus thought to recruit these mRNA export factors to the mRNA during transcription (4, 11). Sub2 is a DEAD-box RNA helicase that was originally identified as a splicing factor (12–14). Subsequently, it has been shown that Sub2 is required for the export of mRNAs derived from intron-containing genes as well as for intronless mRNAs (4, 15–18). Importantly, Sub2 is conserved throughout evolution, and a function in mRNA export also has been shown for its human homologue, UAP56, and its Drosophila melanogaster homologue, HEL (15). Sub2/UAP56, in turn, is thought to recruit Yra1/Aly (also called REF-BP) to the mRNA (17, 18). Yra1/Aly binds directly to the mRNA exporter Mex67-Mtr2 in yeast and Tap-p15 in higher eukaryotes (19–21). The mRNA exporter then transports the mRNP through the nuclear pore complex, which spans the nuclear envelope.
In this article, we show a specific association of Gbp2 and Hrb1 with the TREX complex. Moreover, we show that Gbp2 and Hrb1 are recruited to genes during transcription and bind to the mRNAs transcribed from these genes. Gbp2 and Hrb1 also interact with the C-terminal domain (CTD)-kinase Ctk1. Ctk1 phosphorylates serine 2 of the CTD of RNA polymerase II during the elongation phase of transcription and is required for efficient transcription elongation. Thus, interactions between Gbp2 and Hrb1 with the TREX complex and Ctk1 could provide a mechanism for cotranscriptional recruitment of these SR-like RNA-binding proteins to nascent mRNA transcripts.
Methods
Yeast Strains. Wild-type RS453, SUB2-TAP, HPR1-TAP, and GAL1::YLR454w strains have been described (4). Strains GBP2-TAP, HRB1-TAP, NPL3-TAP, and CTK1-TAP were generated by integration of the tandem affinity-purification (TAP) tag into the genome C-terminal of the GBP2, HRB1, NPL3, and CTK1 genes, respectively, by homologous recombination as described (22). The double knockout strain of GBP2 and HRB1 was generated by first crossing the GBP2 knockout strain obtained from Euroscarf (Frankfurt) with RS453 to obtain a Δgbp2 trp1– ade2– strain. This strain was mated to JBa (23) to obtain Δgbp2 trp1– ade2– ade3–. This strain was then mated to strain Δhrb1 trp1– ade2–, which was generated by crossing Δhrb1 obtained from Euroscarf with RS453, yielding strain Δgpb2 Δhrb1 trp1– ade2– ade3–. Into this strain the TAP tag was integrated C-terminally of the SUB2 and the HPR1 genes, respectively. For hemagglutinin (HA)-tagging of CTK1, the HA tag was integrated C-terminally of CTK1 into the genome of strains RS453, SUB2-TAP, HPR1-TAP, GBP2-TAP, and HRB1-TAP by homologous recombination. GBP2-myc GAL1::YLR454w and HRB1-myc GAL1::YLR454w were generated by integrating a 13xmyc tag C-terminally of GBP2 and HRB1, respectively, into the genome of the GAL1::YLR454w strain. The Δgpb2 ade2 ade3 strain for the synthetic lethal screen with Δmft1 was generated by mating the mft1 knockout strain (7) to Jba (23).
Plasmids. Plasmid pRS315-MFT1 was cloned by amplifying the ORF of MFT1 plus ≈500 base pairs upstream and downstream by PCR, generating SalI and BamHI sites, and cloning this fragment into the same sites of pRS315. The XbaI–SacI fragment of pRS315-MFT1 containing the MFT1 gene was subcloned into the XhoI and SacI sites of pHT4467Δ, generating pHT4467Δ-MFT1.
TAP Purifications. TAP purifications were done essentially as described (22). For RNase A treatment, whole-cell protein extracts were incubated with RNase A for 30 min at room temperature at a final concentration of 50 μg/ml. Mass spectrometry using tryptic digests from Coomassie-stained proteins was performed as described (23). Proteins were identified by using mascot (Matrix Science, London) and the msdb protein database (ftp://ftp.ncbi.nih.gov/repository/MSDB/msdb.nam).
