Abstract
The replication of eukaryotic positive-strand RNA virus genomes occurs through a complex process involving multiple viral and host proteins and intracellular membranes. Here we report a cell-free system that reproduces this process in vitro. This system uses a membrane-containing extract of uninfected plant protoplasts from which the vacuoles had been removed by Percoll gradient centrifugation. We demonstrate that the system supported translation, negative-strand RNA synthesis, genomic RNA replication, and subgenomic RNA transcription of tomato mosaic virus and two other plant positive-strand RNA viruses. The RNA synthesis, which depended on translation of the genomic RNA, produced virus-related RNA species similar to those that are generated in vivo. This system will aid in the elucidation of the mechanisms of genome replication in these viruses.
Numerous viruses contain positive-strand (i.e., messenger-sense) RNA genomes in their virions and replicate by means of negative-strand RNA that is complementary to the genomic RNA. These viruses, referred to as positive-strand RNA viruses, include the plant viruses tobacco mosaic virus, tomato mosaic virus (ToMV), brome mosaic virus (BMV), and turnip crinkle virus (TCV); many pathogenic human and animal viruses, including poliovirus and hepatitis C virus; and certain bacteriophages, such as Qβ. Upon infection, the genomic RNAs of these viruses are translated to generate the proteins that are required for replication of the viral genomes (replication proteins). Qβ replicase that catalyzes the complete cycle of Qβ RNA replication can be purified from infected cells as a soluble enzyme (1) and was shown to be composed of a viral replication protein and host-derived polypeptide subunits (2, 3). As one of the simplest models of genome replication, Qβ RNA replication has been extensively studied using this enzyme, resulting in many important discoveries (4). Inspired by these results, many attempts have been made to purify similar replicase enzymes from a variety of RNA viruses. However, for eukaryotic positive-strand RNA viruses, soluble replicase enzymes that reproduce the in vivo processes have not been obtained because the replication complexes are bound to membranes.
Replication proteins of eukaryotic positive-strand RNA viruses recruit viral genomic RNA templates to the cytoplasmic faces of endomembranes, forming membrane-bound replication complexes. This process is followed by the synthesis of negative-strand RNA and progeny genomic RNA, and in some cases, subgenomic RNAs (5, 6). Accumulating data suggest that viral replication complex formation, the key process in virus infection, requires dynamic molecular interactions among viral replication proteins, viral genomic RNA, and host proteins and membranes (7). To investigate the mechanisms of replication complex formation, a cell-free system that can reproduce this process is indispensable. Such systems have been established for poliovirus and encephalomyocarditis virus that belong to the picorna-like virus supergroup, by using extracts from uninfected mammalian cells (8, 9). In these systems, viral genomic RNAs are translated, replicated, and assembled into virus particles. However, no other instances of in vitro translation-coupled replication of eukaryotic positive-strand RNA viruses have been reported. We describe here the development of a plant cell extract-based in vitro translation-genome replication system applicable to multiple plant positive-strand RNA viruses: ToMV and BMV that belong to the alpha-like virus supergroup, and TCV that belongs to the carmo-like virus supergroup (5).
Materials and Methods
Viruses. ToMV (formerly referred to as TMV-L; ref. 10), BMV-M1 (11, 12) and TCV-B (13) were used. ToMV is a tobamovirus closely related to tobacco mosaic virus.