Chromatin Immunoprecipitation (ChIP) Experiments. ChIP experiments were performed essentially as described (4). Cells were grown in selective media containing raffinose. At logarithmic phase (OD600 0.5–0.8), 2% galactose was added to the culture to induce expression of GAL promoter-driven genes for 1 h. For RNase treatment, RNase A was added to the whole-cell extract to a final concentration of 0.1 mg/ml. The whole-cell extract then was incubated for 30 min at room temperature, spun to remove any precipitates resulting from this treatment, and subjected to immunoprecipitation at room temperature for 3 h.
RNA Immunoprecipitation (RNA-IP) Experiments. Cells with proper tags were grown in selective media containing raffinose to logarithmic phase, and GAL promoter-driven gene expression was induced by the addition of 2% galactose for 1 h. The cells were spun down and washed once with 1× TBS. Cells from 40 ml of culture were resuspended in 1 ml of RNA-IP buffer (25 mM Tris·HCl, pH 7.5/100 mM KCl/0.2% Triton X-100/0.2 mM PMSF) supplemented with 1× protease inhibitor mixture, 5 mM DTT, and 10 units/ml RNasin. The cells were lysed by bead beating three times for 30 seconds. For DNase I treatment, 150 μl of lysate was incubated with 3 μl of DNase I at 30° for 1 h, spun to remove any precipitates, and subjected to immunoprecipitation in a final volume of 1.2 ml at 4° for 3 h. The beads were then washed five times with 1× RNA-IP buffer, and the RNAs in the pull-downs were precipitated according to a standard protocol. One microliter of lysate was processed in parallel to obtain a total input sample. Each RNA pellet was dissolved in 10 μl of H2O (the totals in 80 μl of H2O). Then 2.5 μl of each RNA sample was used for a 20-μl reverse transcription. Of this reverse transcription reaction, 2 μl each was used for a 10-μl analytical PCR.
Sequences of primers used for ChIP and RNA-IP experiments are available on request.
Miscellaneous. The synthetic lethal screen with the mft1 deletion allele was performed according to the method in ref. 19. Purified protein complexes were separated on SDS/11% polyacrylamide or 4–12% gradient gels (Invitrogen) and stained with Coomassie blue. HA-tagged Ctk1 was detected after Western blotting with an anti-HA antibody from rat (Roche Molecular Biochemicals) and goat-anti-rabbit antibody coupled to horseradish peroxidase (Sigma).
Results
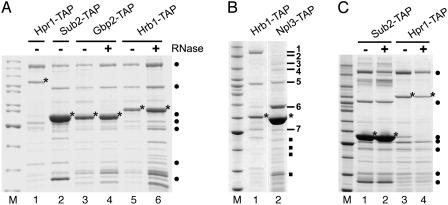
Gbp2 and Hrb1 Specifically Associate with the TREX Complex. Previously, Gbp2 and Hrb1 were detected in purifications of proteins that are known subunits of the TREX/THO complex (4, 5). To determine whether Gbp2 and Hrb1 are bound specifically to the TREX complex, we genomically TAP-tagged Gbp2 and Hrb1 and purified both proteins by using the TAP method (22). As shown in Fig. 1A, both Gbp2 and Hrb1 copurify all known components of the TREX complex (i.e., Tho2, Hpr1, Mft1, Thp2, Tex1, Sub2, and Yra1) in a highly specific manner.
Fig. 1.
Gbp2 and Hrb1 are components of the TREX complex. (A) TAP-tagged Gbp2 (lanes 3 and 4) and Hrb1 (lanes 5 and 6) were purified from Saccharomyces cerevisiae, and copurifying proteins were identified by mass spectrometry. For comparison, purification of TAP-tagged Hpr1 (lane 1) and Sub2 (lane 2) are shown. Tagged proteins are marked with an asterisk. TREX complex components are indicated by a filled circle. From to top bottom: Tho2, Hpr1, Hrb1 (only visible in lane 1), Sub2, Mft1, Gbp2 (all three proteins comigrate), Tex1, Thp2, and Yra1. Yeast lysates were treated with RNase A before purification of Gbp2 (lane 4) and Hrb1 (lane 6). (B) Npl3 does not associate with the TREX complex. TAP-tagged Npl3 was purified from yeast, and copurifying proteins were identified by mass spectrometry (lane 2). For comparison, a purification of Hrb1-TAP is shown (lane 1). Tagged proteins are indicated by an asterisk. Copurifying proteins are numbered and correspond as follows: 1, Kem1; 2, Snt1; 3, Tif-4F; 4, Rat1 and Tif-4F; 5, Sto1/Cbp80; 6, Ssb1, Ded1, and Pab1; and 7, Gbp2. Common contaminants are indicated by a filled square and correspond to a porin from Escherichia coli and ribosomal proteins. (C) Gpb2 and Hrb1 are not necessary for the stability of the TREX complex. Purification of TAP-tagged Sub2 (lanes 1 and 2) and Hpr1 (lanes 3 and 4) from wild-type yeast cells (lanes 1 and 3) and cells lacking both Gbp2 and Hrb1 (lanes 2 and 4). The tagged proteins are indicated by an asterisk, and TREX complex components are indicated by filled circles, as in A. The leftmost lanes show molecular weight markers (M).
Because Gbp2 and Hrb1 contain RRMs and bind to poly(A)+ RNA in vivo (6, 24), we asked whether the association of Gbp2 and Hrb1 with the TREX complex depends on RNA. Whole-cell lysates from cells expressing TAP-tagged Gbp2 or Hrb1 were treated with RNase A before TAP purification of Gbp2 or Hrb1. However, all components of the TREX complex remained bound to Gbp2 and Hrb1 upon incubation with RNase (Fig. 1 A, lanes 4 and 6).
Npl3 is an mRNA-binding protein that plays a role in mRNA export (25). Similar to Gbp2 and Hrb1, Npl3 contains SR and RRM domains. To investigate whether Npl3 is associated with the TREX complex, we TAP-tagged Npl3 and affinity-purified it. Although Npl3 is highly enriched in the TAP purification, this protein is not associated with the TREX complex (Fig. 1B). In contrast, Npl3 copurifies with a different set of proteins, which includes Kem1 (5′–3′ exonuclease), Snt1 (histone deacetylase), Tif4-F (translation initiation factor), Rat1 (5′–3′ exonuclease), Cbp80 (cap-binding complex), Ssb1 (ATPase involved in translation), Ded1 (ATP-dependent DEAD-box RNA helicase), and Pab1 (polyadenylate-binding protein). Moreover, Gbp2 is associated with affinity-purified Npl3 (Fig. 1B), most likely bound to the same mRNA as part of a nuclear mRNP. Taken together, Gbp2 and Hrb1, but not Npl3, specifically associate with the TREX complex in an RNA-independent manner.
Gbp2 and Hrb1 Are Not Required for TREX Complex Assembly. To determine whether Gbp2 or Hrb1 is required for TREX complex assembly, the TREX complex was affinity-purified by means of TAP-tagged Sub2 or Hpr1 from a yeast strain with disrupted GBP2 and HRB1 genes. This yeast strain is viable and not impaired in mRNA export (data not shown). In contrast, deletion of other THO complex components causes a temperature-sensitive growth phenotype, mRNA export defects, and dissociation of the THO complex from Sub2 (4). As shown in Fig. 1C, purification of Sub2-TAP or Hpr1-TAP from yeast cells lacking Gbp2 and Hrb1 still yields the typical TREX complex pattern of proteins. We conclude that Gbp2 and Hrb1 can associate specifically with the TREX complex, but that this interaction is not required for TREX complex formation (see Discussion).