Preparation of the Cell-Free Extract. Removal of vacuoles from protoplasts was performed essentially as described by Sonobe (14). To prepare protoplasts, cells of a suspension-cultured tobacco cell line, BY-2 (15), were treated with a solution of wall-digesting enzymes [1% Cellulase Onozuka RS (Yakult Pharmaceutical, Tokyo)/0.1% Pectolyase Y-23 (Kyowa Chemical Products, Osaka)/0.1% Macerozyme R-10 (Yakult Pharmaceutical) in 0.4 M mannitol, pH 5.8]. One milliliter (packed cell volume) of the protoplasts were mixed with 5 ml of 30% (vol/vol) Percoll (Amersham Pharmacia) solution, overlaid on a 70% (vol/vol) (2 ml) to 40% (vol/vol) (6 ml) stepwise Percoll gradient, and centrifuged at 10,000 × g for 1 h at 25°C in a JS-24 rotor (Beckman Coulter). All Percoll solutions contained 0.7 M mannitol, 20 mM MgCl2, and 5 mM Pipes-KOH (pH 7.4). After centrifugation, evacuolated protoplasts were recovered from the 40–70% Percoll solution interface. The evacuolated protoplasts were suspended in four volumes of TR buffer [30 mM Hepes-KOH, pH 7.4/100 mM potassium acetate/2 mM magnesium acetate/2 mM DTT, 500 μg/ml Bentonite/1 tablet per 10 ml of Complete Mini protease inhibitor mixture (Roche Diagnostics)] and disrupted with ≈70 strokes in a Dounce homogenizer (Wheaton Science Products, Millville, NJ). Nuclei and nondisrupted cells were removed by centrifugation for 10 min at 500 × g at 4°C. The extract was frozen at –80°C in 0.5-ml aliquots until use to assure the elimination of intact cells. We hereafter refer to this evacuolated BY-2 protoplast lysate as BYL.
In Vitro Translation. Each 50-μl BYL translation reaction mixture contained 0.75 mM ATP, 0.1 mM GTP, 25 mM creatine phosphate, 25 μM each of methionine and leucine, 50 μM each of the other 18 amino acids required for translation, 80 μM spermine, 5 μg of creatine phosphokinase (Roche Diagnostics), 40 units of Recombinant RNasin Ribonuclease Inhibitor (Promega), 25 μl of BYL, and 1 μl of RNA [40 ng of Renilla luciferase mRNA (16), 1 μg or 100 ng of ToMV RNA, 100 ng of BMV RNA, or 100 ng of TCV RNA], adjusting the volume with TR buffer. The translation reactions were performed at 25°C. Rabbit reticulocyte lysate (RRL) was obtained from Promega, and wheat germ extract (WGE) (PROTEIOS; ref. 17) was from Toyobo (Tokyo). For RRL and WGE, translation reactions were performed according to the manufacturers' instructions.
In Vitro Viral RNA Replication. Translation reactions were incubated for 1 h, and 40 μl of each reaction was mixed with 10 μl of 5× R buffer [5 mM each of ATP, GTP, and UTP/125 μM CTP containing 20 μCi of [α-32P]CTP; 50 mM DTT/500 μg/ml actinomycin D (Wako Pure Chemicals, Osaka)/17 mM magnesium acetate; 1 Ci = 37 GBq] and incubated at 28°C for an additional 1 h.
Protein and RNA Analysis. Renilla luciferase activity was measured by using a Renilla Luciferase Assay System kit (Promega) and a Lumat LC9507 luminometer (Berthold, Nashua, NH). Western blot analysis was performed by using antibodies specific to the ToMV 130-kDa and 180-kDa proteins as described (18). Total RNA was purified with an Isogen LS kit (Nippon Gene, Tokyo). RNase protection assays for ToMV-related RNAs were performed by using the riboprobes P2M and P2P and an RNase protection kit (Roche Diagnostics) as described (19). A 32P-labeled RNA synthesis product from 8.3 μl of in vitro replication reaction mixture was annealed with 5 pmol of unlabeled P2M RNA for detection of positive-strand RNAs. Likewise, to detect negative-strand RNA, a 32P-labeled RNA synthesis product from 16.6 μl of in vitro replication reaction mixture was annealed with 5 pmol of unlabeled P2P RNA. To estimate the amount of negative-strand RNA, unlabeled RNA product from 50 μl of in vitro replication reaction mixture was annealed with 0.07 pmol of 32P-labeled P2P RNA, and RNase protection assays were performed. As a quantity standard, P2M RNA was used. S1 nuclease treatment (20), inoculation of BY-2 protoplasts with ToMV RNA, and subsequent protein and RNA analyses (19) were performed as described. 3H-labeled RNA bands were visualized by using fluorography (21).