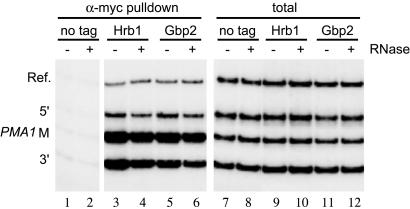
Gbp2 and Hrb1 Are Recruited to Actively Transcribed Genes. The above data are consistent with a model in which the RNA-binding proteins Gbp2 and Hrb1 associate with the TREX complex, which in turn functions as a platform to recruit these proteins to the nascent mRNA during transcription elongation (see below). If so, Gbp2 and Hrb1 should associate with activated genes during elongation. To test this possibility, we performed ChIP experiments with strains carrying myc-tagged Gbp2 or Hrb1. Exponentially growing cells were incubated with formaldehyde to stabilize protein–protein and protein–DNA interactions, and Gbp2 or Hrb1 was immunoprecipitated with an anti-myc antibody. After reversal of crosslinks, coimmunoprecipitated DNA was analyzed by PCR amplification with primers specific for the 5′ end, the middle (M), and the 3′ end of the ORF of the constitutively expressed PMA1 gene. Both Gbp2 and Hrb1 associate with the coding region of the PMA1 gene (Fig. 2, lanes 3 and 5). In contrast, a nontranscribed region is not associated with Gbp2 or Hrb1 (Fig. 2, lanes 3 and 5, Ref.). In addition to the PMA1 gene, Gbp2 and Hrb1 also associate with the transcribed region of other genes, including GAL1::YLR454w, ADH1, RPS5, and HYP2 (data not shown). Previously, we have shown that the THO complex moves along the actively transcribed gene together with RNA polymerase II (4). Thus, our data suggest that Gbp2 and Hrb1 together with the TREX complex move with elongating RNA polymerase II along activated genes.
Fig. 2.
Gbp2 and Hrb1 associate with actively transcribed genes. Gbp2 and Hrb1 associate with the constitutively expressed PMA1 gene. ChIP experiments were performed by using nontagged (no tag), Hrb1-myc-tagged (Hrb1), and Gbp2-myc-tagged (Gbp2) strains in the absence (–) or presence (+) of RNase A. Coimmunoprecipitated DNA was analyzed by PCR using primers specific for the 5′, middle (M), and 3′ regions of the PMA1 gene and a nontranscribed region as negative control (Ref.). As a positive control, PCRs were performed by using whole-cell extract as template (total).
Because Gbp2 and Hrb1 bind to RNA in vivo (6, 24), the association of Gbp2 and Hrb1 with chromatin could be mediated by RNA. To test this possibility, extracts were treated with RNase before ChIP analysis of Gbp2 or Hrb1. Under these conditions, Gbp2 and Hrb1 still are associated with activated chromatin (Fig. 2, lanes 4 and 6). However, association of Gbp2 and Hrb1 with chromatin is slightly (1.6- to 2.1-fold) reduced after treatment with RNase (Fig. 2, compare lanes 3 and 4 and also lanes 5 and 6), indicating that part of the interaction of Gbp2 and Hrb1 with chromatin may be mediated by RNA. Thus, Gbp2 and Hrb1 appear to interact directly with the chromatin but also may bind to the RNA in the vicinity of the chromatin/transcription complex.
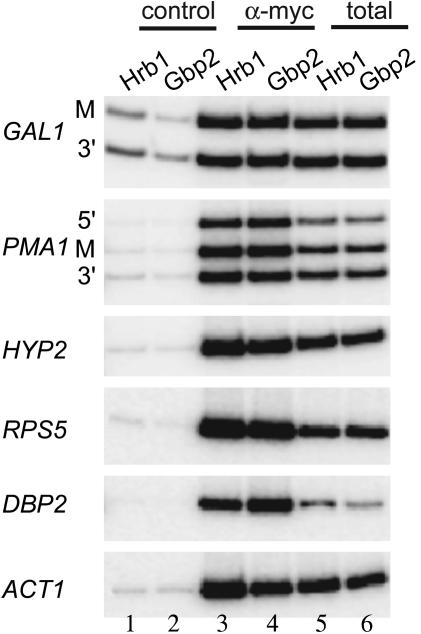
Gbp2 and Hrb1 Are Associated with mRNAs That Are Derived from Their Activated Genes. To investigate whether Gbp2 and Hrb1 are associated with mRNAs derived from the genes Gbp2 and Hrb1 associate with, we developed an RNA-IP assay. In this assay, Gbp2 or Hrb1 was immunoprecipitated from crosslinked cells, the DNA was digested by treatment with DNase, and coimmunoprecipitated mRNAs were analyzed by RT-PCR. Transcripts of several genes were tested, including an inducible gene (GAL1), constitutively expressed genes (PMA1, HYP1, and RPS5), and two intron-containing genes (DBP2 and ACT1). As shown in Fig. 3 (lanes 3 and 4), Gbp2 as well as Hrb1 are bound to all of these mRNAs. Taken together, the data are consistent with a model in which Gbp2 and Hrb1 are transferred from the chromatin to the nascent transcripts emerging from RNA polymerase II during transcription elongation.
Fig. 3.
Gbp2 and Hrb1 bind to transcripts of several genes. For RNA-IP analysis, RNA coimmunoprecipitated with myc-tagged Gbp2 or Hrb1 was reverse-transcribed and quantified by PCR using specific primers to regions of the GAL1, PMA1, HYP2, RPS5, DBP2, and ACT1 genes. As controls, immunoprecipitation with a control antibody (control) and RT-PCRs with whole-cell extracts as template (total) are shown.
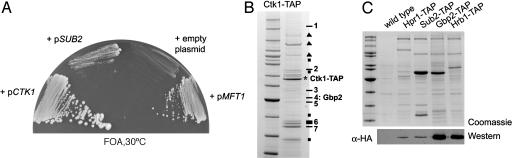
The CTD-Kinase Ctk1 Interacts Genetically with the TREX Component Mft1 and Associates Physically with Gbp2 and Hrb1. To identify additional factors that are crucial for TREX complex function in the living cell, we performed a synthetic lethal screen with the mft1 null mutant (Mft1 is a subunit of the TREX complex). We found that the CTK1 gene complements one of synthetic lethal mutants identified in this genetic screen (Fig. 4A). Moreover, SUB2 partially can suppress the synthetic lethal phenotype between Δmft1 and ctk1 (Fig. 4A and data not shown). Ctk1 is a cyclin-dependent kinase that phosphorylates the CTD of the largest subunit of RNA polymerase II (see ref. 26 and references therein). The CTD consists of heptapeptide repeats with a consensus of YSPTSPS. Serines 2 and 5 of this heptad repeat are phosphorylated and dephosphorylated during the transcription cycle. Ctk1 phosphorylates the CTD on Serine 2, which is the serine that is predominantly phosphorylated during the elongation phase of RNA polymerase II (26, 27). Moreover, Ctk1 associates with the entire ORF region of the gene (26), and the yeast protein as well as its metazoan homologue, CTDK-I, have been implicated in transcription elongation (28, 29). To assess whether the TREX complex interacts physically with Ctk1, we purified TAP-tagged Ctk1 and identified copurifying proteins by mass spectrometry (see Fig. 4B). Consistent with our genetic data, one of the proteins copurifying with Ctk1 is Gbp2. Thus, the physical interaction of Gbp2 and Ctk1 might provide a link between the TREX complex component Gbp2 and the transcription apparatus. To determine whether Hrb1 and other components of the TREX complex also associate with Ctk1, TAP-tagged Gbp2, Hrb1, Sub2, or Hpr1 was purified from yeast carrying HA-tagged Ctk1. A strain expressing HA-tagged Ctk1 in an otherwise wild-type background served as negative control. Copurification of Ctk1 was assessed by Western blotting with anti-HA antibodies. Ctk1 copurifies equally well with Gbp2 and Hrb1 (Fig. 4C, lanes 3 and 4), whereas less Ctk1 is associated with Sub2 and Hpr1 (Fig. 4C, lanes 1 and 2). The binding of Ctk1 to Sub2 and Hpr1 appears to be specific, because no signal is detected when a mock purification is carried out by using the strain carrying only HA-tagged Ctk1. We conclude that Ctk1 physically interacts with TREX complex components, in particular Gbp2 and Hrb1, which could provide a link between the TREX complex and the transcription machinery by means of Ctk1.