Results and Discussion
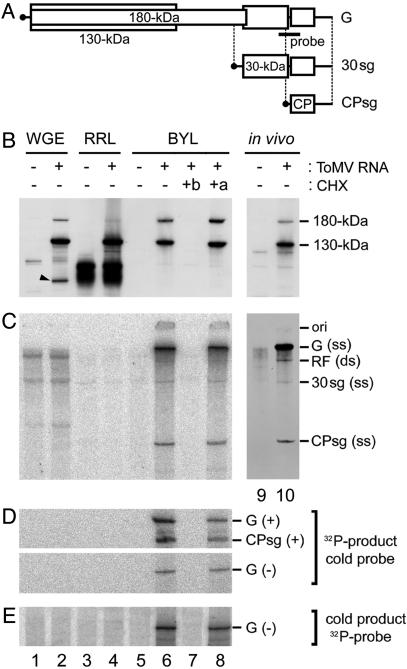
Inability of RRL and WGE to Support ToMV RNA Replication. As a first step to establishing cell-free replication systems for plant viruses, we translated the genomic RNA of ToMV in two commercial cell-free translation systems by using RRL or WGE, and examined whether viral RNA replication occurred. The ToMV genome encodes four proteins: a 130-kDa protein and its read-through product, a 180-kDa protein (these are both involved in viral RNA synthesis), a 30-kDa protein that is necessary for cell-to-cell movement of the virus, and the coat protein. The 130- and 180-kDa replication proteins are synthesized from the genomic RNA, and the other two proteins are synthesized from respective subgenomic RNAs (Fig. 1A) (22). In RRL and WGE, ToMV RNA was translated to produce the 130- and 180-kDa proteins (Fig. 1B, lanes 1–4). However, no viral RNA replication products were detected in either system after adding ribonucleoside triphosphates (Fig. 1 C–E, lanes 1–4). The failure to detect any RNA replication products may result from a lack of endomembranes on which the ToMV replication complex is formed (18), or other necessary host factors, because these translation systems are derived from nonhost organisms.
Fig. 1.
Cell-free translation of ToMV RNA and subsequent RNA synthesis in WGE, RRL, and BYL. (A) Schematic representation of the ToMV genomic RNA (G) and subgenomic RNAs for the 30-kDa protein (30sg) and coat protein (CPsg) (not to scale). The region covered by the RNase protection probes is also indicated and corresponds to probes P2P and P2M described in ref. 18. (B–E) Translation and RNA synthesis reactions were performed by using WGE (lanes 1 and 2), RRL (lanes 3 and 4), and BYL (lanes 5–8). ToMV RNA (1 μg for B and 0.1 μg for C–E) was added to the translation reaction mixtures (50 μl) in lanes 2, 4, 6, 7, and 8, but not in lanes 1, 3, and 5, and incubated for 1 h. Ten micrograms of cycloheximide (CHX) were added before starting (+b, lane 7) or immediately after terminating (+a, lane 8) the translation reaction (50 μl). The translation reaction mixtures (40 μl) were then mixed with 10 μl of 5× R buffer and were incubated for an additional 1 h. For comparison, in vivo samples from mock-inoculated (lane 9) and ToMV-inoculated (lane 10) BY-2 protoplasts cultured in medium containing actinomycin D were simultaneously analyzed in B and C.In B, the translation products were separated by SDS/PAGE using a 4–12% gradient gel, and the 130- and 180-kDa proteins were detected by immunoblotting. The “in vivo” samples (lanes 9 and 10) were harvested 8 h after inoculation. The positions of the 130- and 180-kDa proteins are indicated to the right. In C, 32P-labeled RNA products were separated on a 2.4% polyacrylamide gel containing 8 M urea and visualized by using a Bio Imaging Analyzer (BAS 1000, Fuji). To prepare in vivo samples (lanes 9 and 10), [3H]uridine was added to the culture medium at 6 h after inoculation, and RNA was extracted at 8 h after inoculation and analyzed as described above. 3H-labeled bands were visualized by fluorography. The positions of the genomic, subgenomic, and RF RNAs are indicated to the right of C. ss, single stranded; ds, double stranded. The position of the origin of electrophoresis (ori) is also indicated. In D, the 32P-labeled RNA products visualized in C were subjected to RNase protection assays using the unlabeled (“cold”) probes shown in A; these probes hybridize with the positive- (Upper) or negative- (Lower) strand ToMV RNAs. In E, unlabeled RNA products synthesized in reaction mixtures lacking [α-32P]CTP and containing 1 mM CTP were subjected to RNase protection assays using a 32P-labeled probe (A) that hybridizes with the negative-strand ToMV RNA. In D and E, protected RNA was separated by using 8 M urea/3% PAGE, and 32P-labeled bands were visualized as described above. The positions of the protected bands corresponding to the ToMV genomic RNA [G(+)], coat protein subgenomic RNA [CPsg(+)], and the negative-strand genomic RNA [G(–)] are indicated to the right.
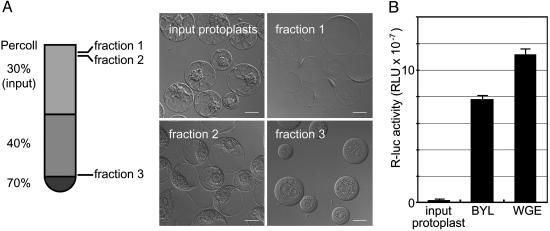
Replication of ToMV RNA in a Cell-Free Extract of Evacuolated Protoplasts. To circumvent the possible problems mentioned above, we formulated endomembrane-containing cell-free extracts from suspension-cultured tobacco BY-2 cells (15), in which ToMV efficiently multiplies (23). Most plant cells have lytic vacuoles that occupy the majority of the intracellular space and contain hydrolytic enzymes such as proteases and ribonucleases, which could inhibit RNA translation or replication (24). Although BY-2 cells have relatively undeveloped lytic vacuoles, an extract derived from the direct disruption of BY-2 protoplasts had poor translation activity (Fig. 2B). To avoid these factors, we removed the vacuoles from BY-2 protoplasts before disrupting the cells, using the method of Sonobe (14), in which protoplasts are subjected to centrifugation in a colloidal silica (Percoll) stepwise density gradient. This method is based on the lower density of the lumen of the vacuoles as compared with that of the cytoplasmic space and nuclei. Upon centrifugation in a density gradient, a protoplast was divided into two types of protoplasts: a low-buoyant-density, vacuole-containing protoplast (Fig. 2 A, fraction 1) and a high-buoyant-density evacuolated protoplast (Fig. 2 A, fraction 3). The evacuolated protoplasts that concentrated at the interface between the 40% and 70% Percoll layers were collected, washed, and disrupted by using a Dounce homogenizer. The nuclei and undisrupted cells were removed by low-speed centrifugation to obtain a cell-free, membrane-containing extract, referred to as BYL. Translation was much more efficient in BYL than in a crude extract of BY-2 protoplasts. Renilla luciferase RNA translation (25°C, 1 h) in BYL reached ≈70% of that in WGE (Fig. 2B). When ToMV RNA was translated in BYL, the 130- and 180-kDa proteins were produced, with a 180- to 130-kDa ratio higher than those in the RRL and WGE systems (Fig. 1 A). This finding may represent either more efficient translational read-through at the termination codon of the 130-kDa protein (25), lower activity of contaminating ribonucleases or proteases in the BYL reaction, or both. The appearance of smaller-than-full-length translation products in the WGE system (the arrowhead in Fig. 1B, lane 2) supports the latter possibility. Incubation of 50 fmol (0.1 μg) of the genomic RNA of ToMV in 50 μl of standard BYL translation reaction mixture (25°C, 1 h) resulted in the production of ≈400 and 40 fmol of the 130- and 180-kDa replication proteins, respectively (Fig. 1B and data not shown).
Fig. 2.
Preparation of evacuolated BY-2 protoplasts. (A) Evacuolation method. Representative Nomarski images of protoplasts are also shown. (Scale bars = 20 μm.) For descriptions of fractions 1 and 3, see Replication of ToMV RNA in a Cell-Free Extract of Evacuolated Protoplasts. The protoplasts in fraction 2 contained both vacuoles and nuclei, suggesting that these cells escaped the evacuolation process during centrifugation. (B) Comparison of the translation activities of an extract of untreated protoplasts (input protoplast), BYL (an extract of “fraction 3” cells), and PROTEIOS WGE. Renilla luciferase RNA (40 ng) (16) was translated for 1 h at 25°Cin50-μl reaction mixtures containing the above extracts. The y axis represents relative light units per μl of reaction mixture. Data are the averages and standard deviations of three experiments using independent batches of extracts.