Fig. 4.
Gbp2 and Hrb1 are recruited to the transcription complex by their interaction with Ctk1. (A) Synthetic lethal interaction between MFT1 and CTK1. One of three synthetic lethal candidates of a synthetic lethal screen with a null allele of MFT1 is shown. Synthetic lethal candidate 20 was transformed with plasmids encoding CTK1, MFT1, SUB2, or an empty plasmid, restreaked onto plates containing 5′-fluoroorotic acid (5′-FOA), and grown for 5 days at 30°C. No growth indicates synthetic lethality. (B) Gbp2 associates with Ctk1 in vivo. TAP-tagged Ctk1 was purified from S. cerevisiae. Copurifying proteins were separated by SDS/PAGE, stained with Coomassie, and identified by mass spectrometry. TAP-tagged Ctk1 is marked by an asterisk. Identified proteins are indicated by numbers and correspond as follows: 1, Kem1; 2, major coat protein, virus L-A; 3, Imd3; 4, Gbp2; 5, Tef2; 6, Ctk2; and 7, Ctk3. Bands indicated by a filled triangle could not be identified unambiguously. Common contaminants are marked by filled squares (heat-shock proteins and ribosomal proteins). (C) TAP-tagged Sub2, Hrp1, Gbp2, or Hrb1 was purified from strains containing Ctk1 tagged with HA. A strain with HA-tagged Ctk1 only served as negative control (wild type). Copurification of Ctk1 was assessed by Western blotting using anti-HA antibodies. (Upper) The SDS/polyacrylamide gel of the EGTA eluate of the TAP purification stained with Coomassie blue. (Lower) The Western blot with anti-HA. The leftmost lanes of A and B show a protein standard.
Discussion
How mRNA-binding proteins are loaded onto their cognate transcripts, which are generated inside the nucleus during transcription, before the export of the mRNP to the cytoplasm, is an important issue of current research. Our work provides a clue as to how this process might occur in the case of the poly(A)+ RNA-binding proteins Gbp2 and Hrb1, which resemble members of the SR family of splicing factors found in higher eukaryotes. Importantly, Gbp2 and Hrb1 specifically bind to the TREX complex and, like the other components of the TREX complex, associate with actively transcribed genes during transcription elongation. Moreover, Gbp2 and Hrb1 are bound to mRNA transcripts that are derived from the genes with which these poly(A)+ RNA-binding proteins associate. According to these findings, TREX-bound Gbp2 and Hrb1 could be transferred from the TREX complex, which follows the elongating RNA polymerase II, to the nascent pre-mRNA during transcription.
Cotranscriptional binding of Gbp2 and Hrb1 to the mRNA might be a more general way to efficiently recruit poly(A)+ RNA-binding proteins to their cognate mRNAs. A later recruitment of these proteins to the mRNA (e.g., recruitment after transcription, splicing, and 3′ end processing of the pre-mRNA or recruitment in the cytoplasm) might be inefficient or unfavorable. Interestingly, a significant portion of Gbp2 and Hrb1 cosediments with polysomes, implicating a role of these mRNA-binding proteins during translation (M. Windgassen and H. Krebber, personal communication). Thus, Gbp2 and Hrb1 may employ a cotranscriptional loading onto the mRNA to ensure their later function as part of the mRNP in the cytoplasm.
We do not know whether Gbp2 and Hrb1 play an active role in mRNP assembly inside the nucleus. The fact that overexpression of Gbp2 impairs export of the mRNA may point to a role in mRNA export but could also result from titration of Gbp2-binding partner(s) crucial for mRNA export (e.g., TREX members). Thus, it is possible that Gbp2 and Hrb1 do not function in mRNA export but passively follow the mRNA to the cytoplasm. Consistent with this possibility is the finding that yeast cells lacking Gbp2 and Hrb2 are viable and do not display any detectable growth and mRNA export defects. In addition, cytoplasmic versions of Gbp2 and Hrb1 require ongoing transcription and the general mRNA export receptor Mex67-Mtr2 for their own export (24).
Notably, Gbp2 and Hrb1 appear to differ from Npl3, another RRM-containing and SR-like protein in yeast, in the way they are recruited to the mRNA. Whereas Npl3 associates with an entire gene, including its promoter region (30), Gbp2 and Hrb1 are absent from the promoter (data not shown). Moreover, Npl3 was shown to bind to the cap-binding complex (ref. 31 and Fig. 1B) and thus could be recruited to the 5′ end of the nascent pre-mRNA, whereas Gbp2 and Hrb1 use the TREX complex to reach the active gene. Consistent with these findings, nuclear export of Gbp2 and Hrb1 requires THO complex members, but nuclear export of Npl3 is THO-independent (24).
Our work also has revealed an unexpected genetic interaction between the TREX complex and Ctk1, a kinase that phosphorylates the CTD of the largest subunit of RNA polymerase II during transcription elongation. The genetic interaction between the THO complex component Mft1 and Ctk1 strongly points to an important functional connection. Indeed, we find that Gbp2 copurifies with TAP-tagged Ctk1, corroborating a Ctk1–Gbp2 interaction detected in a high-throughput screening in yeast (5). In addition to Gbp2, Hrb1, and, to a lesser extent, Sub2 and Hpr1 copurify Ctk1. Thus, Gbp2 and Hrb1 are candidate proteins that physically link the TREX complex to Ctk1 and to the transcription machinery. Taken together, our data suggest that the SR-like poly(A)+ RNA-binding proteins Gbp2 and Hrb1 are recruited to nascent mRNAs through a physical interaction with the TREX complex and/or Ctk1 during transcription elongation.
Acknowledgments
We thank Dr. J. Lechner, Sabine Merker, and Petra Ihrig (Biochemie-Zentrum Heidelberg, University of Heidelberg) for identification of proteins by mass spectrometry; Dr. Andres Aguilera (University of Seville, Seville, Spain) for the Δmft1 strain; and Marisa Oppizzi (Biochemie-Zentrum Heidelberg, University of Heidelberg) for providing the Δmft1 ade2– ade3– strain and plasmid pHT4467Δ-MFT1. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Leibniz-Program) and Fonds der Chemischen Industrie (to E.H.) and from the National Institutes of Health (to R.R.).
Abbreviations: mRNP, messenger ribonucleoprotein; HA, hemagglutinin; TAP, tandem affinity-purification; ChIP, chromatin immunoprecipitation; CTD, C-terminal domain of RNA polymerase II; RNA-IP, RNA immunoprecipitation; RRM, RNA recognition motif; SR, serine-arginine-rich.