After the 1-h translation reaction, [α-32P]CTP and the other RNA precursors were added and incubation was continued at 28°C for an additional 1 h. Fig. 1C (lane 6) shows the resulting pattern of RNA synthesis products, which are similar to those observed in ToMV-infected protoplasts in vivo (Fig. 1 C, lane 10): genomic RNA (single-stranded); two subgenomic RNAs for the 30-kDa protein and the coat protein, respectively (single-stranded); and genome-length double-stranded RNA (replicative form; RF) (22) (also see Fig. 3 for the sensitivity of these RNA species to S1 nuclease, which digests single-strand RNA). This finding suggests that ToMV RNA replication occurs in the BYL system in a manner similar to that in ToMV-infected plant cells. Translation of 10-fold more ToMV RNA (500 fmol/50 μl translation reaction) yielded ≈10-fold more 130- and 180-kDa proteins, but produced similar amounts of RNA synthesis products.
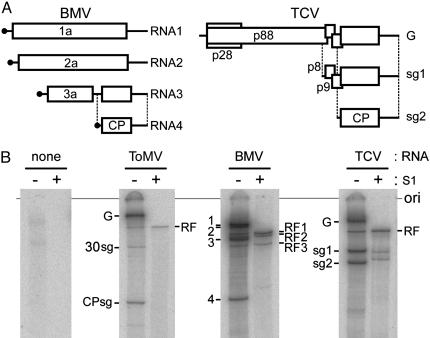
Fig. 3.
Cell-free replication of BMV and TCV RNAs in BYL. (A) Schematic representation of BMV and TCV genomic and subgenomic RNAs (not to scale). (B) Cell-free replication of ToMV, BMV, and TCV RNAs. After translation and RNA synthesis reactions in BYL, total RNA was purified and analyzed as in Fig. 1C. Samples were either treated with S1 nuclease to remove single-stranded RNA (lanes labeled with “+”) or left untreated (lanes labeled with “–”). Positions are indicated for the following RNAs: for ToMV, the genomic RNA (G), the subgenomic RNAs for the 30-kDa protein (30sg) and the coat protein (CPsg), and the RF RNA (RF); for BMV, RNA 1 to 4 (1 to 4) and the RF RNAs for RNA 1 (RF1), RNA 2 (RF2), and RNA 3 (RF3); for TCV, the genomic RNA (G), the subgenomic RNAs for p8-p9 (sg1) and the coat protein (sg2), and the RF RNA. The position of the origin of electrophoresis (ori) is also shown.
A ribonuclease protection assay of the 32P-labeled cell-free reaction products, using the unlabeled probes shown in Fig. 1 A, demonstrated that 32P was incorporated internally in both strands (Fig. 1D, lane 6). Consistent with the fact that higher amount of positive-strand RNA is synthesized than negative-strand RNA in ToMV-infected protoplasts, the amount of 32P in the genomic RNA was 2–10 times greater than that in the negative-strand RNA in this system (data not shown). Similar assay that used unlabeled cell-free translation–replication products and a 32P-labeled probe showed that ≈0.1 fmol of negative-strand RNA was produced in a 50-μl standard reaction mixture containing 40 fmol of input ToMV RNA and 20 μl of BYL (Fig. 1E, lane 6, and data not shown). This level of negative-strand RNA accumulation (0.1 fmol per 20 μl of BYL) is lower than the 1/10 level found for 20 μl of evacuolated protoplast lysate prepared from ToMV-infected protoplasts (data not shown). Although the level of negative-strand RNA accumulated in BYL was low relative to what was observed for infected protoplasts, the new RNA from BYL incubation appears to have resulted from a replication-like process. No RNA products of either polarity were detected when the translation inhibitor cycloheximide was added before the translation reaction (Fig. 1 C–E, lane 7); however, replication products were detected when cycloheximide was added immediately after terminating translation reaction (Fig. 1 C–E, lane 8). These results strongly suggest that the observed ToMV RNA replication requires the production of viral replication proteins.
In the coupled translation–replication system for poliovirus, the increase of RNase-resistant infectivity was observed. This increase depended not only upon the translation but also on the de novo replication of poliovirus RNA, suggesting that encapsidation of poliovirus RNA is coupled to replication and that encapsidation of input RNA does not occur (8). In contrast to the poliovirus system, the coat protein of ToMV likely can encapsidate the input RNA (26). Therefore, in the cell-free translation–replication system of ToMV where the increase in ToMV RNA over the input RNA was estimated to be <1/40 (as mentioned above, ≈0.2 to 1 fmol of ToMV RNA was synthesized from 40 fmol of input ToMV RNA), no assay would be able to detect the increase in infectivity derived from the newly synthesized ToMV RNA.
Replication of BMV and TCV RNAs in BYL. To evaluate the versatility of the BYL system, we tested the replication of genomic RNAs of two additional plant positive-strand RNA viruses, BMV and TCV, after translation in this extract. BMV is a member of the alpha-like virus supergroup, but, unlike ToMV, its genome is divided into three RNA segments (RNA 1–3). Three of the four proteins encoded by the BMV genome are translated from the genomic RNAs, and the coat protein is translated from subgenomic RNA 4, which represents the 3′ proximal part of RNA 3 (Fig. 3A). TCV is a member of the carmo-like virus supergroup, which is not closely related to either the alpha-like virus supergroup or the picorna-like virus supergroup (5). Two of the five proteins encoded by the TCV genome are translated from its genomic RNA, and the rest are translated from two subgenomic RNAs (sg1 and sg2) (Fig. 3B).
Translation and subsequent RNA synthesis reactions using virion RNA from these viruses were performed in the same reaction conditions used for the ToMV RNA. Single-stranded genomic and subgenomic RNAs and genome-length, double-stranded RF RNAs were produced from both BMV and TCV RNAs (Fig. 3C). The virus-related RNA species produced in BYL were similar to those found in plant cells infected with the respective viruses, suggesting that the genomes of BMV and TCV also successfully replicated in this system. The levels of newly synthesized BMV and TCV RNAs over the respective input RNAs were similar to or slightly higher than that observed for ToMV (Fig. 3 and data not shown). Together with the fact that the coat proteins of these viruses are able to encapsidate the cognate input RNA in vitro (27, 28), it is also unlikely that the increase of infectivity that reflects the newly synthesized BMV or TCV RNAs can be detected in the BYL system.
Among the cell-free BMV RNA replication products, the double-stranded RNA 4 was either absent or present as only a faint band (Fig. 3B). This result parallels the in vivo BMV RNA synthesis pattern and suggests that synthesis of negative-strand RNA in this system depends on the replication enhancer element that is necessary for negative-strand RNA synthesis, which is absent in RNA 4 but present in the intercistronic region of RNA 3 (6). This finding is in contrast to the detergent-solubilized BMV RNA polymerase isolated from infected cells, which efficiently synthesizes negative-strand RNA 4 (29).
The results presented here demonstrate that a cell-free extract prepared from plant protoplasts by using a simple evacuolation protocol was able to support translation and subsequent RNA replication of at least three distinct plant viruses: ToMV, BMV, and TCV. This in vitro system will aid in elucidating the mechanisms of replication complex formation and other events in the replication of these viruses.
Acknowledgments
We thank Prof. Y. Okada for helpful discussions, Dr. Y. Hagiwara for a critical reading of the manuscript, K. Ishibashi for technical assistance, and K. Fujiwara for general assistance. We gratefully acknowledge the use of the Radioisotope Laboratory of the Graduate School of Agriculture, Hokkaido University. This work was supported by Core Research for Evolutional Science and Technology of Japan Science and Technology Corporation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ToMV, tomato mosaic virus; BMV, brome mosaic virus; TCV, turnip crinkle virus; RRL, rabbit reticulocyte lysate; WGE, wheat germ extract; RF, replicative form.