References
- 1.Maniatis, T. & Reed, R. (2002) Nature 416, 499–506. [DOI] [PubMed] [Google Scholar]
- 2.Reed, R. (2003) Curr. Opin. Cell Biol. 15, 326–331. [DOI] [PubMed] [Google Scholar]
- 3.Stutz, F. & Izaurralde, E. (2003) Trends Cell Biol. 13, 319–327. [DOI] [PubMed] [Google Scholar]
- 4.Strasser, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A. G., Aguilera, A., Struhl, K., Reed, R. & Hurt, E. (2002) Nature 417, 304–308. [DOI] [PubMed] [Google Scholar]
- 5.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 6.Windgassen, M. & Krebber, H. (2003) EMBO Rep. 4, 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavez, S., Beilharz, T., Rondon, A. G., Erdjument-Bromage, H., Tempst, P., Svejstrup, J. Q., Lithgow, T. & Aguilera, A. (2000) EMBO J. 19, 5824–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rondon, A. G., Jimeno, S., Garcia-Rubio, M. & Aguilera, A. (2003) J. Biol. Chem. 278, 39037–39043. [DOI] [PubMed] [Google Scholar]
- 9.Libri, D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M. & Jensen, T. H. (2002) Mol. Cell. Biol. 22, 8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huertas, P. & Aguilera, A. (2003) Mol. Cell 12, 711–721. [DOI] [PubMed] [Google Scholar]
- 11.Zenklusen, D., Vinciguerra, P., Wyss, J. C. & Stutz, F. (2002) Mol. Cell. Biol. 22, 8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kistler, A. L. & Guthrie, C. (2001) Genes Dev. 15, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Libri, D., Graziani, N., Saguez, C. & Boulay, J. (2001) Genes Dev. 15, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, M. & Green, M. R. (2001) Genes Dev. 15, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatfield, D., Le Hir, H., Schmitt, C., Braun, I. C., Kocher, T., Wilm, M. & Izaurralde, E. (2001) Curr. Biol. 11, 1716–1721. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, T. H., Boulay, J., Rosbash, M. & Libri, D. (2001) Curr. Biol. 11, 1711–1715. [DOI] [PubMed] [Google Scholar]
- 17.Luo, M. L., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M. & Reed, R. (2001) Nature 413, 644–647. [DOI] [PubMed] [Google Scholar]
- 18.Strasser, K. & Hurt, E. (2001) Nature 413, 648–652. [DOI] [PubMed] [Google Scholar]
- 19.Strasser, K. & Hurt, E. (2000) EMBO J. 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stutz, F., Bachi, A., Doerks, T., Braun, I. C., Seraphin, B., Wilm, M., Bork, P. & Izaurralde, E. (2000) RNA 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou, Z., Luo, M. J., Straesser, K., Katahira, J., Hurt, E. & Reed, R. (2000) Nature 407, 401–405. [DOI] [PubMed] [Google Scholar]
- 22.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M. & Seraphin, B. (1999) Nat. Biotechnol. 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- 23.Bassler, J., Grandi, P., Gadal, O., Lessmann, T., Petfalski, E., Tollervey, D., Lechner, J. & Hurt, E. (2001) Mol. Cell 8, 517–529. [DOI] [PubMed] [Google Scholar]
- 24.Hacker, S. & Krebber, H. (December 15, 2003) J. Biol. Chem., 10.1074/jbc.C300522200.
- 25.Lee, M. S., Henry, M. & Silver, P. A. (1996) Genes Dev. 10, 1233–1246. [DOI] [PubMed] [Google Scholar]
- 26.Cho, E. J., Kobor, M. S., Kim, M., Greenblatt, J. & Buratowski, S. (2001) Genes Dev. 15, 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komarnitsky, P., Cho, E. J. & Buratowski, S. (2000) Genes Dev. 14, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jona, G., Wittschieben, B. O., Svejstrup, J. Q. & Gileadi, O. (2001) Gene 267, 31–36. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J. M. & Greenleaf, A. L. (1997) J. Biol. Chem. 272, 10990–10993. [DOI] [PubMed] [Google Scholar]
- 30.Lei, E. P., Krebber, H. & Silver, P. A. (2001) Genes Dev. 15, 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen, E. C., Stage-Zimmermann, T., Chui, P. & Silver, P. A. (2000) J. Biol. Chem. 275, 23718–23724. [DOI] [PubMed] [Google Scholar]