References
- 1.Haruna, I. & Spiegelman, S. (1965) Proc. Natl. Acad. Sci. USA 54, 579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kondo, M., Gallerani, R. & Weissmann, C. (1970) Nature 228, 525–527. [DOI] [PubMed] [Google Scholar]
- 3.Kamen, R. (1970) Nature 228, 527–533. [DOI] [PubMed] [Google Scholar]
- 4.Blumenthal, T. & Carmichael, G. G. (1979) Annu. Rev. Biochem. 48, 525–548. [DOI] [PubMed] [Google Scholar]
- 5.Buck, K. W. (1996) Adv. Virus Res. 47, 159–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz, M., Chen, J., Janda, M., Sullivan, M., den Boon, J. & Ahlquist, P. (2002) Mol. Cell 9, 505–514. [DOI] [PubMed] [Google Scholar]
- 7.Ahlquist, P., Noueiry, A. O., Lee, W.-M., Kushner, D. B. & Dye, B. T. (2003) J. Virol. 77, 8181–8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molla, A., Paul, A. V. & Wimmer, E. (1991) Science 254, 1647–1651. [DOI] [PubMed] [Google Scholar]
- 9.Svitkin, Y. V. & Sonenberg, N. (2003) J. Virol. 77, 6551–6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno, T., Aoyagi, M., Yamanashi, Y., Saito, H., Ikawa, S., Mashi, T. & Okada, Y. (1984) J. Biochem. 96, 1915–1923. [DOI] [PubMed] [Google Scholar]
- 11.Ahlquist, P., Dasgupta, R. & Kaesberg, P. (1984) J. Mol. Biol. 172, 369–383. [DOI] [PubMed] [Google Scholar]
- 12.Ahlquist, P., French, R., Janda, M. & Loesch-Fries, L. S. (1984) Proc. Natl. Acad. Sci. USA 81, 7066–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrington, J. C., Heaton, L. A., Zuidema, D., Hillman, B. I. & Morris, T. J. (1989) Virology 170, 219–226. [DOI] [PubMed] [Google Scholar]
- 14.Sonobe, S. (1990) Protoplasma 155, 239–242. [Google Scholar]
- 15.Nagata, T., Nemoto, Y. & Hasezawa, S. (1992) Int. Rev. Cytol. 132, 1–30. [Google Scholar]
- 16.Chiba, Y., Sakurai, R., Yoshino, M., Ominato, K., Ishikawa, M., Onouchi, H. & Naito, S. (2003) Proc. Natl. Acad. Sci. USA 100, 10225–10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madin, K., Sawasaki, T., Ogasawara, T. & Endo, Y. (2000) Proc. Natl. Acad. Sci. USA 97, 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagiwara, Y., Komoda, K., Yamanaka, T., Tamai, A., Meshi, T., Funada, R., Tsuchiya, T., Naito, S. & Ishikawa, M. (2003) EMBO J. 22, 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa, M., Meshi, T., Ohno, T. & Okada, Y. (1991) J. Virol. 65, 861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quadt, R., Verbeek, H. J. M. & Jaspars, E. M. J. (1988) Virology 165, 256–261. [DOI] [PubMed] [Google Scholar]
- 21.Laskey, R. A. (1980) Methods Enzymol. 65, 363–371. [DOI] [PubMed] [Google Scholar]
- 22.Dawson, W. O. & Lehto, K. M. (1990) Adv. Virus Res. 38, 307–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe, Y., Ohno, T. & Okada, Y. (1982) Virology 120, 478–480. [DOI] [PubMed] [Google Scholar]
- 24.Wink, M. (1993) J. Exp. Bot. 44, 231–246. [Google Scholar]
- 25.Beier, H., Barciszewska, M., Krupp, G., Mitnacht, R. & Gross, H. J. (1984) EMBO J. 3, 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraenkel-Conrat, H. & Williams, R. C. (1955) Proc. Natl. Acad. Sci. USA 41, 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi, Y. G. & Rao, A. L. N. (2003) J. Virol. 77, 9750–9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorger, P. K., Stockley, P. G. & Harrison, S. C. (1986) J. Mol. Biol. 191, 639–658. [DOI] [PubMed] [Google Scholar]
- 29.Quadt, R., Ishikawa, M., Janda, M. & Ahlquist, P. (1995) Proc. Natl. Acad. Sci. USA 92, 4892–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]